Modification of phytochrome photochemistry and behavior by structure-guided mutagenesis provides new tools to study signaling and manipulate photomorphogenesis for potential agricultural benefit.
Abstract
Phytochromes (phys) encompass a diverse collection of biliproteins that enable cellular light perception by photoconverting between a red-light-absorbing ground state (Pr) and a far-red light-absorbing active state (Pfr). Based on the central role of plant phys in controlling numerous agriculturally important processes, their rational redesign offers great promise toward accelerating crop improvement. Employing as templates the available three-dimensional models of the photosensory module within bacterial phys, we report here our initial attempt to apply structure-guided mutagenesis to phy engineering using Arabidopsis (Arabidopsis thaliana) phyB, the dominant isoform in light-grown plants, as the example. A collection of phyB mutants was generated affecting the bilin-binding pocket that altered photochemistry, thermal stability, and/or nuclear localization patterns, some of which also impacted phenotypic outputs. Of particular interest are the Y361F substitution, which created Arabidopsis plants with greatly enhanced light sensitivity, mutants variably altered in Pfr-to-Pr thermal reversion and nuclear aggregation, and the D307A substitution, which failed to photoconvert from Pr to Pfr and display light-induced nuclear aggregation but retained some biological activity and accelerated turnover in red light. Taken together, this collection provides variants potentially useful to agriculture as well as new tools to better understand the molecular mechanisms underpinning phy signaling.
Phytochromes (phys) encompass a unique set of dimeric photoreceptors that use a bilin (or linear tetrapyrrole) chromophore for light detection (Rockwell et al., 2006; Vierstra and Zhang, 2011). They were first discovered in higher plants based on their striking influence on development, including controls of seed germination, stem/petiole/hypocotyl elongation, chloroplast biogenesis, leaf expansion, flowering time, and the shade-avoidance response (SAR; Franklin and Quail, 2010; Rausenberger et al., 2010). In the past decade, they have also become appreciated as important photoregulators within the microbial world, where they have been implicated in pigmentation, enhancing photosynthetic potential, taxis, and sexual development (Auldridge and Forest, 2011; Vierstra and Zhang, 2011). The canonical phy consists of an N-terminal photosensory module (PSM) sequentially containing Per/Arndt/Sim (PAS), cyclic GMP phosphodiesterase/adenylyl cyclase/FhlA (GAF), and Phytochrome (PHY) domains (Karniol et al., 2005). The PSM is followed by an output module (OPM) that houses various signaling activities, the most common of which in microorganisms directs phosphorelays via a two-component-type His kinase domain (Auldridge and Forest, 2011; Vierstra and Zhang, 2011). The bilin is covalently attached to the apoprotein via a thioether bond between a conserved Cys either within the GAF domain (cyanobacteria and plants) or upstream of the PAS domain (eubacteria and fungi) and then cradled within the GAF domain. Through numerous interactions between the GAF domain and the bilin, phys assume two stable photointerconvertible states, typically a red light (R)-absorbing Pr form that represents the ground state and is biologically inactive and a far-red light (FR)-absorbing Pfr form that is generated only upon R absorption and is biologically active. By interconverting between Pr and Pfr, phys act as novel R/FR photoswitches in various transduction cascades. In plants, this switch ultimately impacts a substantial portion of their transcriptomes (Franklin and Quail, 2010).
Given the central position of phys in controlling plant architecture, SAR, and the timing of reproduction, their modification has long been proposed as a strategy to improve agricultural productivity, for example via the creation of mutant plants expressing varied levels of the photoreceptors or with altered activities that impact downstream signaling pathways (Franklin and Quail, 2010; Nagatani, 2010). One striking example illustrating this potential is the highly fluorescent, photochemically compromised Y276H mutant generated with the Arabidopsis (Arabidopsis thaliana) phyB, the most influential isoform in light-grown plants. When used to replace its corresponding wild-type version, phyBY276H induces seedling photomorphogenesis even in the dark, indicative of constitutive phy signaling without R activation (Su and Lagarias, 2007; Hu et al., 2009). Other examples include various missense mutations that render phyB either hypoactive (Wagner and Quail, 1995; Elich and Chory, 1997; Krall and Reed, 2000; Chen et al., 2003; Oka et al., 2008) or hyperactive (Kretsch et al., 2000; Ádám et al., 2011) in planta. The G564E mutation affected in the Arabidopsis phyB-401 allele is particularly notable for its ability to increase seedling sensitivity to R by as much as 1,000-fold (Ádám et al., 2011). These prior approaches mainly exploited random mutagenesis followed by phenotypic selection in the hope of finding phys with altered photochemistry and/or signaling. However, with the advent of remarkably coherent, high-resolution three-dimensional structures of the PSM from numerous bacterial relatives (Wagner et al., 2005, 2007; Yang et al., 2007, 2008, 2011; Cornilescu et al., 2008; Essen et al., 2008; Li et al., 2010; Ulijasz et al., 2010; Bellini and Papiz, 2012; Burgie et al., 2013) combined with extensive mutational analyses of key conserved amino acids (Fischer and Lagarias, 2004; Hahn et al., 2006; von Stetten et al., 2007; Wagner et al., 2008; Ulijasz et al., 2010), opportunities for more predictive redesign are now possible.
Here, we tested this rational approach in which site-directed substitutions of critical amino acids based on the bacterial scaffolds were introduced into the Arabidopsis phyB isoform. The photochemistry of the mutant PSMs was then examined after recombinant assembly with the native chromophore phytochromobilin (PΦB), and the full-length versions were assessed for their phenotypic rescue of the phyB-9 null mutant using the native PHYB promoter and 3′ untranslated region to drive expression. Our goal was not to generate strong phyB mutants that would elicit phenotypes too extreme to be useful agronomically (e.g. constitutive or absent photomorphogenesis) but to compromise the photoreceptor in more subtle ways that might differentially adjust various aspects of photoperception under phyB control. The results collectively demonstrate that various aspects of phy dynamics and signaling can be adjusted, which in some cases generates plants with unique and potentially useful photobehavioral properties.
RESULTS
Rational Design of phyB Variants to Alter Light Signaling
For proof of concept, we examined five phyB PSM mutations (D307A, Y361F, R582A, R352A, and R322A) predicted from the three-dimensional structures of bacterial phys to alter conserved residues surrounding the chromophore that are likely critical for Pr-to-Pfr interconversion and/or signal transmission (Fig. 1, B and C). Based on the scheme presented in Figure 1A, we tested how well the mutants would (1) assemble with PΦB, (2) photointerconvert between Pr and Pfr, (3) revert thermally from Pfr back to Pr, (4) aggregate after R irradiation into nuclear bodies or “speckles” thought to be important for signaling and/or turnover (Chen and Chory, 2011), (5) degrade upon R irradiation, and (6) stimulate several photomorphogenic processes fully or partially controlled by phyB. Phenotypic responses tested included R-stimulated seed germination, hypocotyl growth inhibition under R, effect of end-of-day (EOD) FR on the hypocotyl R response, and leaf epinasty, rosette architecture, and flowering time under a short-day photoperiod (Schafer and Nagy, 2005; Franklin and Quail, 2010; Nagatani, 2010). Both the epinastic response, which reorients the rosette leaves to be more perpendicular to the soil by changing the stem/petiole angle, and the EOD-FR response are components of the SAR (Fankhauser and Casal, 2004). Pfr turnover is likely driven by the ubiquitin/26S proteasome system, based on mutant analyses and its sensitivity to the proteasome inhibitor MG132 (Christians et al., 2012; see below), but where it occurs (i.e. cytoplasm and/or nucleus) is unresolved.
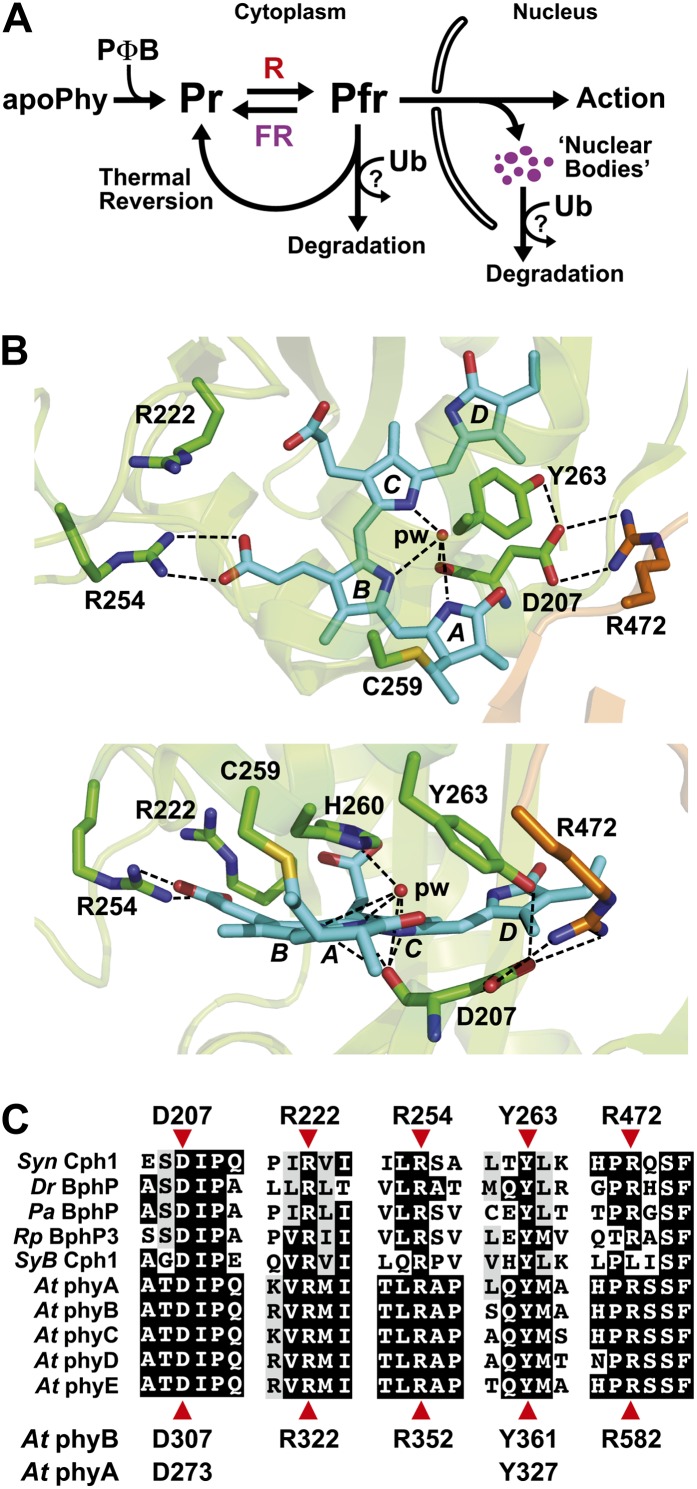
Figure 1.
Scheme of phy action and the three-dimensional relationships of key amino acids within the bilin-binding PSM. A, Scheme depicting the main steps involved in phy assembly, Pr/Pfr photointerconversion, stability, aggregation, and action in higher plants. Photoconversion, thermal reversion, and possibly degradation likely occur in both the cytoplasm and nucleus. B, Top (top) and side (bottom) three-dimensional views of the PSM from Syn-Cph1 (Protein Data Bank code no. 2VEA; Essen et al., 2008) assembled with phycocyanobilin, highlighting the positions of key conserved amino acids surrounding the bilin and the Cys involved in bilin attachment (Cys-259). The residue numbers are those for Syn-Cph1. The GAF domain and PHY hairpin are colored in green and orange, respectively. Phycocyanobilin is in cyan, with the individual pyrrole rings labeled. Sulfur, oxygen, and nitrogen atoms are colored yellow, red, and deep blue, respectively. Important contacts are indicated by dashed lines. pw, Pyrrole water. C, Alignment of the GAF domain and PHY hairpin protein sequences among bacterial phys with available structures and with comparable sequences from the phyA to phyE family in Arabidopsis. Residues pertinent to this study are indicated by red arrowheads; their sequence positions are shown either above for Syn-Cph1 or below for Arabidopsis phyA and phyB. At, Arabidopsis; Pa, Pseudomonas aeruginosa; Rp, Rhodopseudomonas palustris.
The 6His-tagged PSM of all the mutants could be expressed and readily assembled with PΦB in Escherichia coli, indicating that none of the substitutions compromised protein folding or bilin conjugation dramatically (Fig. 2A; Supplemental Fig. S1). The wild-type PSM had Pr and Pfr absorption maxima (663 and 726 nm, respectively) close to expectations (Hanzawa et al., 2001) and rapidly reverted thermally from Pfr back to Pr with a half-life (t1/2) of approximately 110 min at 24°C, with the reaction approaching completion after 24 h (Fig. 2B). These kinetics were slower but more complete than in prior studies using phyB isoforms expressed recombinantly in yeast; the differences potentially reflect the fact that the prior studies used the full-length polypeptides and incorporated phycocyanobilin and not PΦB as the chromophore (Kunkel et al., 1995; Elich and Chory, 1997). With the exception of the PSM from phyBD307A, which is poorly photochromic (see below), all the mutants had reasonably normal Pr and Pfr absorption spectra, except for a slight blue shift in the maxima for several (5–9 nm; Fig. 2A), and had Pr→Pfr and Pfr→Pr photoconversion rates close to that for phyBWT (Supplemental Fig. S2).
Figure 2.
Absorption and FR-R difference spectra and Pfr→Pr thermal reversion rates of wild-type and mutant versions of the purified recombinant PSM from Arabidopsis phyB assembled with PΦB. For SDS-PAGE analysis, see Supplemental Figure S1. A, UV-visible absorption spectra of Pr (solid lines) or following its exposure to saturating R (dashed lines). Difference maxima and minima (Pr minus R) are indicated. B, Rates of Pfr→Pr thermal reversion at 24°C. All rates are expressed as percentages of Pfr in the sample, using the absorption maximum of Pfr near 720 nm for quantification.
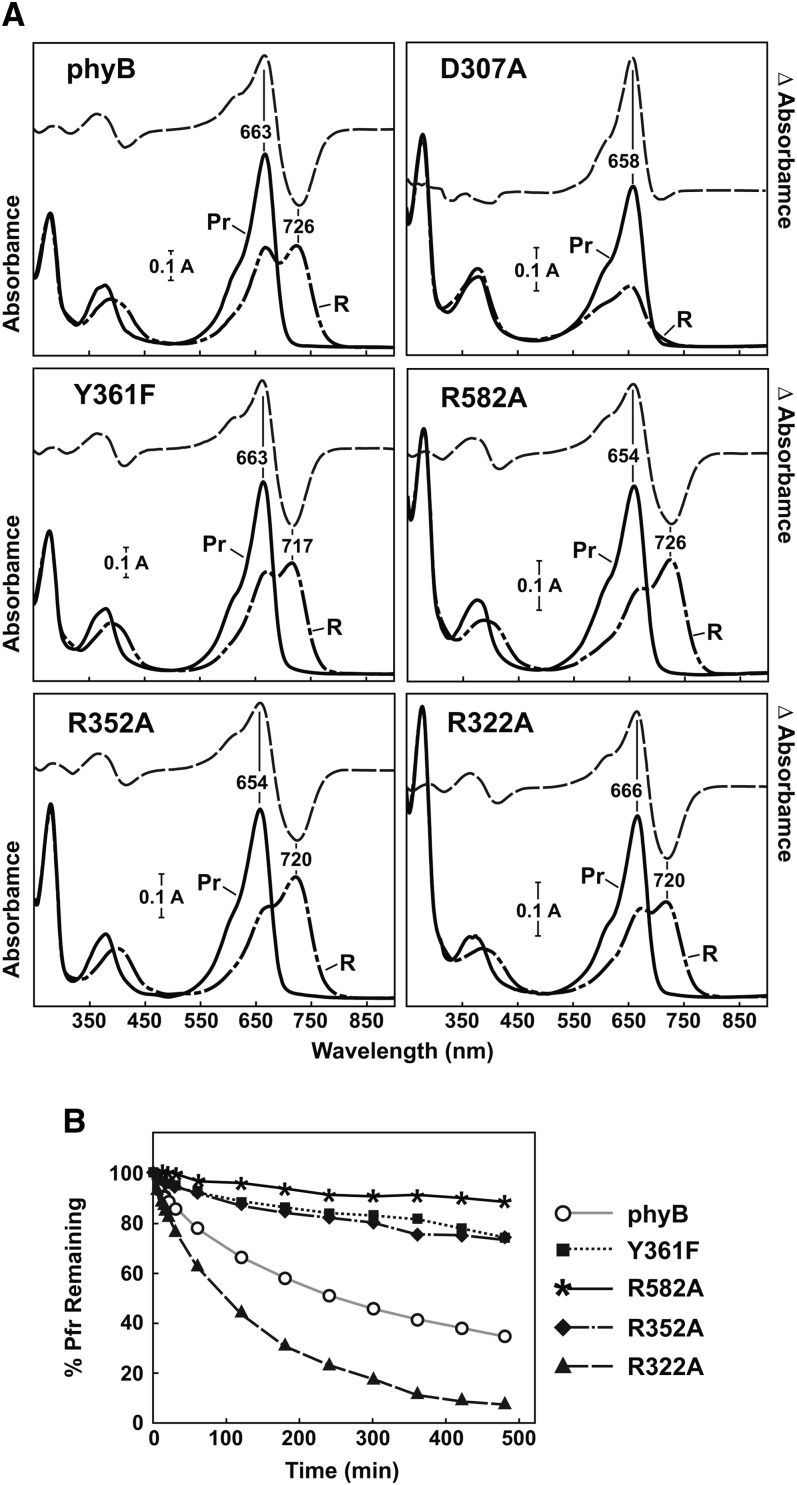
Given that Arabidopsis and other plants are highly sensitive to phy levels (Boylan and Quail, 1991; Wagner et al., 1991; Cherry et al., 1992; Rausenberger et al., 2010), we chose two independently transformed, isogenic phyB-9 lines in the T3 generation that stably expressed either full-length phyBWT or the mutants (appended with a C-terminal 11-amino acid Flag tag sequence; Supplemental Table S1) to levels that matched most closely that in wild-type Columbia (Col-0) plants as judged by immunoblot analysis (Fig. 3D). R582A lines were the only transgenic plants that failed to bracket the levels of phyB detected in the wild type. For simplicity, these homozygous lines are named only by the introduced PHYB transgenes, with the understanding that all the lines were also homozygous for the phyB-9 null allele. It also should be noted that because the level of wild-type phyB drops below detection in light-grown plants due to light-induced degradation (Rausenberger et al., 2010; Christians et al., 2012), we measured the wild-type and mutant chromoprotein levels in etiolated seedlings with the assumption that their relative synthetic rates remained similar in green seedlings, because the same PHYB promoter fragment was used in all cases. Importantly, all of the complemented phyB-9 mutant lines had normal seedling development in the dark, indicating that none of the phyB variants signaled in the absence of photoactivation (Fig. 3B).
Figure 3.
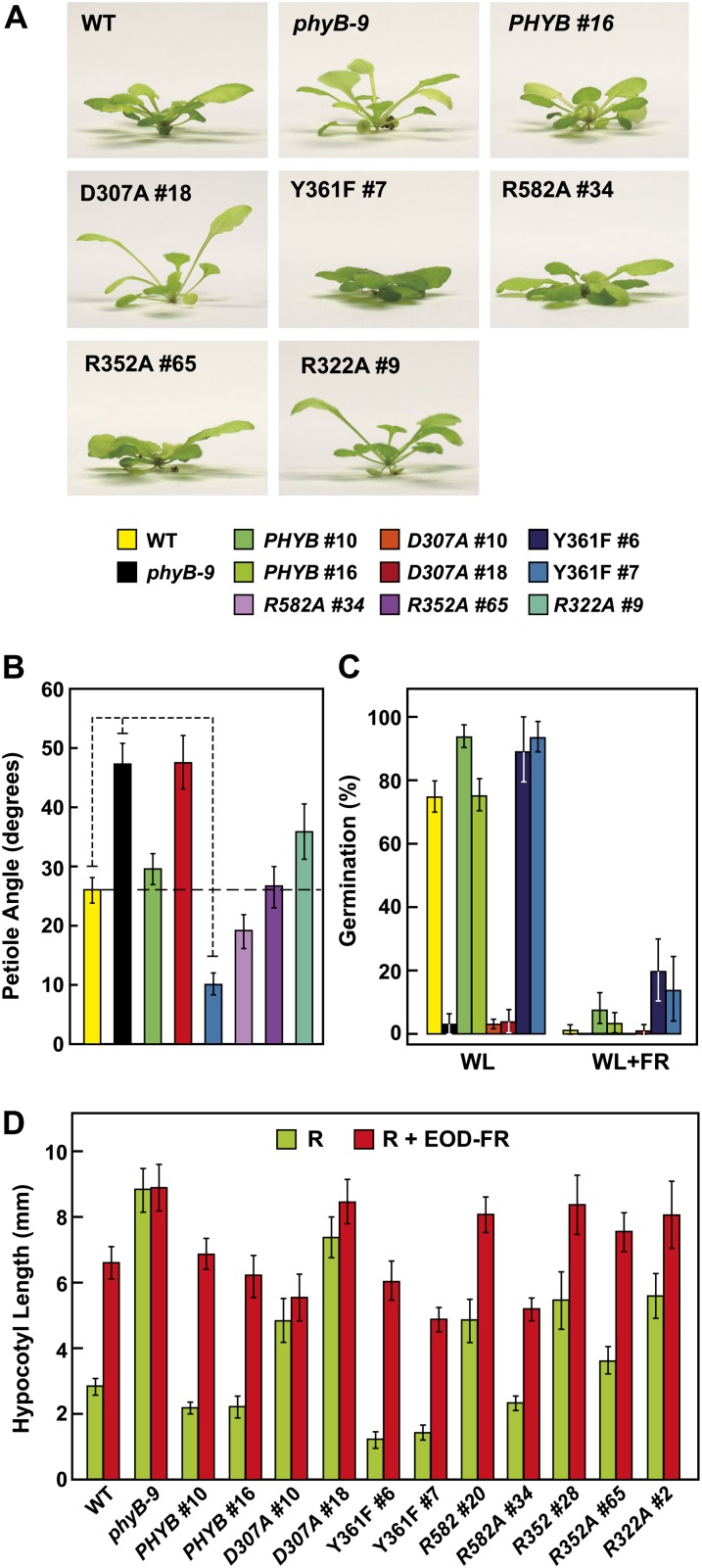
Phenotypes of an Arabidopsis phyB-9 null mutant rescued with transgenes expressing wild-type or mutant versions of full-length phyB. Shown are the wild type (WT), the phyB-9 mutant, and two independent transgenic lines expressing either the wild-type or mutant PHYB cDNAs in the phyB-9 background. A, Representative 3-week-old plants grown in LD. B and C, Representative 4-d-old seedlings either grown in the dark (B) or under continuous 13 μmol m−2 s−1 R (Rc; C). D, Levels of the phyB protein in each of the lines examined above as determined by immunoblot analysis of crude extracts from 5-d-old dark-grown seedlings with an anti-phyB monoclonal antibody. Protein loading was confirmed with anti-PBA1 antibodies.
To examine the ability of the mutants to concentrate in nuclear bodies as Pfr (Chen and Chory, 2011), we also created a parallel set of transgenic lines expressing the phyB mutants fused to the N terminus of yellow fluorescent protein (YFP). The wild-type and mutant phyB-YFP polypeptides also assembled well with PΦB in planta, and in agreement with previous reports (Ádám et al., 2011), their phenotypic activity resembled that of their nontagged counterparts based on their ability (or inability) to suppress hypocotyl elongation in R when introduced into the phyB-9 background (Supplemental Fig. S3). Consistent with previous studies (Kircher et al., 2002; Rausenberger et al., 2010; Ádám et al., 2011), fluorescence from phyBWT-YFP was typically distributed evenly throughout the cytoplasm and the nucleus in hypocotyls from dark-grown plants, as observed by fluorescence confocal microscopy. Upon a 12-h irradiation with a high fluence rate (90 μmol m−2 s−1) but not a low fluence rate (0.25 μmol m−2 s−1) of R, phyB bodies accumulated that were easily seen as numerous intense puncta in the nucleus (see below).
Asp-307 Is Required for Photoconversion, Robust Signaling, and Nuclear Body Formation But Not Turnover
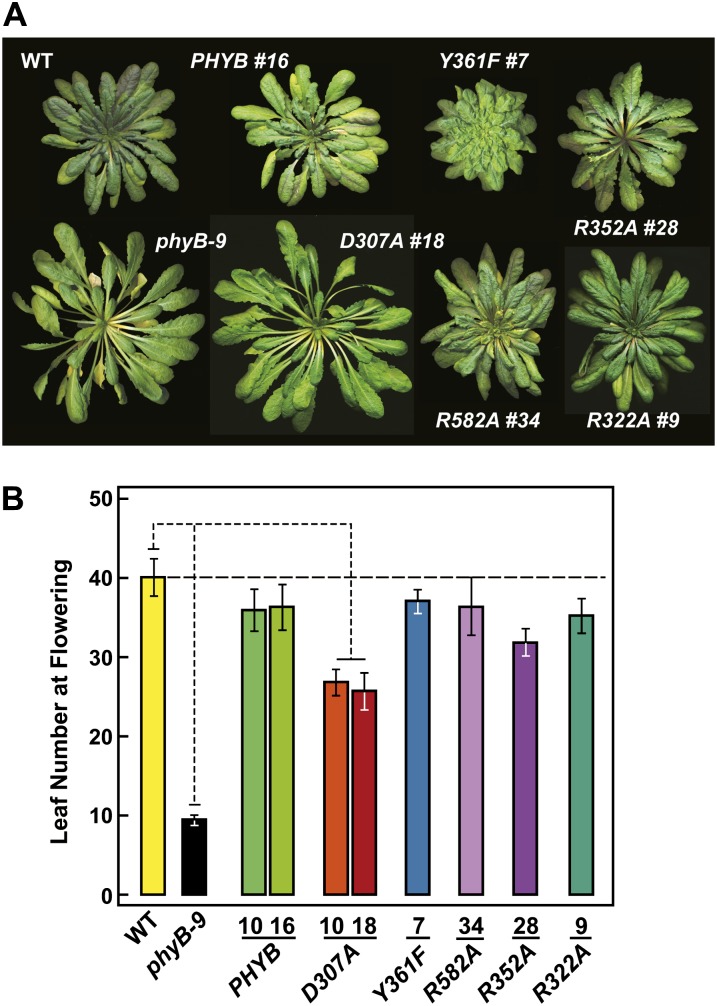
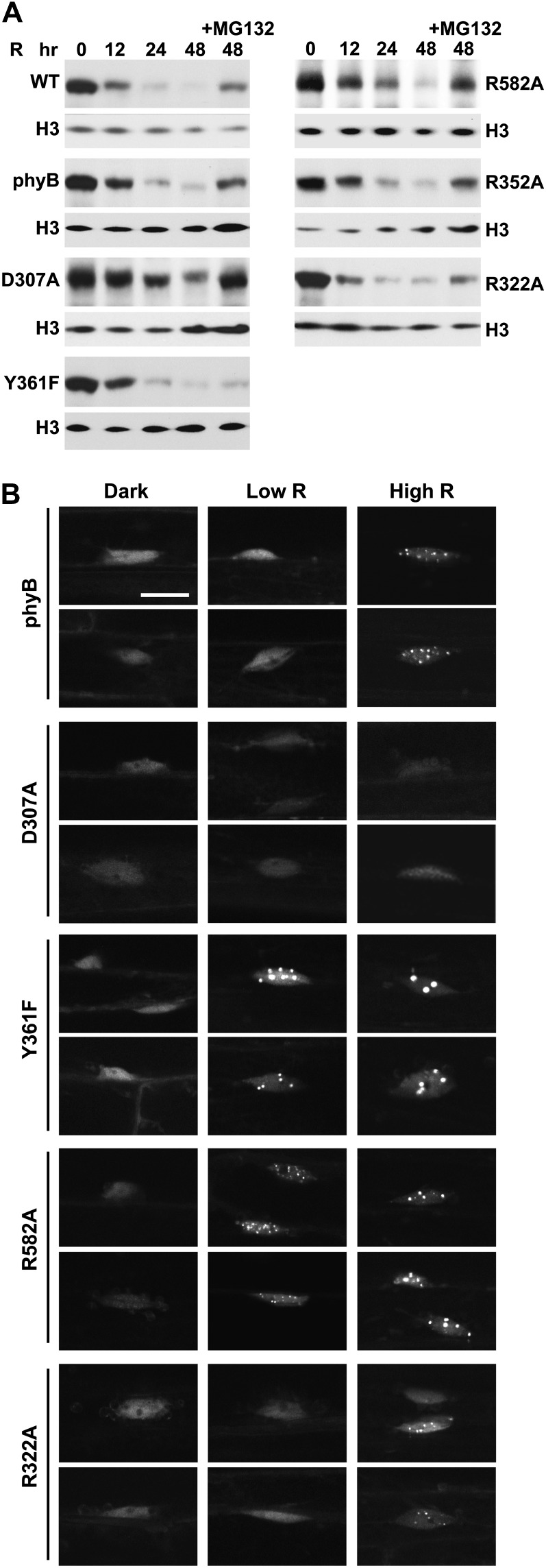
As shown in Figure 1, B and C, the D307A substitution removes a key Asp (Asp-207 in Synechocystis sp. PCC 6803 [Syn]-Cph1) in a near-invariant Asp-Ile-Pro (DIP) motif that participates through its main-chain carbonyl in an extensive and crucial hydrogen-bonding lattice involving the A-C pyrrole rings of PΦB and the centrally positioned pyrrole water (Wagner et al., 2007; Essen et al., 2008). This residue is essential for the Pr→Pfr photoconversion of bacterial phys, as most, if not all, substitutions (including Ala) stall in the deprotonation/protonation cycle following R irradiation and become highly fluorescent (Hahn et al., 2006; von Stetten et al., 2007; Wagner et al., 2008). In line with this importance, we found that the assembled phyBD307A biliprotein readily bound PΦB and generated Pr but failed to photoconvert to Pfr in R (Fig. 2A). Instead, a bleached R-absorbing intermediate appeared that would regenerate Pr upon irradiation with FR or more slowly in prolonged darkness via thermal reversion. Unlike phyBWT, which restored seed germination and suppressed hypocotyl elongation by R and allowed EOD-FR to reverse the effects of R when introduced into the phyB-9 background, the phyBD307A mutant was greatly reduced in phenotypic activity (Figs. 3C, 4A, and 5, C and D). Such compromised signaling was also apparent in more mature plants, as judged by the elongated leaf blades and petioles and strong leaf epinasty (as measured by the large upward angles of petioles) for PHYBD307A plants grown in short days (SD), which better resembled phyB-9 plants as compared with their PHYBWT counterparts (Figs. 5, A and B, and 6A). Consistent with aberrant photochemistry, the accumulation of phyBD307A-YFP in nuclear bodies was also undetectable even after prolonged irradiation with a high fluence rate of R, a condition where the bodies were clearly evident for the wild-type version (Fig. 7B).
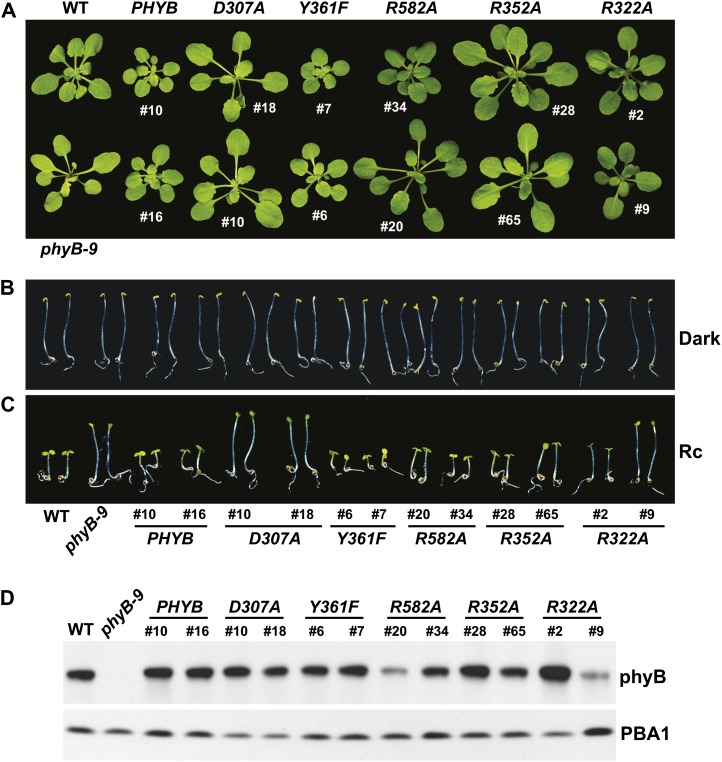
Figure 4.
R sensitivity of hypocotyl elongation for the Arabidopsis phyB-9 null mutant rescued with transgenes expressing wild-type or mutant versions of full-length phyB. Shown are the wild type (WT), the phyB-9 mutant, and two independent transgenic lines expressing either the wild-type or mutant PHYB cDNAs in the phyB-9 background. Hypocotyl length of each line was expressed relative to that measured for dark-grown seedlings. Each data point represents the mean ± se from four independent experiments. A, Sensitivity to a broad range of R fluence rates. B, Sensitivity of the Y361F lines to very low fluence rates of R.
Figure 5.
Sensitivity of phyB-9 plants rescued with various phyB mutants to photomorphogenic processes controlled by phyB. Shown are the wild type (WT), the phyB-9 null mutant, and one or two independent transgenic lines expressing either the wild-type or mutant PHYB cDNAs in the phyB-9 background. For descriptions of the mutant lines, see Figure 3D. A, Side view of 45-d-old seedlings grown in white light under SD illustrating the influence of phyB on leaf epinasty. B, Quantification of leaf epinasty for seedlings in A. Each bar represents the average angle between the soil surface and the petiole for the fourth and fifth leaves of 10 plants (20 total angles). The 95% confidence interval for each average is shown. The values for wild-type, phyB-9, and Y361F lines are significantly different from each other by Student’s t test (P < 0.05). C, Germination efficiency of seeds either treated with a 2-h pulse of white light (WL) alone or followed by a pulse of FR. Germination was assessed after a subsequent 5-d incubation in darkness. Each bar represents the average ± se of five experiments involving at least 60 seeds each. D, Effect of R and EOD-FR on hypocotyl growth. Etiolated seedlings were subjected over 4 d to a 24-h light regime of continuous R (90 µmol m−2 s−1) for 8 h, followed by either darkness (R) or a 10-min pulse of 100 µmol m−2 s−1 FR (R + EOD-FR) and then 16 h of darkness. Each bar represents the average ± se of four experiments involving at least 15 seedlings each. The Y361F #7 line was significantly different from the wild type and PHYB for both R and R + EOD-FR by Student’s t test (P < 0.05).
Figure 6.
Effects of wild-type (WT) and mutant versions of phyB on Arabidopsis rosette organization and flowering time in SD. A, Photographs of representative plants grown in SD just before bolting. B, Flowering time in SD (8 h of light/16 h of dark). Each bar represents the average number of rosette leaves generated before emergence of the inflorescence stem for more than 20 plants. The 95% confidence interval for the averages is shown. The values for the wild type, phyB-9, and D307A are significantly different from each other by Student’s t test (P < 0.05). For descriptions of the mutant lines, see Figure 3D.
Figure 7.
Effect of the phyB mutations on the R-induced degradation and nuclear distribution of the photoreceptor. A, Loss of phyB protein during continuous R irradiation of etiolated seedlings. phyB levels in 4-d-old dark-grown Arabidopsis were measured after various exposure times to 90 µmol m−2 s−1 R by immunoblot analysis using an anti-phyB monoclonal antibody. The seedlings were incubated with 100 μm MG132 or an equivalent volume of dimethyl sulfoxide 12 h before irradiation. Nearly equal protein loading was confirmed with anti-histone H3 antibodies. Lines used include the wild type (WT) and the phyB-9 background expressing phyBWT #10, phyBD307A #18, phyBY361F #06, phyBR582A #34, phyBR352A #65, and phyBR322A #09 (Fig. 3D). B, Light-dependent aggregation of phyB into nuclear bodies. Wild-type phyB or the various mutants were expressed as YFP fusions in the phyB-9 background. Regions surrounding the nucleus were imaged by fluorescence confocal microscopy from hypocotyl cells either kept in the dark or irradiated for 12 h with continuous 0.25 µmol m−2 s−1 (Low R) or 90 µmol m−2 s−1 (High R) R. Expression levels of the fusions and their ability to rescue the phyB-9 phenotype with respect to hypocotyl elongation in R can be found in Supplemental Figure S2. Bar = 10 μm.
However, detailed fluence-response analysis of hypocotyl growth under very high R fluence rates, flowering time in SD, and possibly the effect of EOD-FR on hypocotyl growth revealed that phyBD307A retained some signaling activity despite its inability to photoconvert normally to Pfr (Figs. 4A, 5B, and 6B). For example, early flowering in SD induced by loss of phyB could be nearly fully reversed by expressing the phyBD307A mutant in phyB-9 seedlings. And surprisingly, the phyBD307A protein was still degraded in R, albeit with a slightly slower turnover rate as compared with the wild-type phyB (Fig. 7A). Quantitative analysis of the immunoblots calculated that the t1/2 of R-stimulated phyBD307A degradation was 21 h versus 15 and 14 h for endogenous phyB and the introduced phyBWT chromoprotein, respectively.
phyBY361F Signaling Is Hyperactive
Tyr-361 is part of the aforementioned hydrogen-bonding lattice that holds the side-chain carboxyl group of Asp-307 in place and helps enclose the GAF domain around the bilin (Fig. 1B). Accordingly, the drastic His substitution blocks Pr→Pfr photoconversion and enhances R-stimulated fluorescence of Deinococcus radiodurans (Dr)-BphP (Wagner et al., 2008), whereas the relatively mild Phe replacement in Syn-Cph1 (Tyr-263 in Syn-Cph1) compromises photochemistry more subtly based on a blue shift in Pfr absorption, slower Pr→Pfr photoconversion, and increased fluorescence (Mailliet et al., 2011). Strikingly, the Y361F substitution in Arabidopsis phyB permitted proper photochemistry but made the photoreceptor hyperactive with respect to signaling. Except for a slight blue shift of the Pfr absorption maximum (approximately 9 nm), the recombinant PSM of phyBY361F had normal Pr and Pfr absorption spectra. However, the mutant displayed slightly reduced Pr→Pfr and Pfr→Pr photoconversion rates and a markedly slower rate (4-fold) of Pfr→Pr thermal reversion (t1/2 of approximately 7.6 h at 24°C), indicating that interconversion between the two conformers was perturbed (Fig. 2; Supplemental Fig. S2). Despite these changes in photochemistry for the PSM, the in vivo turnover of full-length phyBY361F as Pfr appeared normal (Fig. 7A).
Contrary to the expectations that some of these photochemical defects might compromise signaling, phyBY361F more effectively directed phyB-mediated responses. Soon after germination, the PHYBY361F seedlings were more sensitive to continuous R with respect to hypocotyl elongation, and as the seedling grew under a long-day photoperiod, they developed more compact rosettes with shorter petioles than wild-type and PHYBWT plants, indicative of light hypersensitivity (Figs. 3, A and C, and 4A). Analyses of hypocotyl elongation at very low fluence rates of R (less than 1 μmol m−2 s−1) estimated that the phyBY361F biliprotein is at least 50 times more active in signaling with respect to this response (Fig. 4B). A modest but statistically significant hypersensitivity to R was also observed for seed germination and the effect of EOD-FR on R-suppressed hypocotyl growth (Fig. 5, C and D). Light hypersensitivity continued for the rosettes of mature plants and dampened the SAR, as judged by increased hyponasty (i.e. more prostrate petiole angles) and the smaller leaves and shorter petioles seen for PHYBY361F plants as compared with PHYBWT plants when grown in SD (Figs. 5, A and B, and 6A). However, for flowering time in SD, introduction of the phyBY361F protein in the phyB-9 background did not lengthen the delay in flowering any further than its wild-type counterpart (Fig. 6B). R hypersensitivity was also observed for nuclear sequestering. Even under a low fluence rate of R (0.25 μmol m−2 s−1), where phyBWT-YFP failed to form such bodies, they were clearly evident for phyBY361F-YFP (Fig. 7B). At the higher fluence of R, there also appeared to be fewer but larger nuclear puncta, suggesting that these phyBY361F-YFP bodies coalesce better into larger aggregates.
The R322A, R352A, and R582A Mutations Poorly Compromise phyB Signaling
A common feature of phys containing sequential GAF and PHY domains is an extraordinary hairpin that extends from the PHY domain to contact the GAF domain and its critical DIP Asp (Essen et al., 2008; Yang et al., 2008; Li et al., 2010; Bellini and Papiz, 2012; E.S. Burgie and R.D. Vierstra, unpublished data). Both its length and juxtaposition to the bilin have implicated this protrusion in Pfr signaling, possibly by helping transmit movements in the bilin from the GAF domain to the PHY domain and ultimately to the OPM. In particular, the highly conserved Arg-582 in Arabidopsis phyB might be crucial for this transmission, given that its counterpart in Syn-Cph1 (Arg-472) forms an interdomain salt bridge with the DIP Asp (Fig. 1B). To test this scenario, we attempted to destabilize this contact via the R582A mutation. The phyBR582A PSM had normal Pr and Pfr absorption spectra (except for a 9-nm blue shift of the Pr absorption maximum) and Pr→Pfr and Pfr→Pr photoconversion rates but was strikingly slower (more than 10-fold) in Pfr→Pr thermal reversion than that of phyBWT (t1/2 of at least 20 h at 24°C), indicating that Arg-582 is not required for photochemistry but helps destabilize the Pfr conformer once formed (Fig. 2; Supplemental Fig. S2). Regardless of its effects on photochemistry, the in vivo turnover of phyBR582A as Pfr was not compromised (Fig. 7A).
Despite these photochemical changes, phyBR582A appeared to signal normally based on the fluence response of hypocotyl growth to continuous R and EOD-FR and its ability to restore normal flowering time in SD (Figs. 3B, 4A, 5D, and 6). If anything, a slight hyperactivity was possible for the phyBR582A chromoprotein based on several phenotypic assays. As shown in Figure 4A, the PHYBR582A-#34 line induced a slightly stronger repression on hypocotyl growth at low R fluence rates (e.g. 1 μmol m−2 s−1) as compared with wild-type and PHYBWT seedlings, despite accumulating similar levels of photoreceptor. Moreover, petiole angles were more prostrate and the rosettes appeared more compact than wild-type and PHYBWT plants in SD (Figs. 5, A and B, and 6A). Surprisingly, even though signaling by phyBR582A appeared reasonably normal, its ability to form nuclear bodies was markedly enhanced. Like phyBY361F-YFP, the phyBR582A-YFP chromoprotein formed obvious nuclear bodies when seedlings were exposed to 0.25 μmol m−2 s−1 R, a fluence rate below that required by the corresponding wild type (Fig. 7B).
The last two mutations (R352A and R322A) potentially eliminated salt bridges between the propionate side chains in PΦB and the bilin-binding pocket, which help restrain the bilin within the photoreceptor (Fig. 1, B and C). Prior studies with bacterial phys (Arg-101 and Arg-133 in Synechococcus sp. OS-B′ [SyB]-Cph1) showed that these Arg residues stabilize and destabilize the Pfr conformer, respectively (Hahn et al., 2006; Cornilescu et al., 2008; Wagner et al., 2008), with their guanidinium side chains undergoing dramatic conformational changes during Pr→Pfr photoconversion (Cornilescu et al., 2008; Wagner et al., 2008; Ulijasz et al., 2010). Arg-352 should directly bridge with the propionate group of pyrrole ring B in PΦB. Its replacement with a Gln in Dr-BphP (R254Q) is one of the few mutations that block the covalent attachment of the bilin (Wagner et al., 2008), whereas the more subtle Arg-to-Ala substitutions in Dr-BphP and SyB-Cph1 effectively inhibit the thermal reversion of Pfr back to Pr (Cornilescu et al., 2008; Wagner et al., 2008). For Arabidopsis phyB directly, previous work showed that Arg-352 is potentially important to phyB signaling, as a Lys substitution was shown to substantially attenuate the R suppression of hypocotyl elongation (Oka et al., 2008).
Here, we found that the R352A substitution has little impact on Pr and Pfr absorption and the photochemistry of phyB, but like its bacterial relatives (Wagner et al., 2008; Ulijasz et al., 2010), the mutation stabilizes Pfr against thermal reversion (Fig. 2; Supplemental Fig. S2). The reversion rate of the PSM was 4-fold slower than that of wild-type phyB, with a t1/2 of approximately 7.4 h at 24°C. When expressed in phyB-9 seedlings, phyBR352A behaved similar to wild-type phyB with respect to its ability to affect hypocotyl growth in R or EOD-FR, restore normal rosette morphology to mature plants, control leaf epinasty and delay flowering in SD, and readily degrade after photoconversion to Pfr (Figs. 3, A and C, 4A, 5, A, B, and D, and 6). At most, phyBR352A was marginally hypoactive phenotypically, as judged by the reduced response of the PHYBR352A-#65 hypocotyls to almost all fluences of R despite accumulating levels of photoreceptor comparable to those in wild-type and PHYBWT plants (Fig. 4A). Regardless of its ability to retard Pfr→Pr thermal reversion, the in vivo turnover of phyBR582A as Pfr did not appear to be accelerated (Fig. 7A).
Analogous to Arg-352, Arg-322 in the GAF domain is predicted to contact PΦB, with the solution NMR structure of SyB-Cph1 showing that the flexibility of its guanidinium side chain allows for transient interactions with the ring C propionate (Cornilescu et al., 2008; Ulijasz et al., 2010). The phyBR322A PSM assembled with PΦB retained relatively normal light absorption (except for a 6-nm shift of the Pfr absorption maximum) and Pr→Pfr and Pfr→Pr photoconversion rates, but unlike the R352A substitution, phyBR322A had a substantially faster rate of Pfr→Pr thermal reversion (2.2-fold) than phyBWT (t1/2 of 50 min at 24°C; Fig. 2). Thus, whereas Arg-322 is not required for phyB photochemistry, it helps stabilize the Pfr conformer once formed.
Phenotypically, phyBR322A behaved similar to phyBWT, as judged by its ability to affect hypocotyl growth in R or EOD-FR, rescue the rosette morphology of mature plants, and control leaf epinasty and delay flowering in SD (Figs. 3, A and C, 4A, 5, A, B, and D, and 6). Like all but the phyBD307A mutant, the phyBR322A photoreceptor was readily degraded upon photoconversion to Pfr in vivo (Fig. 7A). However, its ability to relocalize into nuclear bodies in R appeared to be compromised. Even a high fluence rate of R, which readily induced such nuclear aggregation of phyBWT-YFP, caused only a modest aggregation response for phyBR322A-YFP. Many nuclei accumulated only a few small bodies of phyBR322A-YFP within a diffuse background of YFP fluorescence after a 12-h exposure to 90 μmol m−2 s−1 R (Fig. 7B). Taking the response of the phyBWT, phyBY361F, phyBR582A, and phyBR322A-YFP fusions together, we found an inverse relationship between Pfr→Pr thermal reversion and phyB nuclear body formation (i.e. the slower the thermal reversion rate, the more aggregation).
Analysis of Comparable Mutations in Arabidopsis phyA
Given our ability to modify phyB signaling via the D307A and Y361F substitutions, we also examined whether comparable mutations (D273A and Y327F, respectively; Fig. 1C) would have similar effects on phyA signaling. Phenotypically, phyA is the dominant isoform in etiolated seedlings and in plants exposed to FR-rich environments (Franklin and Quail, 2010). As shown in Supplemental Fig. S4, C and E, phyA activity can be easily measured by its ability to restore FR suppression of hypocotyl elongation in phyA null mutants such as phyA-211 (Reed et al., 1993). As above for phyB, we generated transgenic phyA-211 plants that expressed under the native PHYA promoter either wild-type phyA or the two mutants to levels comparable to that in wild-type plants (Supplemental Fig. S4D). Using this single assay, we found that the D273A mutation also strongly compromises signaling by phyA but that a marginal activity was seen at high FR fluence rates (100 µmol m−2 s−1). In contrast to the Y361F mutation in phyB, the Y327F mutation in phyA did not appear to make the photoreceptor hyperactive, as the response of the PHYAY327F seedlings matched closely that of wild-type and PHYAWT seedlings at all fluence rates tested. When etiolated and more mature plants grown under white light in long days (LD) were examined, we found that the PHYAD273A and PHYAY327F mutants developed similarly to wild-type and PHYAWT plants, suggesting that neither variant interfered with signaling by the other phy isoforms nor stimulated atypical photomorphogenesis (Supplemental Fig. S4). At present, we do not know the consequences of the D273A and Y327F mutations on phyA photochemistry or their effects on phyA turnover and aggregation into nuclear bodies. Consequently, we cannot propose how their phenotypic activities (or lack thereof) correlate with photochemical alterations in the phyA chromoprotein.
DISCUSSION
Given the central importance of phys in regulating plant photomorphogenesis, it has long been proposed that their rational redesign could offer new strategies to better tailor crops to fit specific agricultural environments, including, for example, the improvement of seed germination, attenuation of the SAR, modification of flowering time, and delayed senescence (Ballaré et al., 1994; Izaguirre et al., 2006). While sufficiently resolved three-dimensional structures of plant phys await, we demonstrate here that the bacterial models provide reasonable surrogates with which to predictably modify the photobehavioral properties of the plant versions via structure-guided mutagenesis. For the most part, the photochemical consequences of the panel of five mutants tested here within the PSM of Arabidopsis phyB agreed well with the responses of comparable mutants generated with various eubacterial and cyanobacterial relatives. These effects include the essential role for the Asp within the invariant DIP motif during Pr→Pfr photoconversion (Hahn et al., 2006; von Stetten et al., 2007; Wagner et al., 2008) and the importance of the pair of Arg residues that tether the B- and C-ring propionate side chains to Pfr stability (Hahn et al., 2006; Cornilescu et al., 2008; Wagner et al., 2008; Ulijasz et al., 2010).
The only surprise was the R582A mutation, which might have compromised signaling by eliminating a predicted salt bridge between the PHY domain hairpin and Asp-307 in the GAF domain that also contacts the bilin and the pyrrole water (Fig. 1B). While substitution of Arg-582 with an Ala did not impact photochemistry or substantially alter the ability of the photoreceptor to drive various phenotypic processes directed by phyB, it strongly stabilized the Pfr conformer once formed by slowing thermal reversion. Structural comparisons of the hairpin among the available three-dimensional structures for bacterial PSMs showed that this unique protrusion, while conserved in principle, is quite varied in terms of its secondary structure and in the specific residues that directly participate in the PHY-GAF domain interaction (Essen et al., 2008; Yang et al., 2008; Bellini and Papiz, 2012; E.S. Burgie and R.D. Vierstra, unpublished data). Consequently, it is possible that the hairpin-GAF contact in phyBR582A retained its critical role in plant phy signal transmission by using an ensemble of residues besides Arg-582 to support this connection. One alternative residue in Arabidopsis phyB might be Gly-564, which sits within a highly conserved WGG patch predicted from bacterial structures to be within the stretch of the PHY domain hairpin that interfaces with the GAF domain (Essen et al., 2008). Its importance was revealed by a Glu substitution found by forward genetics of the phyB-401 allele that generates a strongly hyperactive signaling variant (approximately 1,000-fold; Kretsch et al., 2000; Ádám et al., 2011). Clearly, appreciating the specific role(s) of Arg-582 and Gly-564 to phyB signaling will require the three-dimensional structure of phyB PSM.
In addition to being essential for phyB photochemistry, we confirmed that Asp-307 in phyB is also critical for signal transmission. Spectroscopic studies showed that the D307A variant assembles with PΦB to generate a normal Pr conformer but that photoconversion to Pfr by R is stalled, resulting in the accumulation of a bleached R-absorbing intermediate. Surprisingly, this bleached species appears still capable of stimulating phyB-dependent processes, including the suppression of hypocotyl elongation at very high fluence rates of R and the rescue of the early-flowering phenotype of phyB-9 plants in SD, suggesting that it resembles Pfr enough to permit modest signaling. A marginal phenotypic activity for phyA harboring the comparable D273A mutation is also implied by its ability to suppress hypocotyl elongation in high-fluence-rate FR. This “Pfr-like” bleached species of phyBD307A also retains its rapid degradation in continuous R, albeit at a reduced rate, implying that photoconversion all the way to Pfr is not a prerequisite to the light-accelerated turnover of phyB. Surprisingly, phyBD307A also failed to accumulate nuclear bodies. Although we cannot rule out the formation of smaller, poorly detectable aggregates, this disconnection suggests that a fully photoconverted Pfr structure is essential for nuclear aggregation and that the large bodies containing phyB are not prerequisites for phyB ubiquitylation and subsequent breakdown. Together with prior studies on the constitutively active phyBY276H allele (Su and Lagarias, 2007; Hu et al., 2009), it now appears possible to create mutant phys that are capable of stimulating downstream signaling pathways even without photoconversion to the normal Pfr state. An intriguing scenario is that such mutations structurally uncouple chromophore movements from the rest of the PSM and/or the OPM, thus allowing the PSM/OPM to assume the “activated” conformation required for signal transmission and/or breakdown independent of phototransformation.
The most striking mutation analyzed here is Y361F, which generates a phyB variant with enhanced light signal transmission. Unlike the previously described Y276H substitution, which appears to represent a gain-of-function mutation that permits phyB signaling even without light (Su and Lagarias, 2007; Hu et al., 2009), the effect(s) of Y361F are more subtle by generating a hyperactive photoreceptor that still requires light for activation. Why phyBY361F is hyperactive is unclear; it could reflect a greater affinity of the Pfr form for downstream components (e.g. PIFs; Al-Sady et al., 2006), an increase in its kinase activity if plant phys do indeed work as protein kinases (Yeh and Lagarias, 1998; Vierstra and Zhang, 2011), and/or an ability to retain signaling activity even upon conversion back to Pr.
As such, the phyBY361F variant might offer a more practical approach to accentuate phyB-mediated responses in an agricultural setting with fewer side effects. Its replacement of wild-type phyB increased dramatically the sensitivity of hypocotyls to R, generated seeds with a stronger germination response in white light, and for seedlings further accentuated the EOD-FR response and leaf hyponasty in SD but had little impact on flowering time. Given that the phyB-mediated response of hypocotyls to R and EOD-FR and leaf epinasty in low light are connected to the SAR (Franklin and Quail, 2010), we speculate that the increased signaling by phyBY361F attenuates the SAR by enabling the small amounts of Pfr generated by low-fluence R, or the residual Pfr remaining after EOD-FR (or presumably in high FR/R environments), to more effectively promote normal photomophogenesis. A similar repression of SAR was previously observed for transgenic tobacco (Nicotiana tabacum) overexpressing wild-type phyA, including the attenuation of stem elongation in white light, delayed leaf senescence, and repression of SAR elicited by FR-rich light environments or high plant densities (McCormac et al., 1991; Ballaré et al., 1994; Izaguirre et al., 2006).
It is interesting that the comparable Tyr-to-Phe mutation in phyA (Y327F) did not appear to generate a hyperactive variant, potentially in agreement with the distinct signaling mechanism(s) underpinning these phy isoforms and the unique responses they control (Franklin and Quail, 2010). Such a distinction between phyA and phyB was easily seen here by the steeper fluence rate response curves for hypocotyl elongation in FR (phyA driven) versus those in R (phyB driven), indicating that Arabidopsis is exquisitely sensitive to relatively low levels of phyA as Pfr but less so for phyB as Pfr. Regardless of the mechanism(s), such differential responses illustrate the potential of using mutants in specific phy isoforms to drive individual aspects of photomorphogenesis. Thus, by introducing specific structure-guided mutations into the set of phys present in the target species, it might be possible to create next-generation crops better designed to fit their particular cultivation schemes. In particular, the application of hyperactive phyB variants such as Y361F described here and the G564E(phyB-401) variant reported previously (Kretsch et al., 2000; Ádám et al., 2011) might be an attractive approach to minimize the complications of SAR for crops that are planted at high density or to maintain the dwarf stature of ornamental species exposed to low-light environments.
Similar to bacterial phys, we found that altering the GAF domain pocket surrounding the bilin can profoundly alter the thermal reversion of Arabidopsis phyB, with the Y361F, R352A, and R582A mutations strongly stabilizing the Pfr conformer and the R322A mutation destabilizing it. However, with the exception of Y361F, none of these mutations appeared to dramatically enhance or diminish phyB-mediated signaling under high fluence rates of R or in continuous or SD white light, implying that thermal reversion is insignificant to phyB when signaling is robust. However, a differential effect on the control of hypocotyl elongation at very-low-fluence-rate R was possible for PHYBR582A and PHYBR352A seedlings, suggesting that thermal reversion impacts phyB action when signaling is more limited. In addition, none of these mutants substantially affected phyB turnover. Consequently, at least under the high and continuous R fluence rates used here, Pfr thermal reversion does not appear to compete well with Pfr turnover.
Interestingly, we found that this collection of reversion mutants strongly affected nuclear aggregation in an inverse manner. Whereas the phyBR322A mutant, which reverts faster than phyBWT, appeared to show dampened aggregation, the phyBY361F and phyBR582A mutants, which revert much slower, showed more robust aggregation that was even detected at low fluence rates of R. This inverse relationship is also supported by the responses of the phyBG564E mutant, which displays almost no thermal reversion both in vitro and in planta but has a robust aggregation response even in low R (Ádám et al., 2011).
Collectively, the effects of this panel of thermal reversion mutants support the hypothesis that such bodies serve as reservoirs of photoactivated phyB and possibly other isoforms (Rausenberger et al., 2010). Based on the results by Ádám et al. (2011) and those presented here, one can imagine that phyB as Pfr aggregates into these bodies upon nuclear import, but as the pool slowly reverts back to Pr in darkness or under dim R, the bodies disaggregate. Those phyB mutants that revert faster would require higher fluence rates of R to maintain the aggregated pool (phyBR322A), while those that revert more slowly would maintain the aggregated pool even at low fluence rates (phyBY361F, phyBR582A, and phyBG564E mutants). It has also been proposed that these nuclear bodies are the concentrated sites for Pfr ubiquitylation and/or degradation. Using the phyBG564E mutant along with the collection described here, it should now be possible to test this idea via detailed kinetic comparisons of Pfr turnover and nuclear body formation under a range of continuous and pulsed R and FR fluence rates.
MATERIALS AND METHODS
Recombinant phyB Protein Expression, Purification, and Analysis
All the site-directed mutations in PHYB were introduced into the complementary DNA (cDNA) by the Quikchange method (Stratagene). cDNA fragments encoding the PSM (residues 1–624) were appended in frame to the N terminus of the 6His tag (KLHHHHHH) by introduction into the pBAD plasmid (Invitrogen). The PSM plasmids were cotransformed into Escherichia coli BL21 (AI) cells (Invitrogen) with the pPL-PΦB plasmid expressing the Synechocystis sp. PCC 6803 HO1 heme oxygenase and Arabidopsis (Arabidopsis thaliana) HY2 PΦB synthase enzymes (Bhoo et al., 2001; Gambetta and Lagarias, 2001) to direct apoprotein expression and chromophore assembly. Following sequential induction of the HO1/HY2 genes and PHYB genes with isopropyl-β-d-thiogalactopyranoside and Ara, respectively, the cells were disrupted by sonication in extraction buffer (50 mm HEPES-NaOH [pH 7.8], 300 mm NaCl, 30 mm imidazole, 0.1% Tween 20, 10% glycerol, 1 mm 2-mercaptoethanol, and 1 mm phenylmethylsulfonyl fluoride) with the addition of one tablet of protease inhibitor cocktail (Roche) before use. The clarified supernatant was applied to a HisTrap HP column (GE) preequilibrated in extraction buffer, and the column was washed with extraction buffer followed by elution with a 30 to 300 mm imidazole gradient in extraction buffer. The phyB-containing fractions were pooled, dialyzed against 10 mm HEPES-NaOH (pH 7.8), 100 mm NaCl, 5 mm 2-mercaptoethanol, 5 mm Na2EDTA, 50 mm imidazole, and 0.05% Tween 20 overnight and subjected to size-exclusion chromatography using a 24-mL Superose 6 (GE) column preequilibrated with the same buffer. phyB-containing fractions were pooled and stored in 10 mm HEPES-NaOH (pH 7.8), 50 mm NaCl, 1 mm 2-mercaptoethanol, 0.05% Tween 20, and 10% glycerol. Chromophore attachment was assayed by zinc-induced fluorescence of the PSMs following SDS-PAGE (Wagner et al., 2008). Pr/Pfr photointerconversion and Pfr→Pr thermal reversion of each phyB preparation were assayed at 24°C by UV visible absorption spectroscopy using white light filtered through 650- and 730-nm interference filters (Orion) to drive Pr→Pfr and Pfr→Pr phototransformation, respectively. Pfr half-lives in the dark were calculated from initial rates.
Plant Materials and Growth Conditions
All the plant lines were derived from Arabidopsis Col-0 ecotype. The phyB-9 and phyA-211 alleles were as described (Reed et al., 1993, 1994). Seeds were surface sterilized in chlorine gas and stratified in water for 3 d at 4°C before sowing. Unless otherwise noted, seedlings were grown at 22°C under white light in LD (16 h of light/8 h of dark) on 0.7% (w/v) agar medium containing 1× Gamborg’s salts, 2% (w/v) Suc, and 0.5 g L−1 MES (pH 5.7). After 10 d, seedlings were transferred to soil and grown at 22°C under continuous white light in LD or SD.
Plasmid Constructs for Plant Transformation
The full coding regions of PHYA and PHYB (Sharrock and Quail, 1989) were inserted into the pDONR221 plasmid via BP reactions (Invitrogen) to append in frame the coding sequence for the Flag epitope (GGGDYKDDDDK) to their 3′ ends. The PHYA/PHYB promoter, 5′ untranslated regions (2,634 and 1,983 bp upstream from at the ATG translation initiation codon), and 3′ untranslated regions (242 and 27 bp downstream of the translation termination codon) were amplified by PCR from Arabidopsis Col-0 genomic DNA and then sequentially inserted into the pDONR211 plasmids to appropriately flank the coding regions. The complete PHYB and PHYA transgenes were introduced into the pMDC123 plasmid (Invitrogen) via LR reactions. The PHYB-YFP constructs were created by appending the UBQ10 promoter fragment (1,986-bp fragment proximal to the ATG codon) and the cDNA encoding YFP to the 5′ and 3′ ends of the PHYB cDNA in a pDONR211 plasmid, respectively. The complete transgenes were introduced into the pMDC123 plasmid via LR reactions.
Plant Transformation and Selection of Transgenic Lines
The PHYA and PHYB transgenes were introduced into the homozygous Arabidopsis phyA-211 and phyB-9 mutants (Reed et al., 1993, 1994), respectively, via the Agrobacterium tumefaciens-mediated floral dip method using the pMDC123-derived plasmids. Transformed lines were selected by resistance to 10 μg mL−1 BASTA. T2 transgenic plants with a resistance segregation ratio of approximately 3:1 were used to obtain isogenic lines in the T4 or T5 generation for all the biochemical, phenotypic, and localization assays.
Protein Extraction and Immunoblot Analysis
Five-day-old, dark-grown Arabidopsis seedlings were frozen and pulverized at liquid nitrogen temperatures and homogenized in 100 mm Tris-HCl (pH 8.5), 10 mm Na2EDTA, 25% ethylene glycol, 2 mm phenylmethylsulfonyl fluoride, 10 mm N-ethylmaleimide, 5 μg mL−1 sodium metabisulfite, 2% (w/v) SDS, 10 μg mL−1 aprotinin, 10 μg mL−1 leupeptin, and 0.5 μg mL−1 pepstatin. The extracts were heated to 100°C for 10 min and clarified by centrifugation at 13,000g for 10 min. The supernatants were subjected to SDS-PAGE and immunoblot analysis with a monoclonal antibody against phyA (073D; Shanklin et al., 1989), phyB (B1-B7; Hirschfeld et al., 1998), or GFP (Sigma). Anti-PBA1 antiserum (Book et al., 2010) or anti-histone H3 antibodies (Abcam) were used to confirm equal protein loading.
To measure phyB degradation in response to R, seeds were sown in liquid medium containing one-half-strength Murashige and Skoog salts, 0.5 g L−1 MES (pH 5.7), and 10 g L−1 Suc and irradiated with white light (24 h for seeds carrying the PHYBD307A transgene and 12 h for all others) to initiate germination before maintaining the seedlings in the dark for 4 d. Seedlings were collected after various exposure times to continuous 20 μmol m−2 s−1 R at 22°C and subjected to immunoblot analysis as above. Seedlings were incubated for 12 h in the dark with 100 μm MG132 or an equivalent volume of dimethyl sulfoxide before R.
Phenotypic Assays
Germination efficiency was measured according to Oh et al. (2007). The parental plants (five per genotype) were grown side by side at 22°C in LD, and the resulting seeds were harvested as separated seed pools. At least 60 seeds from each pool were sown on 0.7% (w/v) water agar after 20 min of FR irradiation (4 μmol m−2 s−1). The seeds were then exposed to white light for 2 h and either kept in the dark or irradiated with 4 μmol m−2 s−1 FR for 5 min. The plates were kept in darkness for an additional 5 d before measurement of germination, which was scored as emergence of the radicle from the seed coat. For hypocotyl elongation, seeds were sown on solid one-half-strength Murashige and Skoog salts, 0.5 g L−1 MES (pH 5.7), and 0.7% (w/v) agar and irradiated with 12 h of white light. The plates were exposed to either R or FR for 3.5 d using a bank of diodes (E-30LED-controlled environment chamber; Percival) before measurement of hypocotyl length. For measurement of the EOD-FR response, seedlings were irradiated over a 4-d cycle with 90 μmol m−2 s−1 R for 8 h followed by either darkness or a 10-min pulse of 100 μmol m−2 s−1 FR and then darkness for 16 h. The effect on leaf epinasty and flowering time was measured for plants grown under white light in SD (Fankhauser and Casal, 2004). Leaf angles were determined for plants in the middle of the light period.
Confocal Microscopic Analysis
Transgenic seeds expressing wild-type and mutant versions of phyB-YFP were sown on solid medium containing one-half-strength Murashige and Skoog salts, 0.5 g L−1 MES (pH 5.7), 2% (w/v) Suc, and 0.7% (w/v) agar and irradiated for 12 h at 22°C with white light before incubation in the dark for 4 d. Fluorescence of hypocotyl cells, either kept in the dark or irradiated with 0.25 or 90 μmol m−2 s−1 R for 12 h, was imaged immediately after treatment using a Zeiss 510-Meta laser scanning confocal microscope. YFP fluorescence was visualized in the single-track mode by excitation with 488-nm light using the BP 500-530 infrared filter. Images were captured soon after excitation to minimize the effects of phyB photoconversion on localization. Images were processed with the LSM510 image browser (http://www.zeiss.com/lsm).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_100828 (phyA) and NM_127435 (phyB).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Purification and assembly of the PSM from wild-type phyB and the various site-directed mutants.
Supplemental Figure S2. Rates of Pr→Pfr and Pfr→Pr photoconversion for the PSM of phyB and the collection of site-directed mutants.
Supplemental Figure S3. Accumulation and activity of YFP fusions of phyB in Arabidopsis seedlings.
Supplemental Figure S4. Phenotype of the Arabidopsis phyA-211 null mutant rescued with transgenes expressing wild-type phyA or the D273A or Y327F mutant.
Supplemental Table S1. Summary of transgenic plants used in this study.
Acknowledgments
We thank Dr. Peter Quail for the gift of the PHYA and PHYB cDNAs, Dr. J. Clark Lagarias for the PΦB synthesis plasmid, Drs. Ferenc Nagy and Faqiang Li for helpful advice, Dr. Andrew Ulijasz for generating the PHYB PSM construct, and Kai Su and Joseph M. Walker for technical assistance.
Glossary
- phy
phytochrome
- SAR
shade-avoidance response
- PSM
photosensory module
- GAF
cGMP phosphodiesterase/adenylyl cyclase/FhlA
- OPM
output module
- R
red light
- FR
far-red light
- PΦB
phytochromobilin
- EOD
end-of-day
- SD
short days
- t1/2
half-time
- YFP
yellow fluorescent protein
- Syn
Synechocystis sp. PCC 6803
- Dr
Deinococcus radiodurans
- LD
long days
- SyB
Synechoccocus sp. OS-B′
- cDNA
complementary DNA
- t1/2
half-life
References
- Ádám E, Hussong A, Bindics J, Wüst F, Viczián A, Essing M, Medzihradszky M, Kircher S, Schäfer E, Nagy F. (2011) Altered dark- and photoconversion of phytochrome B mediate extreme light sensitivity and loss of photoreversibility of the phyB-401 mutant. PLoS ONE 6: e27250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Auldridge ME, Forest KT. (2011) Bacterial phytochromes: more than meets the light. Crit Rev Biochem Mol Biol 46: 67–88 [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Scopel AL, Jordan ET, Vierstra RD. (1994) Signaling among neighboring plants and the development of size inequalities in plant populations. Proc Natl Acad Sci USA 91: 10094–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini D, Papiz MZ. (2012) Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 20: 1436–1446 [DOI] [PubMed] [Google Scholar]
- Bhoo SH, Davis SJ, Walker J, Karniol B, Vierstra RD. (2001) Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414: 776–779 [DOI] [PubMed] [Google Scholar]
- Book AJ, Gladman NP, Lee SS, Scalf M, Smith LM, Vierstra RD. (2010) Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem 285: 25554–25569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan MT, Quail PH. (1991) Phytochrome a overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc Natl Acad Sci USA 88: 10806–10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Walker JM, Phillips GN, Jr, Vierstra RD. (2013) A photo-labile thioether linkage to phycocyanobilin provides the foundation for the blue/green photocycles in the DXCF-cyanobacteriochromes. Structure 21: 88–97 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Schwab R, Chory J. (2003) Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci USA 100: 14493–14498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JR, Hondred D, Walker JM, Vierstra RD. (1992) Phytochrome requires the 6-kDa N-terminal domain for full biological activity. Proc Natl Acad Sci USA 89: 5039–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hua Z, Lauer TD, Vierstra RD. (2012) The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol 160: 118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Ulijasz AT, Cornilescu CC, Markley JL, Vierstra RD. (2008) Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state. J Mol Biol 383: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elich TD, Chory J. (1997) Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell 9: 2271–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen LO, Mailliet J, Hughes J. (2008) The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci USA 105: 14709–14714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Casal JJ. (2004) Phenotypic characterization of a photomorphogenic mutant. Plant J 39: 747–760 [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Lagarias JC. (2004) Harnessing phytochrome’s glowing potential. Proc Natl Acad Sci USA 101: 17334–17339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta GA, Lagarias JC. (2001) Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA 98: 10566–10571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J, Strauss HM, Landgraf FT, Gimenèz HF, Lochnit G, Schmieder P, Hughes J. (2006) Probing protein-chromophore interactions in Cph1 phytochrome by mutagenesis. FEBS J 273: 1415–1429 [DOI] [PubMed] [Google Scholar]
- Hanzawa H, Inomata K, Kinoshita H, Kakiuchi T, Jayasundera KP, Sawamoto D, Ohta A, Uchida K, Wada K, Furuya M. (2001) In vitro assembly of phytochrome B apoprotein with synthetic analogs of the phytochrome chromophore. Proc Natl Acad Sci USA 98: 3612–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA. (1998) Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Su YS, Lagarias JC. (2009) A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant 2: 166–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre MM, Mazza CA, Biondini M, Baldwin IT, Ballaré CL. (2006) Remote sensing of future competitors: impacts on plant defenses. Proc Natl Acad Sci USA 103: 7170–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Wagner JR, Walker JM, Vierstra RD. (2005) Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem J 392: 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Gil P, Kozma-Bognár L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adám E, Schäfer E, Nagy F. (2002) Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krall L, Reed JW. (2000) The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc Natl Acad Sci USA 97: 8169–8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsch T, Poppe C, Schäfer E. (2000) A new type of mutation in the plant photoreceptor phytochrome B causes loss of photoreversibility and an extremely enhanced light sensitivity. Plant J 22: 177–186 [DOI] [PubMed] [Google Scholar]
- Kunkel T, Speth V, Büche C, Schäfer E. (1995) In vivo characterization of phytochrome-phycocyanobilin adducts in yeast. J Biol Chem 270: 20193–20200 [DOI] [PubMed] [Google Scholar]
- Li H, Zhang J, Vierstra RD, Li H. (2010) Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc Natl Acad Sci USA 107: 10872–10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailliet J, Psakis G, Feilke K, Sineshchekov V, Essen LO, Hughes J. (2011) Spectroscopy and a high-resolution crystal structure of Tyr263 mutants of cyanobacterial phytochrome Cph1. J Mol Biol 413: 115–127 [DOI] [PubMed] [Google Scholar]
- McCormac AC, Cherry JR, Hershey HP, Vierstra RD, Smith H. (1991) Photobiology of tobacco altered by expression of oat phytochrome. Planta 185: 162–170 [DOI] [PubMed] [Google Scholar]
- Nagatani A. (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13: 565–570 [DOI] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G. (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Matsushita T, Mochizuki N, Quail PH, Nagatani A. (2008) Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet 4: e1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J, Hussong A, Kircher S, Kirchenbauer D, Timmer J, Nagy F, Schäfer E, Fleck C. (2010) An integrative model for phytochrome B mediated photomorphogenesis: from protein dynamics to physiology. PLoS ONE 5: e10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer E, Nagy F. (2005) Photomorphogenesis in Plants and Bacteria: Function and Signal Transduction Mechanisms, Ed 3. Springer-Verlag, Dordrecht, The Netherlands [Google Scholar]
- Shanklin J, Jabben M, Vierstra RD. (1989) Partial purification and peptide mapping of ubiquitin phytochrome conjugates in oat. Biochemistry 28: 6028–6034 [Google Scholar]
- Sharrock RA, Quail PH. (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745–1757 [DOI] [PubMed] [Google Scholar]
- Su YS, Lagarias JC. (2007) Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19: 2124–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulijasz AT, Cornilescu G, Cornilescu CC, Zhang J, Rivera M, Markley JL, Vierstra RD. (2010) Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature 463: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD, Zhang J. (2011) Phytochrome signaling: solving the Gordian knot with microbial relatives. Trends Plant Sci 16: 417–426 [DOI] [PubMed] [Google Scholar]
- von Stetten D, Seibeck S, Michael N, Scheerer P, Mroginski MA, Murgida DH, Krauss N, Heyn MP, Hildebrandt P, Borucki B, et al. (2007) Highly conserved residues Asp-197 and His-250 in Agp1 phytochrome control the proton affinity of the chromophore and Pfr formation. J Biol Chem 282: 2116–2123 [DOI] [PubMed] [Google Scholar]
- Wagner D, Quail PH. (1995) Mutational analysis of phytochrome B identifies a small COOH-terminal-domain region critical for regulatory activity. Proc Natl Acad Sci USA 92: 8596–8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Tepperman JM, Quail PH. (1991) Overexpression of phytochrome B induces a short hypocotyl phenotype in transgenic Arabidopsis. Plant Cell 3: 1275–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JR, Brunzelle JS, Forest KT, Vierstra RD. (2005) A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438: 325–331 [DOI] [PubMed] [Google Scholar]
- Wagner JR, Zhang J, Brunzelle JS, Vierstra RD, Forest KT. (2007) High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J Biol Chem 282: 12298–12309 [DOI] [PubMed] [Google Scholar]
- Wagner JR, Zhang J, von Stetten D, Günther M, Murgida DH, Mroginski MA, Walker JM, Forest KT, Hildebrandt P, Vierstra RD. (2008) Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J Biol Chem 283: 12212–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kuk J, Moffat K. (2008) Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci USA 105: 14715–14720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ren Z, Kuk J, Moffat K. (2011) Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479: 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Stojkovic EA, Kuk J, Moffat K. (2007) Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc Natl Acad Sci USA 104: 12571–12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. (1998) Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA 95: 13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]