Altered substrate provision to developing soybeans leads to changes in metabolic pathway fluxes and carbon and nitrogen partitioning.
Abstract
Soybean (Glycine max) seeds store significant amounts of their biomass as protein, levels of which reflect the carbon and nitrogen received by the developing embryo. The relationship between carbon and nitrogen supply during filling and seed composition was examined through a series of embryo-culturing experiments. Three distinct ratios of carbon to nitrogen supply were further explored through metabolic flux analysis. Labeling experiments utilizing [U-13C5]glutamine, [U-13C4]asparagine, and [1,2-13C2]glucose were performed to assess embryo metabolism under altered feeding conditions and to create corresponding flux maps. Additionally, [U-14C12]sucrose, [U-14C6]glucose, [U-14C5]glutamine, and [U-14C4]asparagine were used to monitor differences in carbon allocation. The analyses revealed that: (1) protein concentration as a percentage of total soybean embryo biomass coincided with the carbon-to-nitrogen ratio; (2) altered nitrogen supply did not dramatically impact relative amino acid or storage protein subunit profiles; and (3) glutamine supply contributed 10% to 23% of the carbon for biomass production, including 9% to 19% of carbon to fatty acid biosynthesis and 32% to 46% of carbon to amino acids. Seed metabolism accommodated different levels of protein biosynthesis while maintaining a consistent rate of dry weight accumulation. Flux through ATP-citrate lyase, combined with malic enzyme activity, contributed significantly to acetyl-coenzyme A production. These fluxes changed with plastidic pyruvate kinase to maintain a supply of pyruvate for amino and fatty acids. The flux maps were independently validated by nitrogen balancing and highlight the robustness of primary metabolism.
The legume soybean (Glycine max) is one of the most important oilseed crops, providing large amounts of protein for food and feed applications and supplying a significant proportion of the vegetable oil used for cooking and for chemical feedstocks. Given these demands, there is considerable interest in manipulation of the underlying biochemical pathways that produce oil and protein in the seed. The proportions of these storage reserves are established by both the supply of maternal precursors and the metabolic processes of the developing embryo. Embryos receive sugars and amino acids and convert them into storage reserves through primary metabolism. Thus, the accumulation of protein and oil is controlled by both the maternal (Fabre and Planchon, 2000; Nakasathien et al., 2000; Pipolo et al., 2004) and seed (Wilcox, 1998; Narvel et al., 2000; Hernández-Sebastià et al., 2005) genotypes, which complicates efforts to gain fundamental understanding of the process.
This complexity can be partially circumvented by studying the growth of cultured embryos. In planta, embryos receive carbon and nitrogen apoplastically (Thorne, 1980, 1981). An environment suitable for embryo growth can be recreated within a laboratory setting using known apoplastic nutrients (Hsu et al., 1984; Rainbird et al., 1984). Experimentally providing precise levels of substrates to filling embryos (i.e. reproductive stage R5–R5.5) allows them to grow in a controlled way (Thompson et al., 1977; Obendorf and Wettlaufer, 1984), leads to compositions similar to those in planta (Hsu and Obendorf, 1982; Allen et al., 2009b; Bates et al., 2009), and confines the “maternal” influence to defined substrate concentrations. In this design, the influence of carbon and nitrogen provisions on seed metabolism and the production of storage reserves can be probed.
The impact of carbon and nitrogen availability on protein levels in seeds has been studied in maturing plants, which remobilize leaf nitrogen to increase protein in the seed (Saravitz and Raper, 1995). Likewise, cultured embryos supplied with increasing amounts of nitrogen produce more protein in both the wild type (Saravitz and Raper, 1995; Nakasathien et al., 2000; Pipolo et al., 2004) and high-protein mutants (Hayati et al., 1996). Although plant-supplied substrates impact resource allocation and affect final storage reserve compositions within the soybean seed, there are no existing quantitative flux models that describe how changes in provisions alter the underlying metabolism.
Metabolic flux analysis (MFA) aims to provide a quantitative description of the flow of matter within a biological network and relies on 13C or other isotopes to track, or enrich, metabolites according to biochemical fluxes and pathways (Stephanopoulos et al., 1998). The metabolic reactions in the network rearrange carbon atoms as metabolites are interconverted, without accumulation or depletion of the intermediates. Thus, adherence to the laws of mass conservation allows the calculation of in vivo fluxes. The flux models establish network function (Ratcliffe and Shachar-Hill, 2006) and can occasionally identify unique roles for enzymes (Schwender et al., 2004). Most MFA studies in plants have focused on seeds (Troufflard et al., 2007; Iyer et al., 2008; Allen et al., 2009b; Lonien and Schwender, 2009; Alonso et al., 2010, 2011) because of their pseudo-steady-state metabolism and economic importance, although plant cell suspensions have been used (Rontein et al., 2002; Baxter et al., 2007; Williams et al., 2008; Masakapalli et al., 2010) because of their experimental versatility. Together, these studies have quantified roles for metabolic pathways, such as the use of the tricarboxylic acid cycle (Schwender, 2008; Sweetlove et al., 2008, 2010; Allen et al., 2009b; Kruger and Ratcliffe, 2009) to produce ATP (Alonso et al., 2007a) or to supply citrate for cytosolic acetyl-CoA when operating as an incomplete cycle (Schwender et al., 2006). Additionally, MFA has been used to assess the impact of genetic and environmental perturbations in plants (Rontein et al., 2002; Spielbauer et al., 2006; Alonso et al., 2007b; Junker et al., 2007; Iyer et al., 2008; Williams et al., 2008; Lonien and Schwender, 2009) and, therefore, should be useful in probing the influence of carbon and nitrogen on the resulting composition of the filling embryo.
This study examines how the flux through primary metabolism within developing soybeans is altered by changes in carbon and nitrogen supply. The modeling produced detailed flux maps that described the redistribution of resources within the seed and that matched observed biomass compositions (i.e. amino acid fluxes to biomass were fitted as part of the modeling). Developing seeds were supplied varying levels of substrate (unlabeled or 14C enriched) in different carbon-to-nitrogen (C:N) ratios and were analyzed for the production of oil, protein, and carbohydrate. Next, the flow of carbon through primary metabolism was monitored in separate culture experiments providing Glc, Gln, or Asn labeled with 13C. Three labeling experiments with four to five replicates for each of the three C:N conditions resulted in approximately 40 independent 13C-enriched cultures. The enrichment of amino acids and the direct measurement of biomass were used as inputs for modeling steady-state fluxes of the developing embryo. The analyses indicate the following: (1) soybean protein concentration ranged from approximately 14% to 47% of total biomass and changed consistently as a function of C:N ratio; (2) altered nitrogen supply did not dramatically alter storage protein subunit profiles or affect the relative levels for most amino acids; and (3) Gln supply contributed 10% to 23% of the carbon for biomass production, including 9% to 19% of carbon to fatty acid biosynthesis and approximately 32% to 46% of carbon to amino acids. These results suggest that seed metabolism can accommodate different levels of protein production, although it may constrain total biomass accumulation. Additionally, ATP-citrate lyase and malic enzyme flux changes are offset with plastidic pyruvate kinase to maintain flow through pyruvate and to acetyl-CoA for the C:N conditions. The flux maps have been independently validated by nitrogen balancing and are discussed in the context of metabolic engineering.
RESULTS
C:N Ratio Influence over Seed Growth Rate
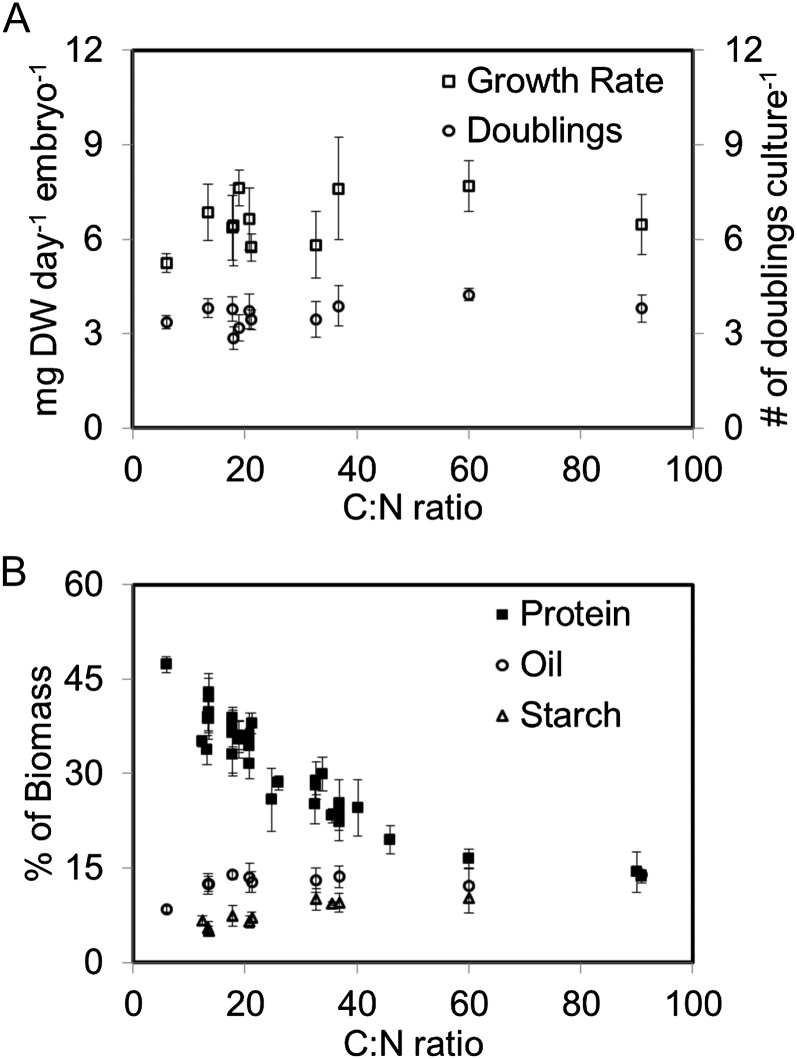
Soybean plants were grown under summer-like conditions to generate seeds for embryo culturing. Once the embryos had finished cell division, started linear seed filling (Egli, 1998), and weighed 30 to 40 mg fresh weight, the embryos were dissected from the seed coat and cultured in medium with unlabeled, 14C-labeled, or 13C-labeled substrates (Table I). Fourteen days of culturing resulted in growth rates between 4.7 and 7.9 mg dry weight per day per embryo and approximately 90 to 100 mg of added biomass (Table II), consistent with both in planta growth measured at 5 to 7 mg dry weight per day per embryo (Rubel et al., 1972; Egli et al., 1985) and previous culturing studies in soybean (Hsu and Obendorf, 1982; Allen et al., 2009b). Biomass production and doubling rates (3.65 ± 0.17 doublings per 14 d of culture) were independent of C:N ratio except possibly in the extreme cases (Fig. 1A; Table II). The embryos provided with the lowest C:N ratio generally grew at a slower rate and, along with the highest C:N ratio cultures (i.e. C:N = 60 and 91), were visually different from other cultures and thus not used for further studies. The light level and temperature for all cultures were maintained at 30 to 35 µmol m−2 s−1 and 26°C to 27°C to minimize changes in metabolism of the growing embryos. These culturing conditions are sufficient to meet metabolic and isotopic steady-state assumptions (Allen et al., 2009b) and, in this study, produced growth that accounted for greater than 93% of all embryo biomass.
Table I. Provision of varied C:N molar ratios in culture medium.
Growth and total biomass compositions were determined from soybean ‘Jack’ embryos grown on medium supplied with various concentrations of Suc, Glc, Gln, and Asn. An example calculation of the resulting C:N ratio, taking into account carbon coming from supplied amino acids, is shown in Figure 2. Culture compositions in boldface were chosen for 13C-metabolic flux analysis. Other culture parameters are described in “Materials and Methods.”
| C:N Ratio | Carbon Supplied as Amino Acids | Substrate Provided |
|||
|---|---|---|---|---|---|
| Suc | Glc | Gln | Asn | ||
| % | mm | ||||
| 6 | 40 | 90 | 45 | 140 | 50 |
| 13 | 18 | 140 | 70 | 83 | 12 |
| 13 | 18 | 140 | 70 | 70 | 25 |
| 18 | 13 | 140 | 70 | 50 | 18 |
| 19 | 13 | 150 | 75 | 50 | 18 |
| 21 | 11 | 150 | 75 | 45 | 16 |
| 33 | 7 | 170 | 85 | 30 | 12 |
| 37 | 6 | 170 | 85 | 25 | 12 |
| 60 | 4 | 200 | 100 | 16 | 10 |
| 91 | 3 | 180 | 90 | 11 | 4 |
Table II. Biomass composition of soybean ‘Jack’ embryos from altered carbon and nitrogen provision.
Protein, oil, and starch content were determined (unless indicated otherwise [ND]), and percentage was calculated as mg/100 mg × 100. Protein was established by C:N analysis, total oil was established with gas chromatography-flame ionization detection, and starch was determined by spectrophotometrically monitoring starch hydrolysis to Glc. Cell wall and soluble metabolites are presumed to represent the remainder of the composition. Boldface C:N ratios were used in further experiments.
| C:N Ratio | Biomass | Doubling No. | Protein | Oil | Starch |
|---|---|---|---|---|---|
| mg dry wt d−1 embryo−1 | % | ||||
| 6 | 5.2 ± 0.3 | 3.4 ± 0.2 | 47.3 ± 1.2 | 8.5 ± 0.4 | ND |
| 13 | 6.9 ± 0.9 | 3.8 ± 0.3 | 41.2 ± 3.0 | 12.5 ± 1.4 | 5.2 ± 0.9 |
| 18 | 6.4 ± 1.2 | 2.8 ± 0.4 | 36.2 ± 1.4 | 13.9 ± 0.7 | 7.4 ± 1.7 |
| 19 | 7.6 ± 0.6 | 3.2 ± 0.4 | 35.8 ± 2.6 | ND | ND |
| 21 | 6.6 ± 1.0 | 3.7 ± 0.6 | 34.1 ± 3.1 | 13.1 ± 2.0 | 6.8 ± 0.9 |
| 33 | 5.8 ± 1.0 | 3.4 ±0.6 | 27.4 ± 2.5 | 13.1 ± 1.9 | 10.0 ± 1.7 |
| 37 | 7.6 ± 1.6 | 3.9 ± 0.6 | 23.8 ± 2.5 | 13.6 ± 1.8 | 9.5 ±1.5 |
| 60 | 7.7 ± 0.8 | 4.2 ± 0.2 | 16.5 ± 1.6 | 12.2 ± 2.6 | 10.3 ± 2.4 |
| 91 | 6.5 ± 0.9 | 3.8 ± 0.4 | 14.0 ± 2.1 | 13.9 ± 0.8 | ND |
Figure 1.
Variation in biomass growth and composition of developing soybean embryos from provision of altered amounts of carbon and nitrogen. The amount of biomass produced (A) and the percentage of storage in the forms of protein, oil, and starch (B) were measured across molar C:N ratios that varied from 6:1 to 91:1. Each point with error bars represents three to six separate culture replicates; in total, 184 cultures were examined. DW, Dry weight.
Effect of C:N Ratio on Protein Concentration
Different levels of carbon and organic nitrogen in the culture medium resulted in different biomass compositions (Fig. 1B; Table II). Embryos receiving more organic nitrogen (i.e. lower C:N ratio; defined in Fig. 2A) produced more protein per unit of biomass, consistent with other reports (Saravitz and Raper, 1995; Hayati et al., 1996). Protein changed from approximately 47% to 14% of total biomass (Table II; Supplemental Data Set S1 as the carbon supplied from amino acids was reduced from 40% to 3% of total (Table I).
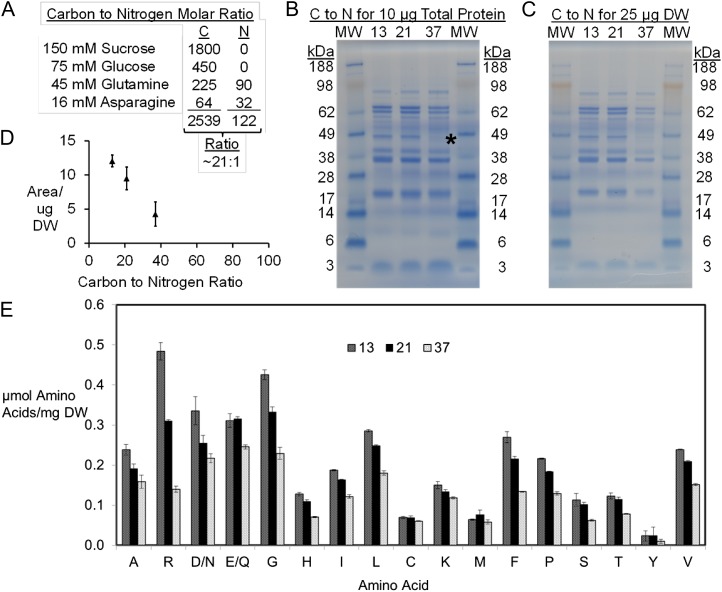
Figure 2.
Fluxes to protein-derived amino acids. The protein composition and amino acid concentrations derived from protein were experimentally determined and used to calculate metabolic fluxes. A, Sample calculation for C:N ratio for the values described. B, Analysis of 10 μg of total protein by SDS-PAGE. The 48-kD missing band described in the text is highlighted by an asterisk. C, Analysis of 25 μg of biomass. D, Analysis of equivalent amounts of dry weight (DW) over increasing C:N ratios by gel densitometry analysis. E, Individual amino acid fluxes relative to the production of 1 mg of biomass.
The profile and subunit composition of storage proteins were inspected by separating equal amounts of either total protein or biomass by SDS-PAGE. Images in Figure 2, B and C, representative of three gels each, revealed a similar storage protein pattern for different C:N ratios. Densitometry analyses confirmed the change in protein levels across C:N ratios (Fig. 2D). At the lowest supplied nitrogen level, the production of a storage protein at 48 kD was diminished. Proteomic analysis of this band identified it as the β-subunit of β-conglycinin (Supplemental Data Set S2). This subunit lacks sulfur-containing amino acids but has similar nitrogen content to other subunits; therefore, its disappearance was unexpected. While different inorganic nitrogen forms have been found to alter the concentration of β-subunits (Paek et al., 1997), the regulation of β-conglycinin is both transcriptional and posttranscriptional (Harada et al., 1989) and specific to individual α-, α′-, and β-subunits. Thus, the variable quality and quantity of subunits in β-conglycinin and glycinin (Wilson, 1987) remain a point of interest (Yaklich, 2001).
The amino acid profiles (micromoles of amino acid relative to the production of 1 mg of biomass) from hydrolyzed protein were compared for three different C:N ratios (13:1, 21:1, and 37:1) and used to calculate amino acid biosynthetic rates per milligram of biomass. Although total protein content increased with decreasing C:N ratios, the overall composition of the protein-bound amino acids did not drastically change, with the exception of Arg (Fig. 2E; Table III; Supplemental Data Set S3). The increased protein levels in embryos cultured in medium with a C:N ratio of 13:1 resulted in larger fluxes for most amino acids (on average, the 13:1 C:N ratio resulted in amino acid biosynthetic fluxes 71% greater than the 37:1 C:N ratio). These data indicated that total protein concentration responded to nitrogen levels but that relative amino acid composition varied more modestly. The protein levels for flux analysis were determined using Jones factors for the measured amino acid compositions as described in “Materials and Methods” and Supplemental Data Set S1 (Salo-Väänänen and Koivistoinen, 1996).
Table III. Fluxes to protein-bound amino acids, acetyl groups, and hexose units.
The fluxes to amino acids from hydrolyzed protein were determined experimentally by a combination of C:N and amino acid analyses, as described in “Materials and Methods,” and resulted in higher flux values for all amino acids in C:N 13:1. The model fluxes were determined relative to the measured fatty acid fluxes determined per two acetyl units (i.e. four acetyl carbons), and carbohydrates representing the remainder of the biomass were determined on a per hexose unit basis. All fluxes are reported as micromoles of metabolite per milligram dry weight.
| Metabolite | 13:1 |
21:1 |
37:1 |
|||
|---|---|---|---|---|---|---|
| Flux | se | Flux | se | Flux | se | |
| Ala | 0.24 | 0.01 | 0.19 | 0.01 | 0.16 | 0.02 |
| Arg | 0.48 | 0.02 | 0.31 | 0.00 | 0.14 | 0.01 |
| Asp + Asn | 0.34 | 0.04 | 0.25 | 0.02 | 0.22 | 0.01 |
| Glu + Gln | 0.31 | 0.02 | 0.32 | 0.01 | 0.25 | 0.01 |
| Gly | 0.43 | 0.01 | 0.33 | 0.01 | 0.23 | 0.01 |
| His | 0.13 | 0.00 | 0.11 | 0.00 | 0.07 | 0.00 |
| Ile | 0.19 | 0.00 | 0.16 | 0.00 | 0.12 | 0.00 |
| Leu | 0.29 | 0.00 | 0.25 | 0.00 | 0.18 | 0.01 |
| Cys | 0.07 | 0.00 | 0.07 | 0.00 | 0.06 | 0.00 |
| Lys | 0.15 | 0.01 | 0.13 | 0.01 | 0.12 | 0.00 |
| Met | 0.06 | 0.00 | 0.08 | 0.01 | 0.06 | 0.01 |
| Phe | 0.27 | 0.01 | 0.22 | 0.01 | 0.13 | 0.00 |
| Pro | 0.22 | 0.00 | 0.18 | 0.00 | 0.13 | 0.00 |
| Ser | 0.11 | 0.02 | 0.10 | 0.01 | 0.06 | 0.00 |
| Thr | 0.12 | 0.01 | 0.11 | 0.01 | 0.08 | 0.00 |
| Tyr | 0.02 | 0.01 | 0.02 | 0.02 | 0.01 | 0.01 |
| Val | 0.24 | 0.00 | 0.21 | 0.00 | 0.15 | 0.00 |
| Carbohydrates | 2.86 | 0.11 | 3.26 | 0.13 | 3.86 | 0.09 |
| Fatty acids | 1.89 | 0.07 | 1.97 | 0.09 | 2.05 | 0.12 |
Seed storage compounds contain different amounts of carbon (i.e. a typical triacylglycerol with triolein composition, C57H104O6, is 77% carbon by weight; Glc polymers in carbohydrates [C6H12O6 minus H2O] are 44% carbon; and protein with the observed amino acid composition is 51% carbon); therefore, 14C labeling was used to monitor the changes in carbon allocation into biomass (Table IV). Soybeans were cultured in medium containing a combination of [U-14C12]Suc, [U-14C6]Glc, [U-14C5]Gln, and [U-14C4]Asn at levels consistent with the molar composition of carbon for each substrate (Goffman et al., 2005). Embryos were grown for 14 d before the biomass was processed as described in “Materials and Methods.” Briefly, oil, ethanol-soluble material, and protein were sequentially extracted. The ethanol fraction was further subjected to ion-exchange chromatography and HPLC to establish its contents. HPLC indicated that the only significant radioactivity in the cationic fraction eluted completely in the column void volume and may have represented a small amount of hydrophobic protein solubilized by the 80% ethanol (i.e. amino acid pools that did not come in the void volume were sensitively detected by UV [254 nm] and were present at low concentrations that did not result in measureable dpm values). Neither sugars, which represent part of the carbohydrate pool, nor organic acids changed with C:N ratio, consistent with their roles as stored intermediates in metabolism that have measureable levels in the vacuole. Organic acids accounted for 3.2% to 3.4% of total biomass (Table IV), indicating possible vacuolar pools of malate or citrate in soybeans as has been reported in plant cells (Adams and Rinne, 1981; Adams et al., 1982; Gout et al., 1993; Martinoia et al., 2007; Sweetlove et al., 2010; Etxeberria et al., 2012).
Table IV. 14C labeling of embryos cultured with altered concentrations of substrates.
Embryos cultured in 14C-labeled substrates were grown for 14 d and then processed (see “Materials and Methods”) to assess biomass composition and production. A comparison of the numbers with those measured by other means is provided in Supplemental Data Set S1.
| C:N Ratio | Biomass Growth | Biomass | Organic Acids | Protein | Oil | Carbohydrates |
|---|---|---|---|---|---|---|
| mg d−1 embryo−1 | dpm | |||||
| 13 | 5.0 ± 1.3 | 59,626 ± 1,497 | 1,915 ± 185 | 27,809 ± 2,684 | 9,279 ± 513 | 20,623 ± 1,791 |
| 21 | 4.8 ± 0.7 | 57,932 ± 6,209 | 2,005 ± 198 | 24,324 ± 4,217 | 10,523 ± 717 | 21,080 ± 3,973 |
| 37 | 5.1 ± 0.4 | 56,942 ± 2,918 | 1,955 ± 191 | 20,860 ± 1,070 | 14,163 ± 1,273 | 19,963 ± 2,151 |
Total dry weight accumulation was not dependent on C:N conditions (4.8–5.1 mg of biomass per day per embryo) and resulted in similar amounts of total 14C incorporation (56,942–59,626 dpm mg−1 dry weight; Table IV). Quantitative extraction of protein required urea (detergents alone were insufficient), which coextracts some carbohydrates and results in modestly overestimated protein levels but reduced differences between them for C:N conditions (Supplemental Data Set S1). Together, the measurements of dry weight accumulation and composition (Table II) and 14C-labeling experiments (Table IV) indicated that seed filling is capable of sustaining increased protein production but may restrict the total stored carbon in biomass.
13C Labeling in Biomass
13C-labeling experiments were performed to assess the carbon sources used for protein production. In each culturing experiment, only one labeled substrate was provided, while the other carbon substrates remained unlabeled (i.e. isotopic natural abundance). Therefore, any change in 13C abundance in amino acids derived from hydrolyzed protein reflected the fractional contribution of a particular carbon source. The amount of labeled, protein-derived amino acids was examined by gas chromatography-mass spectrometry (GC-MS), except for Arg, Lys, and Cys, which were approximated by levels of other amino acids that share the same precursors (i.e. Glu, Ile, and Ser, respectively). Together, the average isotopic enrichments and the measured flux to amino acids were used to determine what fraction of each carbon source flows to which amino acids (Supplemental Data Set S4). Amino acids were grouped by family for simple interpretation (Fig. 3), with Ile and Lys split between the pyruvate and Asp families, consistent with their known biosynthetic origins.
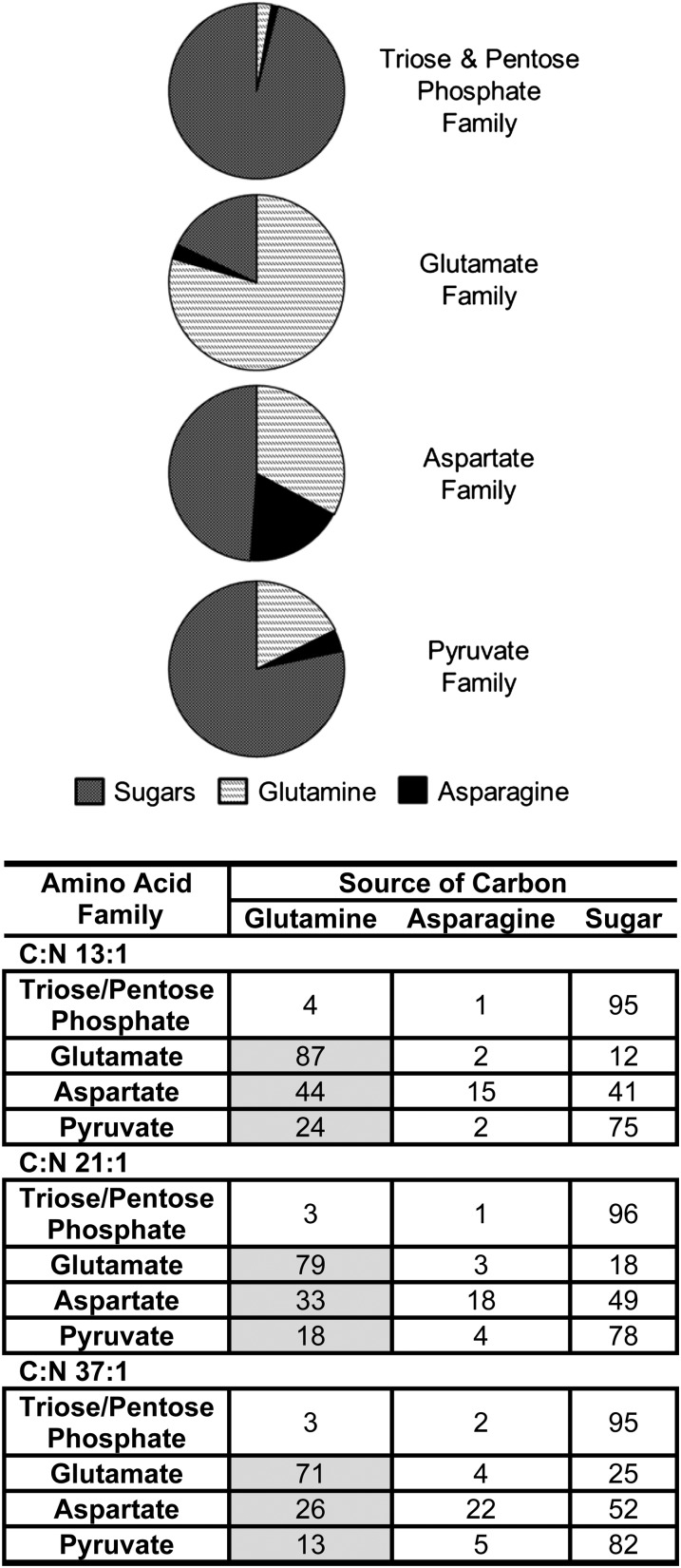
Figure 3.
Carbon source allocation into amino acid families. Provision of fully labeled substrates (Gln or Asn) resulted in the redistribution of label to amino acids and was used to establish the source of carbon for each amino acid family. Distribution of label to each amino acid along with carbon composition and corresponding biosynthetic fluxes were used to calculate the fractional contributions of different carbon sources to amino acids (for details, see Supplemental Data Set S4). Amino acids were grouped according to biosynthetic families by averaging. The results of the 21:1 C:N ratio culture are provided in the top panel, with the 13:1 and 37:1 C:N ratios that indicated similar trends provided in the table. The contributions of Gln-derived carbon to the Glu, Asp, and pyruvate amino acid families are highlighted by the shaded regions in the table.
Labeling experiments for the C:N ratios of 13:1, 21:1, and 37:1 revealed the carbon allocation into the four amino acid families (Fig. 3, with only 21:1 shown in the top panel; Supplemental Data Set S4). Carbon partitioning varied between the three C:N ratios (Fig. 3, bottom panel; Supplemental Data Set S4), with sugars providing more carbon at higher C:N ratios. The contribution of each carbon source to a particular amino acid family relative to other carbon sources is indicated by looking across the rows (i.e. the sum across each row is 100; Fig. 3). Triose/pentose phosphate-derived amino acids were made predominantly from sugars (95%–96% of their carbon composition), with small contributions to this family from Gln (3%–4%) and Asn (1%–2%) for the biosynthesis of Gly, consistent with our previous findings (Allen et al., 2009b). The low labeling in His, Ser, and Gly from the [U-13C]Gln provision (Supplemental Data Set S5) indicated that the phosphoenolpyruvate carboxykinase and gluconeogenic pathways are less active in embryo sink tissues. Furthermore, the absence of gluconeogenic activities across all three C:N ratios suggested that this form of metabolic activity was not induced or regulated by Gln, which serves as a precursor of organic acids in developing seeds.
Gln provided 32% to 46% of carbon for amino acids (shaded cells in the table in Fig. 3). Gln was a significant source of carbon for the Glu (71%–87%), Asp (26%–44%), and pyruvate (13%–24%) amino acid families and provided significantly more carbon to amino acids as the C:N ratio decreased. Since Gln contributed significantly to the biosynthesis of three of the four amino acid families, we examined the specific allocation of Gln-derived carbon (Supplemental Data Set S6). Gln partitioned predominantly into Glu-derived amino acids (64%–66% of supplied carbon) and to a lesser extent into Asp (18%–19%) and pyruvate (14%–16%) amino acid families. The relative allocation varied by less than 3% among the C:N conditions (Supplemental Data Set S6). Thus, the increased use of Gln-derived carbon (relative to sugars) at low C:N ratios reflected a concomitant increase in flux from Gln to each amino acid family. Amino acids derived from pyruvate (Ala, Val, Leu, Ile, and Lys) had significant incorporation of carbon from the Gln source (i.e. varied levels from 6% to 18% of total carbon; Fig. 4). Likewise, fatty acids derived from pyruvate consisted of 9% to 19% Gln-derived carbon (Fig. 4), a doubling through the range of the C:N experiments. Computationally fitted uptake fluxes indicated that Gln supply provided between 10% and 23% of all carbon for biomass, changing inversely with C:N conditions. Coordinated increases in the absolute values (i.e. unchanged relative flux values) have been reported in the primary metabolism of plant tissues where other environmental perturbations were considered (Williams et al., 2008) and emphasizes the plasticity of primary metabolism.
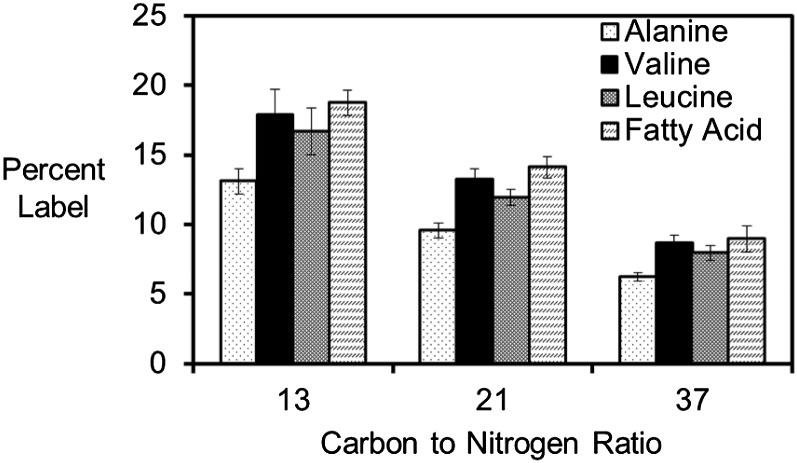
Figure 4.
Fatty acid synthesis utilizes Gln-derived carbon in soybeans. [U-13C5]Gln supplied to filling embryos resulted in pyruvate-derived metabolic products that were 13C labeled. The average proportion of labeled carbon was measured in pyruvate-derived amino acids and terminal acetate groups of fatty acids. The percentage of carbon labeled was similar between the two pyruvate-derived compound classes at each C:N ratio. The amount of incorporated carbon derived from Gln increases with decreasing C:N ratio.
Generation of Flux Maps for Three C:N Ratios from Multiple Labeling Experiments
MFA was used to investigate changes in the storage reserve composition of filling soybean embryos. MFA is a computational approach that mathematically balances the flow of carbon through metabolite pools in a given stoichiometric network. The network description included the carbon atom rearrangements catalyzed by enzymes and, therefore, accounted for isotope label redistribution through different metabolic pathways. Through a χ2 fitting process, the values for fluxes (i.e. the computational variables) were optimized to recapitulate both the experimentally measured label and flux data, resulting in “best estimates” of global flux values. The combination of isotopic labeling experiments with MFA provided a quantitative description of the flow of metabolites through biochemical pathways.
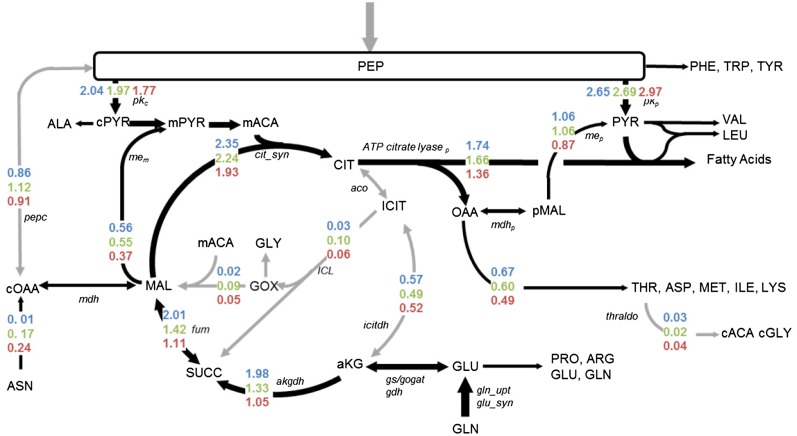
Three independent labeling experiments using [U-13C4]Asn, [U-13C5]Gln, or [1,2-13C2]Glc were used to assess central carbon metabolism under variable carbon and nitrogen supply. Measurements used for modeling were taken from label deposition into amino acids from hydrolyzed protein, which is commonly evaluated for flux analysis (Dauner et al., 2001). In total, between the three to five replicates for each of the three conditions, 6,337 isotopomer measurements were recorded (i.e. 152 distinct mass isotopomers for multiple conditions and replicates; Supplemental Data Set S5). Each model comprised 63 metabolites with 70 net and 20 exchange fluxes. The optimized flux network for each of the three culture variations (Fig. 5; Table V; Supplemental Data Set S7) was based upon a previous model (Allen et al., 2009b) that represented the key carbon transitions in primary metabolism and was expanded with information from a third labeling experiment. Figure 5 and Table V depict the fluxes that varied significantly with C:N ratio, while Supplemental Data Set S7 provides the entire description of the model and fluxes. Fluxes that increased most dramatically with the highest provision of Gln (i.e. C:N of 13:1) are designated by thick black arrows (Fig. 5).
Figure 5.
Metabolic flux maps of developing soybean embryos supplied three different C:N ratios. The values have units of micromoles of metabolite per milligram dry weight produced. For easy visualization, arrows that are increased in thickness and darkness emphasize differences in fluxes. Colored numbers indicate the fluxes from the 13:1 (blue), 21:1 (green), and 37:1 (red) C:N ratios. Flux values that changed most significantly are provided in Table V, and a complete list of all fluxes, 95% CIs, and a flux map that includes metabolism into phosphoenolpyruvate is provided in Supplemental Data Set S7. Metabolite abbreviations are in uppercase letters, and key enzymes are indicated in italics.
Table V. Net fluxes of pathway intermediates determined through metabolic flux analysis in developing soybean embryos provided three different C:N ratios.
Flux value rates are shown in units of micromoles per milligram dry weight produced. The 95% CIs are shown in brackets. A complete list of all fluxes including exchange fluxes is given in Supplemental Data Set S6.
| Reaction Description | 13:1 |
21:1 |
37:1 |
|||
|---|---|---|---|---|---|---|
| Flux Value | 95% CI | Flux Value | 95% CI | Flux Value | 95% CI | |
| Phosphoenolpyruvate carboxylase/carboxykinase | 0.86 | [0.75, 0.96] | 1.12 | [1.02, 1.19] | 0.91 | [0.80, 1.00] |
| Mitochondrial malic enzyme | 0.56 | [0.50, 0.62] | 0.55 | [0.50, 0.61] | 0.37 | [0.32, 0.42] |
| Plastidic malic enzyme | 1.06 | [1.01, 1.11] | 1.06 | [1.02,1.10] | 0.87 | [0.81, 0.92] |
| Cytosolic pyruvate kinase | 2.04 | [1.95, 2.15] | 1.97 | [1.89, 2.05] | 1.77 | [1.71, 1.86] |
| Plastidic pyruvate kinase | 2.65 | [2.54, 2.75] | 2.69 | [2.60, 2.79] | 2.97 | [2.86, 3.08] |
| Glu production | 2.11 | [2.02, 2.20] | 1.33 | [1.30, 1.67] | 0.80 | [0.77, 0.88] |
| Glu dehydrogenase | 3.51 | [3.35, 3.67] | 2.17 | [2.11, 2.54] | 1.33 | [1.28, 1.42] |
| Citrate synthase | 2.35 | [2.22, 2.48] | 2.24 | [2.14, 2.35] | 1.93 | [1.86, 2.01] |
| ATP-citrate lyase | 1.74 | [1.67, 1.81] | 1.66 | [1.58, 1.73] | 1.36 | [1.29, 1.42] |
| Isocitrate dehydrogenase | 0.57 | [0.44, 0.70] | 0.49 | [0.43, 0.58] | 0.52 | [0.47, 0.57] |
| 2-Oxoglutarate dehydrogenase | 1.98 | [1.78, 2.16] | 1.33 | [1.25, 1.45] | 1.05 | [0.98, 1.12] |
| Fumarase | 2.01 | [1.85, 2.18] | 1.42 | [1.35, 1.52] | 1.11 | [1.05, 1.17] |
| Thr aldolase | 0.03 | [0.02, 0.04] | 0.02 | [0.01, 0.03] | 0.04 | [0.03, 0.05] |
| Asparaginase/Asn aminotransferase | 0.01 | [−0.04, 0.06] | 0.17 | [0.09, 0.21] | 0.24 | [0.20, 0.28] |
| Thr biosynthesis | 0.34 | [0.33, 0.36] | 0.30 | [0.28, 0.31] | 0.24 | [0.23, 0.25] |
| Carbon dioxide production | 10.76 | [9.60, 11.94] | 8.84 | [8.12, 9.69] | 8.37 | [7.48, 9.38] |
| Isocitrate lyase | 0.03 | [0.00, 0.08] | 0.10 | [0.07, 0.12] | 0.06 | [0.04, 0.08] |
| Glyoxylate-Gly transaminase | 0.01 | [−0.08, 0.03] | 0.01 | [−0.06, 0.02] | 0.00 | [−0.09, 0.01] |
| Malate synthase | 0.02 | [0.00, 0.12] | 0.09 | [0.06, 0.15] | 0.05 | [0.03, 0.14] |
Differences in label incorporation in Ala and other pyruvate-derived amino acids were modeled as separate pyruvate kinase and malic enzyme activities in the cytosol and plastid. The existence of nonequilibrated pyruvate pools in developing seeds is supported through isotopic labeling experiments (Schwender et al., 2006; Williams et al., 2008). Furthermore, plastidic pyruvate kinase-based production of pyruvate is an important metabolic step for fatty acid biosynthesis (Andre et al., 2007). The data also indicated that citrate transport and cleavage by ATP-citrate lyase and malic enzyme supply carbon for amino and fatty acids. Small fluxes through Thr aldolase and the glyoxylate cycle were supported by labeling data but did not change with the C:N conditions. Labeling in other parts of the metabolism was inspected for further evidence of compartmentation (Wahrheit et al., 2011; Zamboni, 2011). However, additional subcellular details, such as vacuolar pools or the duplication of cytosolic and plastidic components of glycolysis or pentose phosphate pathways, were not necessary to develop models that passed statistical criteria (sum of squared residuals [SSR] of 221, 228, and 209 for 13:1, 21:1, and 37:1 that are less than the upper 95% confidence cutoff of 298) and, therefore, were not included.
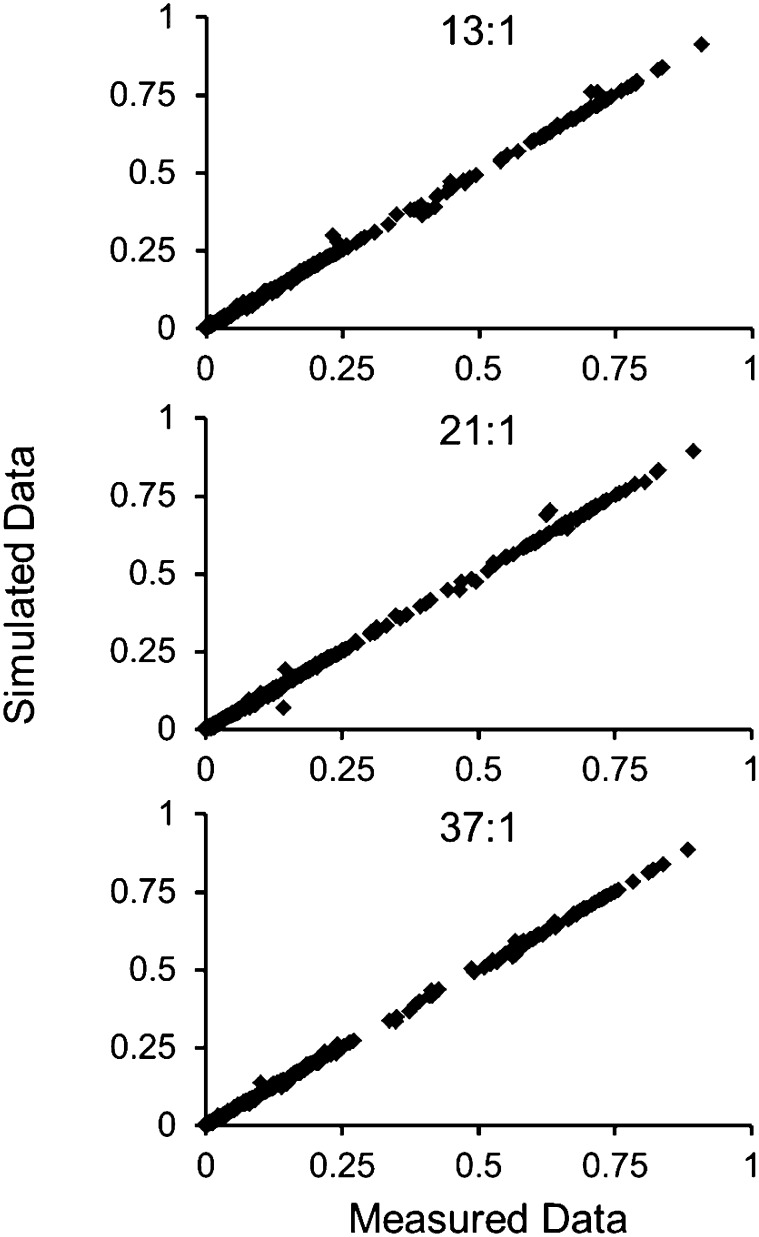
A comparison of fitted and simulated data indicated no gross errors (Fig. 6). The largest deviations were from measurements of Pro and C1 efflux. The evaluation of standards and reports by others (Antoniewicz et al., 2007) have indicated that Pro is less trustworthy. C1 efflux was loosely established from differences in its production and utilization that do not sensitively contribute to other flux estimates. The statistical agreement between fitted and measured data supported the accuracy of the model. Further additions to the network resulted in overparameterization that was indicated by SSR values significantly below the cutoff value (less than noise-based error) and by large confidence intervals (CIs). Advanced experimental techniques (Allen et al., 2012) or alternative labeling strategies that probe particular paths of interest would be necessary to further increase the network complexity (Allen et al., 2009a).
Figure 6.
Measured versus fitted data. The experimentally determined mass isotopomers were compared with the simulated values from metabolic flux analysis for each of the three flux maps. Measurements were in agreement, indicating that gross errors were not present and that the models were a reasonable description of metabolism.
The Effect of Combined Labeling Experiments on CIs for Flux Estimates
The coordinated modeling of multiple independent labeling experiments has been done previously (Schwender et al., 2006; Allen et al., 2009b; Masakapalli et al., 2010) and has resulted in confident flux estimates. We combined different labeling data sets into our simulations for central metabolism. The CIs (Antoniewicz et al., 2006; Young et al., 2008) established from one, two, or all three labeling experiments were compared with the flux values from which they were derived along with the degrees of freedom and the SSR (Table VI). The number of fluxes (out of 90 modeled) that had a CI less than the magnitude of the flux estimate were counted. The use of three labeling experiments generated 60 fluxes with CIs less than the flux magnitude. As the number of labeling experiments used in the simulation decreased, the system became poorly parameterized and fewer fluxes were sensitively determined (Table VI; i.e. their CIs became larger than their flux value). Sorting the data by the highest percentage of confidently established net fluxes (Table VI, last column) revealed that inclusion of the Gln labeling experiment better determined a greater number of fluxes than either of the other two experiments (Table VI).
Table VI. Additional labeling experiments affect flux sensitivity.
Fluxes were determined through the combination of one, two, or three labeling experiments. The 95% CIs were calculated from the flux values using a parameter continuation method described elsewhere (Antoniewicz et al., 2006). The CIs were then compared with the value of their corresponding flux. The number of net fluxes that have CIs that are smaller than the magnitude of the flux are presented along with the SSR and degrees of freedom (i.e. overdetermination). All simulations were based upon the presented model (90 fluxes, including 70 net fluxes and 20 exchange fluxes) and using measurements from C:N 21:1. Nineteen net fluxes, although not constrained, represent fluxes to biomass (e.g. amino acid biosynthetic fluxes to protein) and therefore have smaller measured errors and are established regardless of labeling. Therefore, the number of highly confident fluxes calculated minus the 19 biosynthetic fluxes are shown in parentheses. The number of fluxes determined with high confidence increases with the number of labeling experiments and the degrees of freedom.
| Simulated Experiments | Simulation Results (90 Fluxes) |
||
|---|---|---|---|
| Degrees of Freedom | SSR | No. of Fluxes with 95% CI Less Than the Flux Value | |
| [1,2-13C]Glc, [U-13C]Gln, [U-13C]Asn | 252 | 221 | 60 (41) |
| [1,2-13C]Glc, [U-13C]Gln | 157 | 163 | 56 (37) |
| [U-13C]Gln, [U-13C]Asn | 157 | 151 | 55 (36) |
| [1,2-13C]Glc, [U-13C]Asn | 157 | 71 | 53 (34) |
| [U-13C]Gln | 62 | 106 | 50 (31) |
| [U-13C]Asn | 62 | 26 | 33 (14) |
| [1,2-13C]Glc | 62 | 19.5 | 32 (13) |
Flux through Plastidic Pyruvate in Developing Soybean Embryos
Each of the models reported significant flux through glycolysis from hexose to pyruvate (plastidic pyruvate kinase flux was 70%–76% of total flux to pyruvate), indicating that sugars are an important source of carbon for fatty acid metabolism. Together, the actions of glycolysis and pyruvate dehydrogenase provide two reducing equivalents and one ATP that are stoichiometrically required for the incorporation of an acetyl-CoA group in fatty acid biosynthesis. However, sugars are not the only source of carbon, since supplied Gln can also be used for the biosynthesis of pyruvate-derived compounds. Changes in the supplied C:N ratio affected the contribution of Gln as a source of carbon for C4 dicarboxylic acids (such as malate), plastidic pyruvate, pyruvate-derived amino acids (Val and Leu), and fatty acids. These four classes of metabolites incorporated more Gln-derived carbon when less sugar was supplied (Fig. 4).
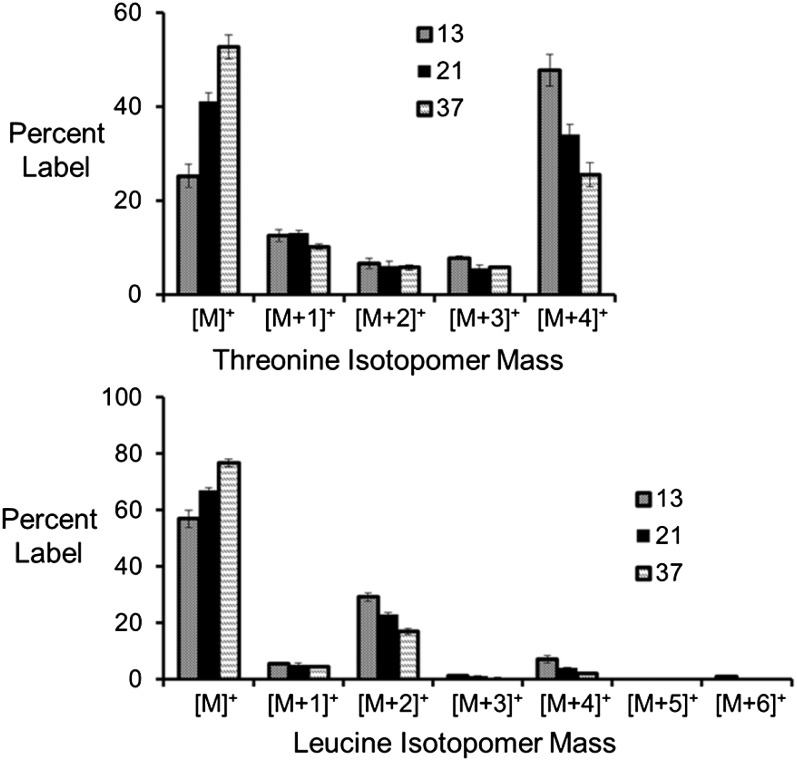
Significant changes in dicarboxylic acid-derived products were seen as the C:N ratio decreased and the soybean embryos relied more heavily on Gln as their carbon source (Fig. 7). Results from [13C]Gln labeling experiments revealed that the labeling of all four carbons in Thr increased with the decreased C:N ratios, indicating a significant proportion of fully labeled precursors such as C4 dicarboxylic acids like oxaloacetate or malate. Changes were also seen in pyruvate-derived amino acids (Fig. 7). Leu displayed an enrichment in [M+2]+ and [M+4]+ isotopomers, indicating derivation from acetyl-CoA that was labeled in both carbon positions. This double labeling of acetyl-CoA is consistent with malic enzyme and pyruvate dehydrogenase using malate that was labeled in multiple positions. The flux models indicated that citrate cleaved by ATP-citrate lyase provides 33% to 46% of the acetyl groups for fatty acid metabolism and can supply dicarboxylic acids used to generate pyruvate with malic enzyme (Fig. 5). Together, ATP-citrate lyase and plastidic malic enzyme contribute 43% to 51% of the carbon used for the biosynthesis of pyruvate-derived products (i.e. pyruvate-derived amino acids and fatty acids). Based upon the relative contributions of plastidic malic enzyme and pyruvate kinase to the pyruvate supply, malic enzyme and ATP-citrate lyase provide 47% to 59% of the carbon for acetyl-CoA production. Flux from α-ketoglutarate to malate increased by 90% with a change from high to low C:N ratio (1.05–1.98 μmol mg−1 dry weight), resulting in the utilization of more labeled organic acids for amino and fatty acid biosynthesis relative to plastidic pyruvate kinase (Fig. 5; Table V; Supplemental Data Set S7).
Figure 7.
13C label in Thr and Leu. [U-13C5]Gln was provided to developing embryos in different amounts to reflect three different C:N ratios (13:1, 21:1, 37:1). The mass isotopomer distribution in Thr and Leu, which are made from C4 dicarboxylic acids and acetyl-CoA, are presented. Labeling of all four carbons in Thr (i.e. [M+4]+) indicated a significant amount of labeling in organic acid precursors, such as malate and citrate. Leu was labeled in even-weighted mass isotopomers, indicating that these fractions were derived from acetyl-CoA, which was labeled in both carbons.
Using MFA to Distinguish Amino Acid Biosynthetic Precursors
While Asn in the apoplastic space of developing soybean embryos represents the second largest source of nitrogen for the developing seed (Rainbird et al., 1984), flux analysis and labeling experiments indicated that it made a smaller contribution to metabolism. Asn uptake was approximately 9% to 37% of Gln uptake flux. Nonetheless, 13C-labeled Asn was also an informative tracer for probing metabolism and deciphering between alternative network descriptions. Feeding labeled Asn resulted in Met and Thr labeling percentages that significantly differed from the combination of labeled Asp and Asn (ASX; Supplemental Data Set S8). Supplying labeled [13C]Asn (at 21:1 C:N) resulted in a 5-fold greater final label of ASX over Met or Thr (Supplemental Data Set S8, black bars). Conversely, labeled Gln experiments resulted in a 2-fold lower level of labeled ASX (Supplemental Data Set S8, hatched bars). These data indicate that Asp and Asn are not in isotopic equilibrium and that Asp is not derived predominantly from supplied Asn. The data also support ATP-citrate lyase-based generation of oxaloacetate in the plastid, as the cytosolic pool of oxaloacetate could not be fitted as a precursor to labeling in Thr, Ile, or ASX (Supplemental Data Set S9) when all three labeling experiments were considered.
The use of [U-13C4]Asn did not result in labeled products of phosphoenolpyruvate or pyruvate-derived metabolites. Aromatic amino acids were less than 1% labeled, and pyruvate-derived amino acids were labeled at only low levels (less than 5%) due to malic enzyme activity (Supplemental Data Set S5). These observations resulted in optimized flux maps with limited phosphoenolpyruvate carboxykinase (or reversible phosphoenolpyruvate carboxylase) activity (exchange flux of 3%–21% of net phosphoenolpyruvate carboxylase flux), consistent with sink metabolism in developing seeds lacking gluconeogenesis.
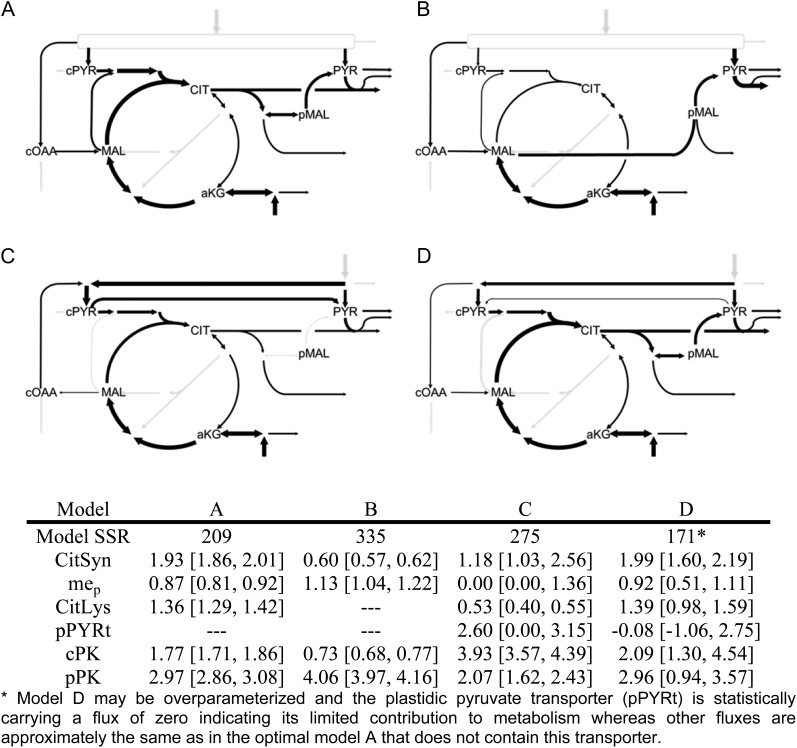
Multiple network descriptions were considered to describe carbon allocation for amino and fatty acid metabolism. We observed that [U-13C5]Gln labeling resulted in asymmetric enrichment in Asp family amino acids such as Thr and Ile, a pattern further confirmed qualitatively by inspection of Met. Previously observed reversibility in isocitrate dehydrogenase (Schwender et al., 2006; Allen et al., 2009b) was required to create citrate with higher mass isotopomer fractions. Four example networks that represent different alternatives are summarized in Figure 8 with additional description provided in Supplemental Data Set S9. The table in Figure 8 indicated that the SSR were minimized when the model included ATP-citrate lyase accompanied by reversible isocitrate dehydrogenase. Provision of [U-13C5]Gln resulted in [M+3]+ mass isotopomers of Thr and Ile that could not be simulated through malate transport (Fig. 8B) and resulted in a higher SSR. A network containing compartmentalized phosphoenolpyruvate with a pyruvate transporter (Fig. 8C) resulted in incompatibility between pyruvate-derived amino acids from different subcellular locations (i.e. Ala and Val) and also gave a higher SSR value. When pyruvate transport and phosphoenolpyruvate subcellular descriptions were together included with the optimized model (Fig. 8D), the flux estimates remained consistent with the optimized model (Fig. 8A) and indicated that pyruvate transport was not significant. These steps further confirmed the optimized model description (Fig. 8A). The small pyruvate transport flux was not added to the final model because genes for plastidial pyruvate transporters in oilseeds have not been established (Linka and Weber, 2010) and the flux is poorly determined (CI in the table was larger for plastidic pyruvate kinase in model D). Similarly, steps to describe hypothetical vacuolar pools were not included, as they were not necessary to fit the data (Fig. 6) and would require further experimental labeling descriptions to avoid low-confidence determination (Allen et al., 2009a).
Figure 8.
Alternative pathways for carbon import into plastids for amino and fatty acid biosynthesis. Development of the optimal model was the result of considering many network descriptions with the three combined labeling experiments for C:N of 37:1: A, Optimal model as described in the text. B, Export of malate. C, Export of citrate but with additional compartmentation of phosphoenolpyruvate and a hypothetical pyruvate transporter but without reversible isocitrate dehydrogenase. D, Optimal model plus inclusion of phosphoenolpyruvate compartmentation and a pyruvate transporter (cPYR→PYR is positive flux), which results in overfitting. The table includes the SSR of each model and the flux values for six key enzymes along with their CIs in brackets. cPYR, Cytosolic pyruvate; MAL, malate; cOAA, cytosolic oxaloacetate; aKG, α-ketoglutarate; CIT, citrate; PYR, plastidic pyruvate; pMAL, plastidic malate; CitSyn, citrate synthase; mep: plastidic malic enzyme; CitLys, citrate lyase; pPYRt, pyruvate transporter; cPK, cytosolic pyruvate kinase.
Oxidative and Nonoxidative Activities in Pentose Phosphate Metabolism
Experiments with [1,2-13C2]Glc resulted in similar quantities of [M+1]+ (12%–15%) and [M+2]+ (11%–14%) mass isotopomers in His (Supplemental Data Set S10). These labeled species of His support the existence of both an oxidative and a reversible, nonoxidative operation of the pentose phosphate pathway or alternative sources of pentoses. The [M+2]+ labeling would result from the conversion of Glc-6-P (hexose phosphate) into ribulose-5-phosphate using trans-ketolase (tk1/tk2) and aldolase (ta) enzymes, both of which maintain an intact bond between the first and second carbons of labeled hexose. On the other hand, oxidative metabolic steps break this bond and produce [M+1]+ labeling. The presence of both [M+1]+ and [M+2]+ isotopomers may indicate partial pentose phosphate pathway activities in both the plastid and cytosol that are connected by pentose or other transported molecules (Linka and Weber, 2010; Weber and Linka, 2011), as the oxidative and reductive pathways are controlled by multiple levels of regulation (Buchanan, 1980; Buchanan and Luan, 2005). Given the presence of Rubisco in green oilseeds (Ruuska et al., 2004), the detailed operation of pentose phosphate pathways will require more labeling experiments. Because the differences in His labeling between C:N ratios were small (Supplemental Data Set S10) and not significant, the poorly determined fluxes in this part of the metabolism were not further considered.
DISCUSSION
Primary metabolism results in diverse storage compositions in seeds. The creation of organic nitrogen and more highly reduced carbon compounds is energetically costly for a plant; therefore, the optimal use of elements benefits plant growth. Given both the importance and the differing contributions of carbon and nitrogen to various storage reserves, we altered the provision of these compounds as one parameter: the C:N ratio. We varied the molar ratio of carbon to nitrogen (provided as sugars and amino acids) and found altered production of storage biomass, with protein going from 14% to 47% of total biomass. The change in protein and associated metabolic fluxes indicated that seeds are not merely a receptacle for stored reserves but are dynamically involved in influencing final composition. Gln is a major source of both carbon and nitrogen for developing soybeans. Our culture experiments revealed that approximately 10% to 23% of all carbon (32%–46% of amino acid carbon and 9%–19% of carbon in fatty acids) and 63% to 91% of all nitrogen comes from Gln, values that are greater than in other plant systems. Furthermore, these data support the hypothesis that seed nitrogen demand is greater than the available supply and may limit protein yield (Sinclair and de Wit, 1975; Egli and Bruening, 2007; Rotundo et al., 2009; Rotundo and Westgate, 2009).
Storage reserves are generated by primary metabolism, implying that carbon, energy, and redox allocation are regulated differently in different plant seeds (e.g. maize [Zea mays], soybean, oilseed rape [Brassica napus]). Correlations between the two pairs of protein and yield and of protein and other reserves in soybeans have been discussed (Wilcox and Shibles, 2001; Clemente and Cahoon, 2009), although associated fluxes through primary metabolism have not considered variations in carbon and nitrogen. While we have previously used 13C metabolic flux analysis to generate flux map descriptions of soybeans ‘Amsoy’, this study probed the influence of substrates on seed-filling metabolism (in soybean ‘Jack’) in order to better assess fluxes that direct seed reserve composition.
Limitations and Validation
Solving flux analysis problems is both a forward and inverse mathematical problem, involving relationships between parameters (fluxes) that can be nonlinear with high degrees of branching. The derivation of the maximum information about a flux map involves a priori sensitivity analysis. When possible, different labeling experiments are performed, but this is limited by the commercial availability of labeled metabolites and the biological relevance of substrates in vivo (Metallo et al., 2009). Past work dedicated to optimal experimental design (Möllney et al., 1999; Libourel et al., 2007; Masakapalli et al., 2010) has specified that the choice and number of labeling experiments are important considerations that depend upon whether all or only specific fluxes are of interest. Performing a combined labeling experiment (i.e. multiple labeled substrates per experiment) can provide nearly as much information as separate labeling experiments and minimizes the number of experimental replicates (Libourel et al., 2007). However, the multiple labeling sources result in more complex labeling patterns that are less intuitive and challenging to evaluate.
Investigations using isotopic labeling contribute to our understanding of metabolism and, when coupled with metabolic flux analysis, can help uncover new enzyme activities or describe unique pathway regulation (Schwender et al., 2004). MFA is also used to evaluate the impact of environmental (Williams et al., 2008) and genotypical (Spielbauer et al., 2006; Alonso et al., 2007b; Lonien and Schwender, 2009) changes. However, conclusions from flux analyses remain a function of the assumptions and conditions used in modeling and experiments. Namely, the estimation of fluxes requires pseudo-steady-state metabolism over the labeling duration with a consistent environment for each set of experiments. Therefore, diurnal cycles and temperature variations are not reflected in these experiments. Additionally, the media provided to embryo cultures may lack some micronutrients received in planta. These considerations are presumed to be less critical, since the use of substrates at levels similar to those measured in planta (Hsu et al., 1984; Rainbird et al., 1984) combined with salts and vitamins (Obendorf et al., 1978) can capably mimic the production of storage reserves (Allen et al., 2009b). Embryo cultures have been used to study the metabolism of developing soybeans (Thompson et al., 1977; Obendorf et al., 1979; Hsu and Obendorf, 1982; Sriram et al., 2004; Allen et al., 2007, 2009b; Iyer et al., 2008). Furthermore, the changes in protein levels reported here are consistent with other studies that investigated nitrogen sources (Saravitz and Raper, 1995; Nakasathien et al., 2000; Pipolo et al., 2004). Therefore, flux estimates should be consistent with general features of in planta seed metabolism and relevant for metabolic engineering efforts.
Additionally, inferred fluxes are derived numerically by probing the mathematically feasible solution space posed through network constraints and, therefore, represent “best estimates.” Evaluation of all points within a continuous solution space is not possible. However, by providing label measurements, the models consistently and rapidly converge to the same solution from many different starting points. Reassurance in flux estimates was also achieved by independently validating the balance of nitrogen. Nitrogen, like carbon, must be conserved, but it is not used as a constraint for modeling. From modeled flux values, we found that nitrogen uptake ranged from 86% to 93% of the nitrogen used in metabolite generation. The balance exceeded 100% in some cases when the se in measured fluxes was considered (Supplemental Data Set S11).
ATP Production Changes to Accommodate Protein Biosynthetic Demands
Protein polymerization is an energetically expensive process requiring approximately 4.3 units of ATP per amino acid added to an elongating peptide (Stephanopoulos et al., 1998). The change in amino acid production and the accompanying ATP expense of protein polymerization were compared between the altered fluxes (Fig. 5, thicker arrows), which resulted from altered Gln uptake. The altered fluxes provided an additional 7.3 to 8.6 µmol ATP mg−1 dry weight depending upon the presumed phosphate:oxygen relationship (Hinkle, 2005). The biosynthetic fluxes to amino acids increased by 1.5 µmol mg−1 dry weight (between the 37:1 and 13:1 C:N ratios). This increased amino acid polymerization required an additional 6.45 µmol of ATP (i.e. 1.50 × 4.3). Although cellular maintenance and futile cycles are not considered, the altered ATP demand (6.45 µmol mg−1 dry weight) was less than the calculated increase in ATP production (7.3–8.6 µmol mg−1 dry weight), indicating that metabolism can accommodate the increase in protein production.
Carbon Partitioning through Pyruvate for Oil and Protein
Fatty acid labeling derived from Gln doubles with increased nitrogen provision (Fig. 4). The flux maps indicated that this was a consequence of changes in plastidic pyruvate kinase activity relative to plastidic malic enzyme and citrate lyase activities (Fig. 5; Table V; Supplemental Data Set S7). The flux through pyruvate and to acetyl-CoA may be an important point of regulation that can partially account for the reported negative correlations between oil and protein (Wilcox and Shibles, 2001), because carbon reallocated from fatty acid biosynthesis to pyruvate-derived amino acid production could result in more dramatic changes in protein levels. Additionally, acetyl-CoA is a central node of metabolism important to a number of secondary metabolites. Some acetyl-CoA could be used to produce saponins, sterols, and mevalonate-derived products (Berhow et al., 2006; Kim et al., 2006) or to decorate amino sugars, histones, or other proteins and polymers.
The role of Gln in soybean carbon metabolism has been noted (Allen et al., 2009b). Others have recently suggested the importance of malic enzyme for redox provision to fatty acids in maize (Alonso et al., 2011) and of ATP-citrate lyase in fatty acid elongation (Schwender et al., 2006) or fatty acid biosynthesis of soybean (Nelson and Rinne, 1975; Nelson and Rinne, 1977a, 1977b) and oilseed rape (Rangasamy and Ratledge, 2000). In comparison with other plant tissues analyzed by flux analysis (Table VII; Rontein et al., 2002; Schwender et al., 2006; Alonso et al., 2007a, 2010, 2011; Williams et al., 2008; Allen et al., 2009b; Lonien and Schwender, 2009; Xiong et al., 2010), modeled malic enzyme in soybean provides more carbon for oil (23%–29% of flux to plastidic pyruvate) than in other developing seeds. Thus, systems that take up less Gln may have alternate roles for anaplerotic reactions that reflect evolved differences in carbon utilization strategies. In our models, soybean has both fluxes for malic enzyme that are comparable to the rate of Gln uptake and higher ratios of Gln consumption relative to fatty acid biosynthesis, indicating that both are important for pyruvate-derived products (Table VII). Furthermore, an increased demand for nitrogen through aminotransferase reactions will generate organic acids that can serve as sources of carbon, such as malate and citrate. Thus, the transport and cleavage of citrate along with the activities of pyruvate kinase and malic enzyme coordinate both glycolysis and sugar utilization with the uptake of amino acids.
Table VII. Comparison of malic enzyme with other fluxes in other seeds and plant cells.
The flux through plastidic malic enzyme [mep] or total malic enzyme [metot] was compared with other fluxes as a percentage (e.g. 100 × [plastidic malic enzyme flux/plastidic pyruvate biosynthesis]). [fa], Fatty acid biosynthesis; [gln_upt], Gln uptake; [pyrp], plastidic pyruvate biosynthesis. Fluxes for calculated ratios were extracted from the literature as described in the footnotes.
| Plant Species | [mep]/[pyrp] | [metot]/[gln_upt] | [gln_upt]/[fa] |
|---|---|---|---|
| % | |||
| Rapeseeda | – | 53 | 5 |
| Arabidopsis (Arabidopsis thaliana) cellsb | 0 | – | – |
| Chlorella spp. (microalgae)c | 0 | – | – |
| Arabidopsis seeds (Columbia)d | 2 | 36 | 9 |
| Arabidopsis seeds (Wassilewskija)d | 3 | 61 | 8 |
| Sunflower (Helianthus annuus)e | 7 | 128 | 31 |
| Maize endospermf | 7 | 167 | 24 |
| Tomato (Solanum lycopersicum) cellsg | 8 | – | – |
| Soybean ’Amsoy’h | 19 | 81 | 36 |
| Maize germi | 9 | 391 | 9 |
| Soybean C:N 37j | 23 | 119 | 25 |
| Soybean C:N 21j | 28 | 98 | 42 |
| Soybean C:N 13j | 29 | 67 | 64 |
Schwender et al. (2006). bWilliams et al. (2008). cXiong et al. (2010). dLonien and Schwender (2009). eAlonso et al. (2007a). fAlonso et al. (2011). gRontein et al. (2002). hAllen et al. (2009b) [pyruvate supply not compartmentalized]. iAlonso et al. (2010). jThis study.
Malic enzyme, in combination with pyruvate dehydrogenase, yields acetyl-CoA and two reducing equivalents, which match the stoichiometric demands necessary to sustain fatty acid biosynthesis. Additionally, an increase in the production of pyruvate-derived amino acids (Val, Ile, Lys, and Leu) for protein could come at the expense of oil biosynthesis but relies less on pyruvate dehydrogenase and utilizes less NADPH. An increase in protein production could be compensated for by malic enzyme activity. However, increased flux through malic enzyme to fatty acids would require an additional supply of ATP and reduce carbon use efficiency by the generation of CO2. The calculated carbon conversion efficiencies of 85% to 87% for our C:N models are similar to previous embryo studies (Allen et al., 2009b) and consistent with examples of carbon refixation in soybean reproductive tissues (Quebedeaux and Chollet, 1975; Sambo et al., 1977; Satterlee and Koller, 1984; Sugimoto et al., 1987; Willms et al., 1999; Allen et al., 2009b) and other legumes (Flinn et al., 1977; Furbank et al., 2004). Tissues that limit CO2 escape, such as the seed coat or pod wall, may serve to concentrate inorganic carbon and enhance its refixation via phosphoenolpyruvate carboxylase or Rubisco, although these carbon-recovery mechanisms frequently require additional ATP. To date, no studies have considered the combined fluxes through multiple tissues.
CONCLUSION
Three metabolic flux models associated with isotopic labeling experiments over diverse carbon and nitrogen conditions demonstrated the plasticity of central carbon metabolism. Maternally provided substrates coordinated with the operation of primary metabolism resulted in the production of different oil, protein, and carbohydrate concentrations. Changes in protein levels were accommodated by more modest differences in fluxes through metabolic steps. Notably, the fluxes from Gln to C4 acids and other amino acids generally increased as protein levels increased, indicating the use of additional carbon from Gln supplied by the media. The increased fluxes from Gln to C4 acids, amino acids, and pyruvate-derived products, such as oil, further imply that sink strength does not limit protein production in soybeans. The elasticity of central carbon metabolism represents a significant impediment to metabolic engineering because the changes needed to achieve biotechnological goals may be subtle. Conversely, this work implies that small changes in multiple enzymatic steps can have a dramatic impact at the phenotypic level, consistent with theories of distributed control of metabolism.
MATERIALS AND METHODS
Labeled Isotopes
[U-13C5]Gln, [1,2-13C2]Glc, and [U-13C4]Asn were purchased from Sigma [U-14C6]Glc and [U-14C5]Gln were purchased from Moravek Biochemical. [U-14C12]Suc and [U-14C4]Asn were purchased from American Radiolabeled Chemicals.
Plant Material, Growth, and Culturing Conditions
Soybean seeds (Glycine max ‘Jack’) were obtained from the U.S. Department of Agriculture-Agricultural Research Service National Plant Germplasm System. Three soybeans were planted in a 1-gallon pot containing Fafard 4M and maintained in a greenhouse or growth chambers mimicking summer in St. Louis. Pots were thinned to two seedlings after several weeks, and the remaining seeds were grown at 25°C to 27°C/21°C to 23°C day/night temperatures and maintained at more than 35% humidity. Sunlight was supplemented with additional light (400–1,000 W m−2) to maintain a 14-h-day/10-h-night photoperiod. Plants were watered daily and received Jack’s 15-16-17 (JR Peters) fertilizer three times per week. Early in the R5 reproductive stage of embryo development (i.e. early seed-filling stage), pods were harvested and directly placed on ice.
The pods were immediately surface sterilized by washing in 5% bleach followed by sterile water. Embryos (approximately 30–40 mg fresh weight and 10 mg dry weight) were removed from pods, dissected from seed coats, and transferred to sterile culture medium containing 150 mm Suc, 75 mm Glc, 45 mm Gln, and 16 mm Asn, which represents the carbon and nitrogen sources for the C:N ratio of 21:1. Other C:N ratios were varied as reported elsewhere in this article. For each labeling experiment, the labeled substrate was provided as 100% of that particular carbon source (i.e. in the case of Gln labeling, all unlabeled Gln was supplanted by [U-13C5]Gln while other carbon sources remained unlabeled). A modified Linsmaier and Skoog medium (Thompson et al., 1977; Hsu and Obendorf, 1982) was supplied along with Gamborg’s vitamins (Sigma) and 5 mm MES buffer adjusted to pH 5.8. The salts and vitamins were identical for all cultures. Each embryo was cultured in a 250-mL Erlenmeyer flask with a foam plug for 14 d at approximately 30 to 35 µmol m−2 s−1 continuous light at 26°C to 27°C. At the conclusion of the culturing period, embryos were washed and then quickly sliced into pieces, frozen in liquid nitrogen, lyophilized to dryness, and stored at −80°C until further processing.
Quantification of Protein, Starch, and Lipid
Protein was quantified by elemental C:N ratio analysis. The composition of carbon and nitrogen in biomass was determined at the Duke Environmental Stable Isotope Laboratory at Duke University by dry combustion using a CE Instruments NC2100 elemental analyzer (ThermoQuest Italia). From each seed, 2 to 4 mg of tissue was accurately weighed and combusted at 900°C in an elemental analyzer in the presence of chemical catalysts to produce CO2 and NOx. The measured nitrogen fraction was converted to protein by multiplication using a factor for all cultures within the initial C:N curve as described elsewhere in this article. This multiplication factor is commonly used for reporting protein and therefore appropriate for biological comparisons; however, for flux analysis, more accurate factors that reflect the actual amino acid composition of soybeans were used. Factors of 5.05, 5.30, and 5.55 for the cultured C:N variations of 13:1, 21:1, and 37:1, respectively, were determined experimentally from amino acid analysis of molar composition. This calculation assumes an equivalent amount of Gln to Glu and of Asn to Asp, which is consistent with their prevalence in soybean storage protein amino acid sequences. Amino acid compositional analysis was established by converting amino acids to their AccQ-Tag derivatives (Waters) and measuring peaks relative to external standards.
Amino acid quantification was performed by the Proteomics and Mass Spectrometry Facility within the Donald Danforth Plant Science Center. Samples were hydrolyzed for 24 h at 116°C in the presence of 6 n HCl containing 0.5% phenol in sealed glass tubes, dried, and resuspended in 20 mm HCl (20 μL). Derivatization with the AccQ-Tag reagent was performed per the manufacturer’s instructions (Waters), including the addition of 70 μL of AccQ-Fluor Borate buffer and 20 μL of derivatization reagent to 10 μL of each sample. Following incubation for 1 min at room temperature, samples were heated at 55°C for 10 min, separated and identified with standards, and quantified by UV detection at 260 nm using reverse-phase ultra-HPLC (UHPLC).
UHPLC was performed using a Waters Acquity Ultra Performance liquid chromatograph with the 2996 PDA UV detector. A 9.5-min UHPLC method used the Waters proprietary AccQ-Tag Ultra column (2.1 × 100 mm) with gradient using 5% Waters proprietary buffer A in milli-Q water and 100% Waters proprietary buffer B at a flow rate of 0.7 mL min−1. The gradient was initiated with 99.9% buffer A/0.1% buffer B and held for 0.54 min, buffer A was decreased to 90.9% in 5.2 min, further decreased to 78.8% in 2 min, then decreased to 40.4% in 0.3 min, held for 0.64 min, after which the column was regenerated and equilibrated.
Lipids were analyzed by transesterification to fatty acid methyl esters with subsequent quantification using triheptadecanoin internal standard (Li et al., 2006). Briefly, seed biomass of approximately 5 to 10 mg was weighed and combined with 5% sulfuric acid in methanol prepared directly before use. Butylated hydroxyltoluene (25 μL of a 0.2% solution in methanol) was added to prevent oxidation. The reactions were incubated at 95°C for 2 h with occasional vortexing. After cooling, 1.5 mL of 0.9% NaCl (w/v) was added. Hexane was used to extract the methyl esters from the reaction liquid. Fatty acid methyl esters were concentrated by nitrogen evaporation and then quantitated by gas chromatography-flame ionization detection. Gas chromatography employed a DB23 column (30 m, 0.25-mm i.d., 0.25-μm film; J&W Scientific). The gas chromatograph was operated in a split mode (40:1), and the flame ionization detector temperature was 260°C with an oven program that ramped from 150°C to 240°C at a rate of 10°C min−1 followed by a programmed hold time of 5 min. Peak area comparison with the internal standard was used for quantification.
Starch was quantified enzymatically by measuring spectroscopic Glc absorbance according to the manufacturer’s protocol (Megazyme), a modified version of AOAC International Official method 996.11. Alternatively, biomass that had been extracted for oil and protein measurements was hydrated by incubation in 0.1 m sodium acetate buffer, pH 4.8 to 5.0, at 121°C for 1 h. Amyloglucosidase, amylase, and isoamylase were added from a fresh stock solution of 10 mg each in 15 mL of acetate buffer or using reagents provided by the manufacturer (Megazyme). After incubation for 1 h at 55°C, 1 mL of ethanol was added, and enzymes were heat denatured for 15 min at 95°C. The solution was centrifuged, and the supernatant was removed and extracted twice more with 80% ethanol before spectroscopic measurement.
SDS-PAGE Analysis
Proteins from 2 mg mL−1 dry weight of cultured soybeans were extracted with 2× Laemmli sample buffer + 5% β-mercaptoethanol and boiled for 15 min. Gels were processed either for equivalent amounts of dry weight (25 μg) or for equivalent amounts of protein loaded per lane (10 μg, as determined by prior total nitrogen analysis of dry material). Samples and SeeBlue_2 Mr markers (Invitrogen) were separated on a 10-well, 4% to 12% Nu-PAGE gel with MOPS buffer run at 200 V constant for 35 min. Protein bands were visualized with Simply Blue Coomassie blue stain (Invitrogen) for 2 h and destained with water. Bands were quantified using Genetix Gelpix analyzer.
Radiolabeled Embryo Culturing and Processing
Soy embryos were aseptically cultured for 14 d with [U-14C12]Suc, [U-14C6]Glc, [U-14C5]Gln, and [U-14C4]Asn provided in proportional representation of the molar carbon composition of the substrates in media. Embryos were removed from cultures, washed briefly with water, quickly sliced into small pieces, frozen with liquid nitrogen, lyophilized, and then pulverized into a fine powder with a bead mill (Retsch) and a 4-mm stainless-steel ball. Dry powder (10–15 mg) was placed into a 2-mL plastic screw cap tube and sequentially extracted using a bead mill and centrifugation at each step. All extractions were repeated four times for quantitative recovery, and supernatants were combined unless indicated otherwise. Initially, the pellet was extracted with 80% ethanol. The 80% ethanol extract was separated into water and oil phases with the addition of equal volumes of chloroform and water. The oily chloroform phase was later combined with additional oil fractions, including two extractions with hexane:isopropanol:chloroform (2:1:3, v/v/v) followed by two extractions of hexane-isopropanol (v/v). The 80% ethanol fraction was further fractionated to separate unmetabolized neutral sugars, Gln and Asn taken up from the media from organic acids, remaining amino acids, and solubilized hydrophobic proteins. Cationic, anionic, and neutral fractions were separated by ion-exchange chromatography (Yazdi-Samadi et al., 1977) using Dowex 50 and Dowex 1 columns (Sigma), with HPLC used to inspect the cationic fraction for hydrophobic proteins and amino acids. Protein was extracted with 7.5 m urea, 0.07% β-mercaptoethanol, and 10 mm Tris, pH 8.1, twice, followed by two further extractions excluding the urea; aliquots were combined. The remaining pellet, which represents carbohydrates, was solubilized with ScintiGest (Fisher Scientific) overnight at 50°C. Each sample (0.5 mL) was counted by scintillation using Hionic-Fluor (Perkin-Elmer).
GC-MS Analyses of Isotopic Labeling
Protein was extracted from biomass using a buffer containing 10 mm Tris, pH 8.5, 138 mm NaCl, 2.7 mm KCl, 295 mg L−1 Na-EDTA, 700 µL of β-mercaptoethanol, 202 mg L−1 sodium azide, and 125 mg L−1 SDS. Extracts were precipitated by the addition of one-tenth volume of 50% TCA (w/v) and incubated on ice for 30 min. The supernatant was discarded, and the pellet was carefully washed in 1:1 ethanol:ether. After drying with nitrogen, proteins were hydrolyzed by the addition of 6 n HCl in the presence of a nitrogen head space at 100°C for 24 h. HCl was evaporated under a stream of nitrogen. The amino acids were dried and derivatized with N-methyl-N-(tertbutyldimethylsilyl)-trifluoroacetamide (100 μL) in the presence of an equal volume of acetonitrile. The derivatization ensued for 1 h at 120°C, after which an aliquot was analyzed by GC-MS. Accepted fragments for label quantification were based upon standards and previous analyses (Antoniewicz et al., 2007).
For fatty acid labeling, dried seed tissue was ground in liquid nitrogen using a mortar and pestle, and a 10- to 15-mg sample was weighed and homogenized with hexane:isopropanol (2:1, v/v) on a Retsch MM 301 bead mill at a frequency of 3,000 per min for 5 min. After centrifugation at 12,000g, the supernatant was decanted and the extraction process was repeated three additional times. Pooled extracts were dried and derivatized to butylamides for labeling analysis as described previously (Allen et al., 2007). Briefly, lipids were amidated by reacting at 100°C with 2 mL of n-butylamine (Sigma) and 3 mL of hexane as a cosolvent. Reactions were allowed to proceed for several days and quenched with the addition of concentrated HCl, resulting in phase partitioning of unreacted butylamine-HCl and amide products. The hexane phase was collected and analyzed by GC-MS as detailed by Allen et al. (2007) using the same mass spectrometer as for amino acid label evaluation.
Metabolic Modeling
A flux model was built based upon biochemical reaction networks for central carbon metabolism in developing seeds with slight modification of a previous soybean model to account for additional labeling experiments with [U-13C4]Asn. Implementation of the isotopomer model that included all carbon rearrangements within reactions, mass conservation of metabolic intermediates, parameter (flux) fitting, and statistical and sensitivity analyses were performed using the INCA software platform under development (J.D. Young, Vanderbilt University; Young et al., 2008). The INCA software platform is MATLAB based, utilizing statistical and optimization toolboxes but with additional in-house-developed algorithms. Identical to other isotopic labeling-based metabolic flux analysis software, INCA requires the provision of a stoichiometric description of the network model, individual experiments that indicate the form of the proffered label, mass spectrometer isotopic labeling measurements, and direct flux measurements. INCA can perform Monte Carlo or parameter continuation-based assessment of nonlinear CIs and is being developed for further extensions.
Flux model values were constrained by a network topology that imposed stoichiometric relationships between molecules and the measured biomass composition of embryos, including protein and oil, with the remainder attributed to carbohydrates such as cell wall and starch. 13C isotopic descriptions provided additional constraining information through the fitting process that established the best estimates for fluxes from minimization of the sum of squared differences of all label measurements. The choice of free variables was randomly established through the software, and feasible starting points were assigned by orthogonal projection of random flux values into the flux solution space (Schuster and Schuster, 1993; Young et al., 2008; Allen et al., 2009b). Labeled isotopomer measurements and labeled substrates entered into the model were corrected for the natural abundance of heteroatoms (i.e. nitrogen, oxygen, sulfur, silicon) as well as carbon that was not part of the labeling experiment (i.e. carbon in the derivative; Lee et al., 1991; Fernandez et al., 1996).
Statistical Analysis
Each experiment was performed with five biological replicates (n = 5). For each variant in C:N ratio, three labeling experiments were performed that complement each other, for a total of 15 experiments per condition. However, biological replication assesses the precision of measurement but not necessarily biases in processing or instrument measurement that impact measurement accuracy. To assess the accuracy of the measurements, standards of unlabeled metabolites were measured, and fragments from GC-MS were chosen accordingly and in agreement with previous descriptions of acceptable measurements from our laboratory (Allen et al., 2009b) as well as others (Dauner and Sauer, 2000; Antoniewicz et al., 2007). Model simulations were performed by assigning a minimal 0.25% error for all measurements that had greater precision than this. CIs are reported for each flux estimate (95%) in accordance with described methods (Antoniewicz et al., 2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Data Set S1. Changes in protein levels and fatty acid composition.
Supplemental Data Set S2. Proteomic analysis of soybean storage protein subunits.
Supplemental Data Set S3. Amino acid concentrations of cultured soybeans.
Supplemental Data Set S4. Calculation of 13C labeling in biomass fractions.
Supplemental Data Set S5. 13C-labeling measurements.
Supplemental Data Set S6. Carbon allocation from Gln.
Supplemental Data Set S7. Metabolic flux maps, values, CIs, and reaction descriptions.
Supplemental Data Set S8. Labeling in C4 dicarboxylic acids.
Supplemental Data Set S9. Description of the modeling process.
Supplemental Data Set S10. Labeling in His.
Supplemental Data Set S11. Balance of nitrogen.
Acknowledgments
We gratefully acknowledge James Gierse for excellent technical help with experiments and the Proteomics and Mass Spectrometry Facility at the Donald Danforth Plant Science Center for amino acid analysis. We also thank multiple anonymous reviewers for helpful comments that improved the manuscript. Any product or trademark mentioned here does not imply a warranty, guarantee, or endorsement by the U.S. Department of Agriculture over other suitable products.
Glossary
- MFA
metabolic flux analysis
- C:N
carbon-to-nitrogen
- SSR
sum of squared residuals
- CI
confidence interval
- ASX
combination of labeled Asp and Asn
- UHPLC
ultra-HPLC
- GC-MS
gas chromatography-mass spectrometry
References
- Adams CA, Broman TH, Rinne RW. (1982) Use of [3,4-C]glucose to assess in vivo competition for phosphoenolpyruvate between phosphoenolpyruvate carboxylase and pyruvate kinase in developing soybean seeds. Plant Cell Physiol 23: 959–965 [Google Scholar]
- Adams CA, Rinne RW. (1981) Interactions of phosphoenolpyruvate carboxylase and pyruvic kinase in developing soybean seeds. Plant Cell Physiol 22: 1011–1021 [Google Scholar]
- Allen DK, Laclair RW, Ohlrogge JB, Shachar-Hill Y. (2012) Isotope labelling of Rubisco subunits provides in vivo information on subcellular biosynthesis and exchange of amino acids between compartments. Plant Cell Environ 35: 1232–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DK, Libourel IGL, Shachar-Hill Y. (2009a) Metabolic flux analysis in plants: coping with complexity. Plant Cell Environ 32: 1241–1257 [DOI] [PubMed] [Google Scholar]
- Allen DK, Ohlrogge JB, Shachar-Hill Y. (2009b) The role of light in soybean seed filling metabolism. Plant J 58: 220–234 [DOI] [PubMed] [Google Scholar]
- Allen DK, Shachar-Hill Y, Ohlrogge JB. (2007) Compartment-specific labeling information in 13C metabolic flux analysis of plants. Phytochemistry 68: 2197–2210 [DOI] [PubMed] [Google Scholar]
- Alonso AP, Dale VL, Shachar-Hill Y. (2010) Understanding fatty acid synthesis in developing maize embryos using metabolic flux analysis. Metab Eng 12: 488–497 [DOI] [PubMed] [Google Scholar]
- Alonso AP, Goffman FD, Ohlrogge JB, Shachar-Hill Y. (2007a) Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus L.) embryos. Plant J 52: 296–308 [DOI] [PubMed] [Google Scholar]
- Alonso AP, Raymond P, Hernould M, Rondeau-Mouro C, de Graaf A, Chourey P, Lahaye M, Shachar-Hill Y, Rolin D, Dieuaide-Noubhani M. (2007b) A metabolic flux analysis to study the role of sucrose synthase in the regulation of the carbon partitioning in central metabolism in maize root tips. Metab Eng 9: 419–432 [DOI] [PubMed] [Google Scholar]
- Alonso AP, Val DL, Shachar-Hill Y. (2011) Central metabolic fluxes in the endosperm of developing maize seeds and their implications for metabolic engineering. Metab Eng 13: 96–107 [DOI] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C. (2007) A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19: 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. (2006) Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng 8: 324–337 [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. (2007) Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem 79: 7554–7559 [DOI] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M. (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter CJ, Liu JL, Fernie AR, Sweetlove LJ. (2007) Determination of metabolic fluxes in a non-steady-state system. Phytochemistry 68: 2313–2319 [DOI] [PubMed] [Google Scholar]
- Berhow MA, Kong SB, Vermillion KE, Duval SM. (2006) Complete quantification of group A and group B soyasaponins in soybeans. J Agric Food Chem 54: 2035–2044 [DOI] [PubMed] [Google Scholar]
- Buchanan BB. (1980) Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol Plant Mol Biol 31: 341–374 [Google Scholar]
- Buchanan BB, Luan S. (2005) Redox regulation in the chloroplast thylakoid lumen: a new frontier in photosynthesis research. J Exp Bot 56: 1439–1447 [DOI] [PubMed] [Google Scholar]
- Clemente TE, Cahoon EB. (2009) Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol 151: 1030–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauner M, Bailey JE, Sauer U. (2001) Metabolic flux analysis with a comprehensive isotopomer model in Bacillus subtilis. Biotechnol Bioeng 76: 144–156 [DOI] [PubMed] [Google Scholar]
- Dauner M, Sauer U. (2000) GC-MS analysis of amino acids rapidly provides rich information for isotopomer balancing. Biotechnol Prog 16: 642–649 [DOI] [PubMed] [Google Scholar]
- Egli DB (1998) Seed Biology and the Yield of Grain Crops. CAB International, New York [Google Scholar]
- Egli DB, Bruening WP. (2007) Accumulation of nitrogen and dry matter by soybean seeds with genetic differences in protein concentration. Crop Sci 47: 359–366 [Google Scholar]
- Egli DB, Guffy RD, Meckel LW, Leggett JE. (1985) The effect of source sink alterations on soybean seed growth. Ann Bot (Lond) 55: 395–402 [Google Scholar]
- Etxeberria E, Pozueta-Romero J, Gonzalez P. (2012) In and out of the plant storage vacuole. Plant Sci 190: 52–61 [DOI] [PubMed] [Google Scholar]
- Fabre F, Planchon C. (2000) Nitrogen nutrition, yield and protein content in soybean. Plant Sci 152: 51–58 [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. (1996) Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom 31: 255–262 [DOI] [PubMed] [Google Scholar]
- Flinn AM, Atkins CA, Pate JS. (1977) Significance of photosynthetic and respiratory exchanges in the carbon economy of the developing pea fruit. Plant Physiol 60: 412–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, White R, Palta JA, Turner NC. (2004) Internal recycling of respiratory CO2 in pods of chickpea (Cicer arietinum L.): the role of pod wall, seed coat, and embryo. J Exp Bot 55: 1687–1696 [DOI] [PubMed] [Google Scholar]
- Goffman FD, Alonso AP, Schwender J, Shachar-Hill Y, Ohlrogge JB. (2005) Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiol 138: 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout E, Bligny R, Pascal N, Douce R. (1993) 13C nuclear magnetic resonance studies of malate and citrate synthesis and compartmentation in higher plant cells. J Biol Chem 268: 3986–3992 [PubMed] [Google Scholar]
- Harada JJ, Barker SJ, Goldberg RB. (1989) Soybean β-conglycinin genes are clustered in several DNA regions and are regulated by transcriptional and posttranscriptional processes. Plant Cell 1: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayati R, Egli DB, Crafts-Brandner SJ. (1996) Independence of nitrogen supply and seed growth in soybean: studies using an in vitro culture system. J Exp Bot 47: 33–40 [Google Scholar]
- Hernández-Sebastià C, Marsolais F, Saravitz C, Israel D, Dewey RE, Huber SC. (2005) Free amino acid profiles suggest a possible role for asparagine in the control of storage-product accumulation in developing seeds of low- and high-protein soybean lines. J Exp Bot 56: 1951–1963 [DOI] [PubMed] [Google Scholar]
- Hinkle PC. (2005) P/O ratios of mitochondrial oxidative phosphorylation. Biochim Biophys Acta 1706: 1–11 [DOI] [PubMed] [Google Scholar]
- Hsu FC, Bennett AB, Spanswick RM. (1984) Concentrations of sucrose and nitrogenous compounds in the apoplast of developing soybean seed coats and embryos. Plant Physiol 75: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu FC, Obendorf RL. (1982) Compositional analysis of in vitro matured soybean seeds. Plant Sci Lett 27: 129–135 [Google Scholar]
- Iyer VV, Sriram G, Fulton DB, Zhou R, Westgate ME, Shanks JV. (2008) Metabolic flux maps comparing the effect of temperature on protein and oil biosynthesis in developing soybean cotyledons. Plant Cell Environ 31: 506–517 [DOI] [PubMed] [Google Scholar]
- Junker BH, Lonien J, Heady LE, Rogers A, Schwender J. (2007) Parallel determination of enzyme activities and in vivo fluxes in Brassica napus embryos grown on organic or inorganic nitrogen source. Phytochemistry 68: 2232–2242 [DOI] [PubMed] [Google Scholar]
- Kim S-L, Berhow MA, Kim J-T, Chi H-Y, Lee S-J, Chung I-M. (2006) Evaluation of soyasaponin, isoflavone, protein, lipid, and free sugar accumulation in developing soybean seeds. J Agric Food Chem 54: 10003–10010 [DOI] [PubMed] [Google Scholar]
- Kruger NJ, Ratcliffe RG. (2009) Insights into plant metabolic networks from steady-state metabolic flux analysis. Biochimie 91: 697–702 [DOI] [PubMed] [Google Scholar]
- Lee WNP, Byerley LO, Bergner EA, Edmond J. (1991) Mass isotopomer analysis: theoretical and practical considerations. Biol Mass Spectrom 20: 451–458 [DOI] [PubMed] [Google Scholar]
- Li YH, Beisson F, Pollard M, Ohlrogge J. (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Libourel IGL, Gehan JP, Shachar-Hill Y. (2007) Design of substrate label for steady state flux measurements in plant systems using the metabolic network of Brassica napus embryos. Phytochemistry 68: 2211–2221 [DOI] [PubMed] [Google Scholar]
- Linka N, Weber APM. (2010) Intracellular metabolite transporters in plants. Mol Plant 3: 21–53 [DOI] [PubMed] [Google Scholar]
- Lonien J, Schwender J. (2009) Analysis of metabolic flux phenotypes for two Arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiol 151: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Maeshima M, Neuhaus HE. (2007) Vacuolar transporters and their essential role in plant metabolism. J Exp Bot 58: 83–102 [DOI] [PubMed] [Google Scholar]
- Masakapalli SK, Le Lay P, Huddleston JE, Pollock NL, Kruger NJ, Ratcliffe RG. (2010) Subcellular flux analysis of central metabolism in a heterotrophic Arabidopsis cell suspension using steady-state stable isotope labeling. Plant Physiol 152: 602–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metallo CM, Walther JL, Stephanopoulos G. (2009) Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol 144: 167–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möllney M, Wiechert W, Kownatzki D, de Graaf AA. (1999) Bidirectional reaction steps in metabolic networks. IV. Optimal design of isotopomer labeling experiments. Biotechnol Bioeng 66: 86–103 [DOI] [PubMed] [Google Scholar]
- Nakasathien S, Israel DW, Wilson RF, Kwanyuen P. (2000) Regulation of seed protein concentration in soybean by supra-optimal nitrogen supply. Crop Sci 40: 1277–1284 [Google Scholar]
- Narvel JM, Fehr WR, Ininda J, Welke GA, Hammond EG, Duvick DN, Cianzio SR. (2000) Inheritance of elevated palmitate in soybean seed oil. Crop Sci 40: 635–639 [Google Scholar]
- Nelson DR, Rinne RW. (1975) Citrate cleavage enzymes from developing soybean cotyledons: incorporation of citrate carbon into fatty acids. Plant Physiol 55: 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Rinne RW. (1977a) In vivo citrate studies with developing soybean cotyledons. Plant Cell Physiol 18: 399–404 [Google Scholar]
- Nelson DR, Rinne RW. (1977b) The role of citrate in lipid synthesis in developing soybean cotyledons. Plant Cell Physiol 18: 1021–1027 [Google Scholar]
- Obendorf RL, Rytko GT, Byrne MC. (1978) Soya bean seed growth and maturation by in vitro pod culture. Ann Bot 51: 217–227 [Google Scholar]
- Obendorf RL, Wettlaufer SH. (1984) Precocious germination during in vitro growth of soybean seeds. Plant Physiol 76: 1024–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek NC, Imsande J, Shoemaker RC, Shibles R. (1997) Nutritional control of soybean seed storage protein. Crop Sci 37: 498–503 [Google Scholar]
- Pipolo AE, Sinclair TR, Camara GMS. (2004) Protein and oil concentration of soybean seed cultured in vitro using nutrient solutions of differing glutamine concentration. Ann Appl Biol 144: 223–227 [Google Scholar]
- Quebedeaux B, Chollet R. (1975) Growth and development of soybean (Glycine max [L.] Merr.) pods: CO2 exchange and enzyme studies. Plant Physiol 55: 745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbird RM, Thorne JH, Hardy RWF. (1984) Role of amides, amino acids, and ureides in the nutrition of developing soybean seeds. Plant Physiol 74: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy D, Ratledge C. (2000) Genetic enhancement of fatty acid synthesis by targeting rat liver ATP:citrate lyase into plastids of tobacco. Plant Physiol 122: 1231–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe RG, Shachar-Hill Y. (2006) Measuring multiple fluxes through plant metabolic networks. Plant J 45: 490–511 [DOI] [PubMed] [Google Scholar]
- Rontein D, Dieuaide-Noubhani M, Dufourc EJ, Raymond P, Rolin D. (2002) The metabolic architecture of plant cells: stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J Biol Chem 277: 43948–43960 [DOI] [PubMed] [Google Scholar]
- Rotundo JL, Borrás L, Westgate ME, Orf JH. (2009) Relationship between assimilate supply per seed during seed filling and soybean seed composition. Field Crops Res 112: 90–96 [Google Scholar]
- Rotundo JL, Westgate ME. (2009) Meta-analysis of environmental effects on soybean seed composition. Field Crops Res 110: 147–156 [Google Scholar]
- Rubel A, Rinne RW, Canvin DT. (1972) Protein, oil, and fatty-acid in developing soybean seeds. Crop Sci 12: 739–741 [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. (2004) The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol 136: 2700–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo-Väänänen PP, Koivistoinen PE. (1996) Determination of protein in foods: comparison of net protein and crude protein (N × 6.25) values. Food Chem 57: 27–31 [Google Scholar]
- Sambo EY, Moorby J, Milthorpe FL. (1977) Photosynthesis and respiration of developing soybean pods. Aust J Plant Physiol 4: 713–721 [Google Scholar]
- Saravitz CH, Raper CD. (1995) Responses to sucrose and glutamine by soybean embryos grown in vitro. Physiol Plant 93: 799–805 [Google Scholar]
- Satterlee LD, Koller HR. (1984) Response of soybean fruit respiration to changes in whole plant light and CO2 environment. Crop Sci 24: 1007–1010 [Google Scholar]
- Schuster R, Schuster S. (1993) Refined algorithm and computer program for calculating all non-negative fluxes admissible in steady states of biochemical reaction systems with or without some flux rates fixed. Comput Appl Biosci 9: 79–85 [DOI] [PubMed] [Google Scholar]
- Schwender J. (2008) Metabolic flux analysis as a tool in metabolic engineering of plants. Curr Opin Biotechnol 19: 131–137 [DOI] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. (2004) Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782 [DOI] [PubMed] [Google Scholar]
- Schwender J, Shachar-Hill Y, Ohlrogge JB. (2006) Mitochondrial metabolism in developing embryos of Brassica napus. J Biol Chem 281: 34040–34047 [DOI] [PubMed] [Google Scholar]
- Sinclair TR, de Wit CT. (1975) Photosynthate and nitrogen requirements for seed production by various crops. Science 189: 565–567 [DOI] [PubMed] [Google Scholar]
- Spielbauer G, Margl L, Hannah LC, Römisch W, Ettenhuber C, Bacher A, Gierl A, Eisenreich W, Genschel U. (2006) Robustness of central carbohydrate metabolism in developing maize kernels. Phytochemistry 67: 1460–1475 [DOI] [PubMed] [Google Scholar]
- Sriram G, Fulton DB, Iyer VV, Peterson JM, Zhou RL, Westgate ME, Spalding MH, Shanks JV. (2004) Quantification of compartmented metabolic fluxes in developing soybean embryos by employing biosynthetically directed fractional 13C labeling, two-dimensional [13C,1H] nuclear magnetic resonance, and comprehensive isotopomer balancing. Plant Physiol 136: 3043–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos G, Aristidou A, Nielsen J (1998) Metabolic Engineering: Principles and Methodologies. Academic Press, San Diego [Google Scholar]
- Sugimoto T, Tanaka K, Momma M, Saio K. (1987) Photosynthetic activity in the developing cotyledon of soybean seeds. Agric Biol Chem 51: 1227–1230 [Google Scholar]
- Sweetlove LJ, Beard KFM, Nunes-Nesi A, Fernie AR, Ratcliffe RG. (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Fell D, Fernie AR. (2008) Getting to grips with the plant metabolic network. Biochem J 409: 27–41 [DOI] [PubMed] [Google Scholar]
- Thompson JF, Madison JT, Muenster AME. (1977) In vitro culture of immature cotyledons of soya bean (Glycine max L Merr). Ann Bot (Lond) 41: 29–39 [Google Scholar]
- Thorne JH. (1980) Kinetics of C-photosynthate uptake by developing soybean fruit. Plant Physiol 65: 975–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne JH. (1981) Morphology and ultrastructure of maternal seed tissues of soybean in relation to the import of photosynthate. Plant Physiol 67: 1016–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troufflard S, Roscher A, Thomasset B, Barbotin JN, Rawsthorne S, Portais JC. (2007) In vivo 13C NMR determines metabolic fluxes and steady state in linseed embryos. Phytochemistry 68: 2341–2350 [DOI] [PubMed] [Google Scholar]
- Wahrheit J, Nicolae A, Heinzle E. (2011) Eukaryotic metabolism: measuring compartment fluxes. Biotechnol J 6: 1071–1085 [DOI] [PubMed] [Google Scholar]
- Weber APM, Linka N (2011) Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu Rev Plant Biol 62: 53–77 [DOI] [PubMed] [Google Scholar]
- Wilcox JR. (1998) Increasing seed protein in soybean with eight cycles of recurrent selection. Crop Sci 38: 1536–1540 [Google Scholar]
- Wilcox JR, Shibles RM. (2001) Interrelationships among seed quality attributes in soybean. Crop Sci 41: 11–14 [Google Scholar]
- Williams TCR, Miguet L, Masakapalli SK, Kruger NJ, Sweetlove LJ, Ratcliffe RG. (2008) Metabolic network fluxes in heterotrophic Arabidopsis cells: stability of the flux distribution under different oxygenation conditions. Plant Physiol 148: 704–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms JR, Salon C, Layzell DB. (1999) Evidence for light-stimulated fatty acid synthesis in soybean fruit. Plant Physiol 120: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RF (1987) Soybeans: Improvement, Production and Uses. American Society of Agronomy, Madison, WI [Google Scholar]
- Xiong W, Liu L, Wu C, Yang C, Wu Q. (2010) 13C-tracer and gas chromatography-mass spectrometry analyses reveal metabolic flux distribution in the oleaginous microalga Chlorella protothecoides. Plant Physiol 154: 1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklich RW. (2001) β-Conglycinin and glycinin in high-protein soybean seeds. J Agric Food Chem 49: 729–735 [DOI] [PubMed] [Google Scholar]
- Yazdi-Samadi B, Rinne RW, Seif RD. (1977) Components of developing soybean seeds: oil, protein, sugars, starch, organic acids, and amino acids. Agron J 69: 481–486 [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. (2008) An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng 99: 686–699 [DOI] [PubMed] [Google Scholar]
- Zamboni N. (2011) 13C metabolic flux analysis in complex systems. Curr Opin Biotechnol 22: 103–108 [DOI] [PubMed] [Google Scholar]