A likely regulator of tomato ripening is identified from a gene network, its function is validated in transgenic plants, and an orthologous gene is shown to play a similar role in pepper.
Abstract
Carotenoids represent some of the most important secondary metabolites in the human diet, and tomato (Solanum lycopersicum) is a rich source of these health-promoting compounds. In this work, a novel and fruit-related regulator of pigment accumulation in tomato has been identified by artificial neural network inference analysis and its function validated in transgenic plants. A tomato fruit gene regulatory network was generated using artificial neural network inference analysis and transcription factor gene expression profiles derived from fruits sampled at various points during development and ripening. One of the transcription factor gene expression profiles with a sequence related to an Arabidopsis (Arabidopsis thaliana) ARABIDOPSIS PSEUDO RESPONSE REGULATOR2-LIKE gene (APRR2-Like) was up-regulated at the breaker stage in wild-type tomato fruits and, when overexpressed in transgenic lines, increased plastid number, area, and pigment content, enhancing the levels of chlorophyll in immature unripe fruits and carotenoids in red ripe fruits. Analysis of the transcriptome of transgenic lines overexpressing the tomato APPR2-Like gene revealed up-regulation of several ripening-related genes in the overexpression lines, providing a link between the expression of this tomato gene and the ripening process. A putative ortholog of the tomato APPR2-Like gene in sweet pepper (Capsicum annuum) was associated with pigment accumulation in fruit tissues. We conclude that the function of this gene is conserved across taxa and that it encodes a protein that has an important role in ripening.
Tomato (Solanum lycopersicum) is a climacteric fruit where ripening is initiated and coordinated by ethylene (Alexander and Grierson, 2002). It is the model system for studying ripening in fleshy fruits because of the exceptional genetic and molecular resources that are available, including well-characterized mapping populations (Lippman et al., 2007), numerous single-gene mutants, routine transformation, and a fully annotated genome sequence (Tomato Genome Consortium, 2012). The repertoire of well-characterized mutations in tomato has permitted the identification of genes that encode proteins that govern the ripening process. These have included Never-ripe (Nr), ripening-inhibitor (rin), nonripening (nor), and Colorless nonripening (Cnr). Mutations at these loci can completely abolish normal ripening (Lanahan et al., 1994; Vrebalov et al., 2002; Manning et al., 2006). The NR, RIN, CNR, and NOR gene products, along with those from tomato HD-Zip homeobox protein1 (LeHB1), Tomato AGAMOUS-LIKE1 (TAGL1), APETALA2 (AP2; Lin et al., 2008; Itkin et al., 2009; Vrebalov et al., 2009; Chung et al., 2010; Karlova et al., 2011), and others govern the onset and progression of the ripening. Despite a growing understanding of this high-level regulatory network, the links to hormonal cues, plastid signals, and downstream effectors mediating alterations in color, texture, and flavor are still poorly understood.
In tomato, the colored carotenoid compounds provide a ready source of phytochemicals in the human diet (Goff and Klee, 2006; Klee and Giovannoni, 2011) and have a protective role against cardiovascular disease and certain cancers (Fraser and Bramley, 2004). During ripening, chloroplasts in the fruit tissues are converted into chromoplasts, and this conversion influences not only the fruit color but also its nutrient and flavor composition (Klee and Giovannoni, 2011). There are many factors responsible for regulating the number and size of plastids in the fruit and other plant tissues.
In tomato, a small number of fruit high-pigment mutants have been described. These include high-pigment1 (hp1), which harbors a lesion in UV-DAMAGED DNA-BINDING PROTEIN1 (DDB1; Lieberman et al., 2004; Liu et al., 2004), and hp2, a mutation in tomato DEETIOLATED1 (DET1; Mustilli et al., 1999). These genes are involved in the suppression of light responses in the absence of light, and for DET1, this is by a molecular mechanism involving chromatin remodeling (Davuluri et al., 2004). The DDB1 and DET1 genes encode proteins that form a complex with Cullin4 (CUL4), a ubiquitin-conjugating E3 ligase to target proteins for proteolysis (Wang et al., 2008). Down-regulation of DET1, DDB1, or CUL4 leads to enhanced levels of chlorophylls, carotenoids, and other pigmentation in the fruits. The effects on carotenoid accumulation were initially believed to be mediated through an enhanced plastid compartment area including increased plastid abundance (Davuluri et al., 2004; Wang et al., 2008). However, integrating information from a range of “omic” data sets has identified the coordinated up-regulation of core metabolic processes as the progenitor of the high-pigment phenotype in the DET1 lines (Enfissi et al., 2010). A further mutation, hp3, shows an increase in plastid number and size and accumulates 30% more carotenoids than wild-type fruits. This effect has been mapped to a lesion in the zeaxanthin epoxidase gene, which results in a block in the biosynthesis of xanthophylls and greatly reduced levels of abscisic acid (ABA), xanthophylls being an essential precursor in the biosynthesis of this plant hormone (Galpaz et al., 2008). All of these hp mutations have deleterious effects on plant development, and because their effects are not fruit specific, they are difficult to harness effectively for crop improvement.
In Arabidopsis (Arabidopsis thaliana) leaf tissue, Golden2-like (GLK2) transcription factors are necessary to coregulate and synchronize the expression of a suite of nuclear photosynthetic genes, and GLK-overexpressing lines in Arabidopsis accumulate higher amounts of chlorophyll (Waters et al., 2008, 2009). Recently, the up-regulation of a GLK2-like transcription factor in tomato has also been shown to elevate levels of chlorophyll and carotenoids in tomato, and a lesion in this gene is responsible for the uniform ripening phenotype (Powell et al., 2012). In this work, we report the discovery of a novel ripening-related transcription factor that influences pigmentation and ripening in tomato and shows strong sequence similarity to ARABIDOPSIS PSEUDO RESPONSE REGULATOR2 (APRR2), a gene related to, but distinct from, tomato GLK2.
RESULTS AND DISCUSSION
Identification of a Putative Fruit Color Regulator
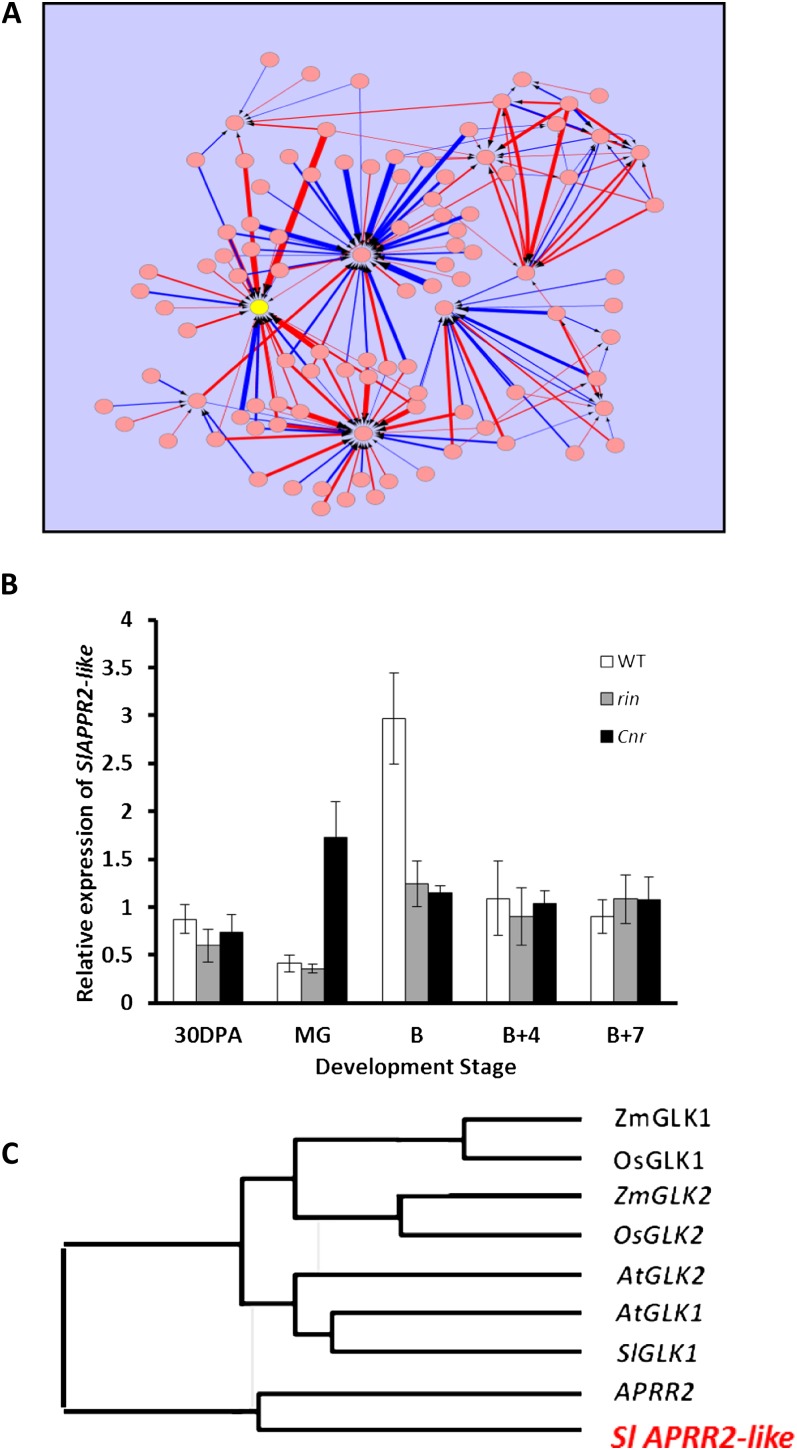
In this study, we used Affymetrix GeneChip transcriptomic data from two separate fruit development and ripening time series to construct gene regulatory networks by artificial neural network (ANN) inference. ANNs have previously been shown to have strong modeling capabilities when correctly applied to the analysis of complex biological data across a range of domains and applications (Lancashire et al., 2010) The ANN method works by training a model using a subset of the available data. In this investigation, we initially looked for novel fruit-ripening regulators by using 551 transcription factor genes present on an Affymetrix Tomato GeneChip and then reducing the number of genes in the data set to the 100 strongest interactions (Lancashire et al., 2005, 2010; Matharoo-Ball et al., 2007). The outcome of the ANN method was an inferred network of interactions based on these top 100 interacting genes. An initial data set, when analyzed by ANN, provided a network of transcription factors with several major highly interacting nodes, including one (Fig. 1A; Supplemental Fig. S1; Supplemental Data Set S1) occupied by a gene annotated as having some similarity to the Arabidopsis GLK sequence on the GeneChip array that can be found on the tomato genome assembly (Tomato Genome Consortium, 2012) as Solyc08g077230 (GenBank accession no. KC147634). The same gene was also represented as a node with a high number of edges in a further network constructed from a much more extensive ripening time series but using the same 100 genes as before (data not shown). Quantitative real-time (qRT)-PCR using wild-type cv Ailsa Craig fruits and near-isogenic lines containing the nonripening mutations rin and Cnr confirmed that this tomato gene had a ripening-related pattern of expression with a strong increase in expression at the breaker stage. In contrast, its expression was suppressed in all the nonripening mutants, except for in Cnr at the mature green (MG) stage, where the levels of expression were highly elevated (Fig. 1B).
Figure 1.
Identification of a novel ripening-related transcription factor by neural network inference analysis. A, Transcription factor network generated from an analysis of MG and breaker + 7 stages of ripening in wild-type tomato fruits. Genes are shown as pink circles (for full details of all gene identifiers, see Supplemental Fig. S1 and Supplemental Data Set S1), positive and negative interactions are designated by red and blue arrows, respectively, and the width of the line is proportional to the relative strength of the interaction. The APRR2-Like gene is shown in yellow. B, Expression of the APRR2-Like gene during ripening in the wild type (WT) and fruits of the nonripening mutants rin and Cnr. B, Breaker stage. C, Phylogenetic tree showing the relationship between the tomato APPR2-Like gene (SlAPRR2-Like) and related sequences in Arabidopsis.
A phylogenetic analysis (Fig. 1C) revealed that the putative tomato GLK sequence showed the closest similarity to the APRR2 gene. In Arabidopsis, this gene has been partially characterized, and it is a likely transcriptional activator that comprises a GARP DNA-binding domain and a receiver-like domain at the N terminus, similar to that encoded by the ARR-B genes (Fitter et al., 2002). However, APRR2 lacks the important AREAEAA motif conserved in the GLK genes, so while it is considered to be related to GLKs, it is distinct from the GLK group (Fitter et al., 2002). The function of the APRR2 transcriptional activator in Arabidopsis has yet to be determined. The position of the tomato APPR2-Like gene as a hub (i.e. with high vertex degree) in our ANNs and its ripening-related expression pattern, however, are suggestive of an important role in the ripening process. Also, the degree of sequence similarity between our tomato gene and GLK-like genes suggested that its function could be related to plastid metabolism and, therefore, color development.
Functional Analysis of the Tomato APRR2-Like Gene in Transgenic Plants
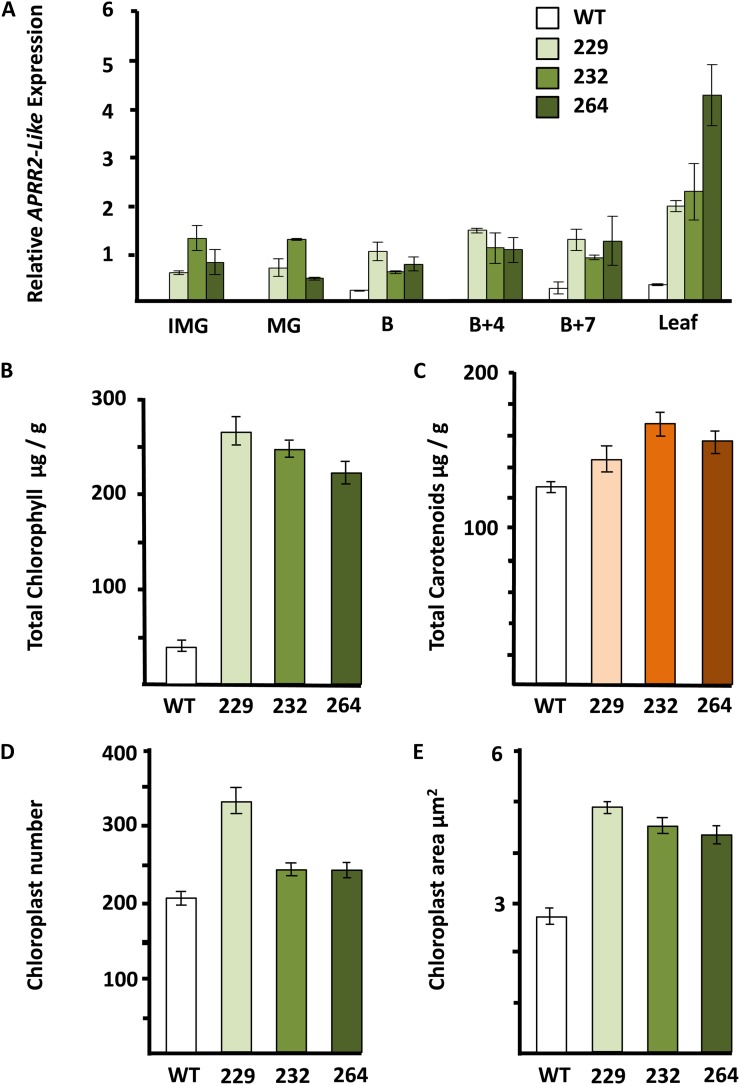
The tomato APRR2-Like coding sequence was placed downstream of the 35S promoter in order to constitutively overexpress this gene in transgenic tomato plants. cv Micro-Tom was chosen to test the gene function, as this miniature tomato can be efficiently transformed and grown to fruiting more quickly than large globe tomato lines. The initial transformation experiment yielded 38 individual positive transformants. Of these, 20 showed a striking fruit color phenotype but no apparent visual changes in other parts of the plant. We selected three independent lines for more detailed analysis. T1 progeny from the three independent transformation events were analyzed for target gene expression in leaves and during fruit development. All three overexpressing lines showed changes in pigmentation that were apparent in the developing and ripening fruits. Unripe fruits of the transgenic lines had a more intense green coloration, and ripe fruits had a more intense red coloration (Fig. 2). The level of APRR2-Like gene expression in leaf and fruit tissues in the transgenic lines was substantially higher than in the wild type, as determined by quantitative PCR (qPCR), with up to a 70-fold increase at MG (Fig. 3A), and this was consistent with alteration in pigment composition. The leaf tissues of both transgenic and wild-type cv Micro-Tom lines had similar total chlorophyll levels (Supplemental Fig. S2). In striking contrast, the fruit tissues of the transgenic lines showed significantly (P < 0.001) enhanced chlorophyll levels in developing fruits and increased carotenoid content in ripe fruits (Fig. 3, B and C). This analysis was entirely consistent with the observed visual alterations in fruit color. Measurements of individual pigment metabolites revealed marked increases in lycopene and β-carotene in the pericarp of the overexpressor lines (Table I).
Figure 2.

Phenotypes of the wild type and cv Micro-Tom lines overexpressing the tomato APPR2-Like gene. The fruit stages are as follows: IMG, immature green; B, breaker; B+4, breaker + 4 d; B+7, breaker + 7 d. Wild-type fruits are shown in column 1, and cv Micro-Tom lines overexpressing the tomato APPR2-Like gene are shown in column 2.
Figure 3.
Effects of APPR2-Like overexpression in transgenic cv Micro-Tom lines on pigment accumulation, chloroplast number, and area. A, Relative APRR2-Like expression. Fruit stages are as defined in Figure 2. B, Total chlorophyll at MG. C, Total carotenoids at breaker + 7 d. D, Chloroplast number at MG. E, Chloroplast area at MG. These parameters are shown for the wild type (WT) and three transgenic lines (229, 232, and 264). Error bars indicate se; n = 3 fruits from each line at each developmental stage.
Table I. Carotenoid and tocopherol contents.
Pigment levels in the wild type and APRR2-Like overexpressor lines (229, 232, and 264) are shown at breaker + 7 d. Data represent average levels from three independent fruit ± sd. Values in boldface indicate significant (P < 0.05) increases in comparison with the wild type.
| Line | Phytoene | Phytofluene | Lycopene | β-Carotene | Lutein | Tocopherol |
|---|---|---|---|---|---|---|
| Wild type | 98.5 ± 39 | 38.8 ± 15 | 439.6 ± 133 | 64.15 ± 19 | 26.3 ± 9.0 | 1.4 ± 0.2 |
| 229 | 95.1 ± 18 | 44.6 ± 5 | 645.1 ± 140 | 89.1 ± 17 | 35.4 ± 5.0 | 1.6 ± 0.5 |
| 232 | 122.4 ± 42 | 44.6 ± 12 | 656.7 ± 246 | 54.7 ± 6.0 | 27.1 ± 6.9 | 1.2 ± 0.23 |
| 264 | 131.3 ± 15 | 51.4 ± 2 | 700.3 ± 370 | 83.3 ± 29 | 31.1 ± 10.0 | 1.4 ± 0.5 |
The APRR2-Like Gene Increases Plastid Compartment Size in Tomato
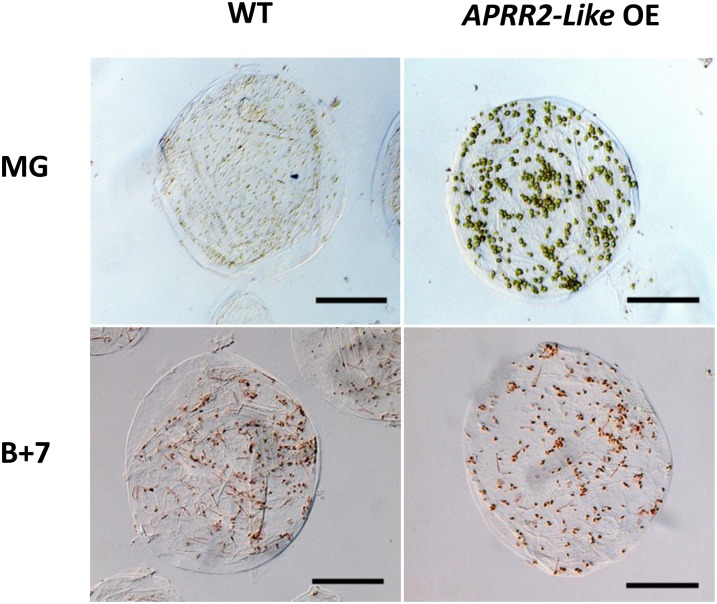
We imaged the cells of the overexpression lines from the inner and outer pericarp tissue and determined chloroplast number, area in the outer pericarp, and plastid numbers in ripe fruits. Similar results were obtained from both outer and inner pericarp tissues. There were significantly (P < 0.001) higher numbers of plastids in the outer pericarp of the transgenic fruits at both the MG and ripe stages in comparison with the wild type (Fig. 3D). The area of the individual outer pericarp plastids was greater in the transgenic lines than in wild-type fruits at MG (Fig. 3E), but there were no apparent differences in ripe fruits. In both the inner and outer pericarp tissues of the transgenic fruits, the plastids were more intensely colored at the MG and red ripe stages (Fig. 4). These phenotypic features resemble those in the hp mutants. The DET1 (hp2) mutation leads to an increase in the number of plastids and total plastid area per cell (up to 3-fold; Enfissi et al., 2010). Like DET1, constitutive silencing of DDB1 (hp1) also leads to elevated plastid number and size and involves the CUL4-based E3 ligase complex, suggesting a possible involvement of ubiquitin-mediated degradation of these organelles (Wang et al., 2008). In the hp3 mutant, higher concentrations of carotenoids and chlorophyll were also measured in both leaves and the pericarp of green fruit. ABA deficiency in hp3 leads to an enlargement of plastid compartment size, probably by increasing plastid division, thus enabling greater biosynthesis and a higher storage capacity for the pigments (Galpaz et al., 2008). Our data indicate that overexpression of the tomato APRR2-Like gene is mediating its effect by both increasing the plastid size in the cells and the levels of chlorophyll and carotenoids. These phenotypic features are also similar to the recently published effects of tomato GLK2 overexpression (Powell et al., 2012).
Figure 4.
Plastid phenotypes in the wild type and transgenic cv Micro-Tom lines overexpressing the tomato APPR2-Like gene. Individual outer pericarp cells from MG and breaker + 7 d (B+7) fruits show differences in both plastid number and color intensity between the wild type (WT) and an APRR2-Like overexpressor line (APRR2-Like OE).
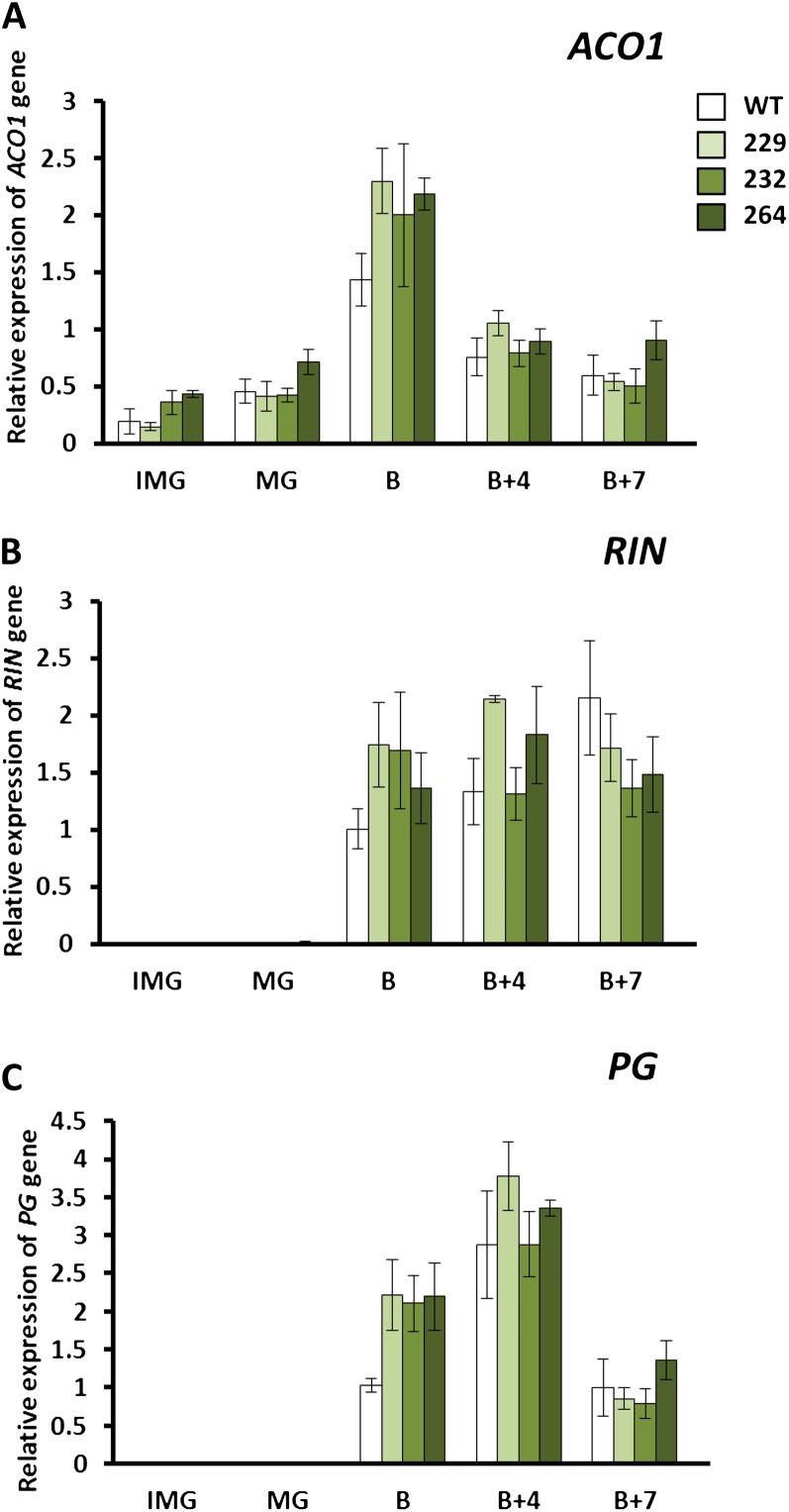
Ripening-Related Gene Expression Is Altered in Tomato Lines Overexpressing the APRR2-Like Gene
qPCR experiments indicated an effect of APRR2-Like overexpression on selected aspects of ripening-related gene expression, although whether this is a direct or indirect effect requires further investigation. The gene encoding the MADS box transcription factor underlying the rin locus (LeMADS-RIN), which is known to be a master regulator of ripening, showed elevated expression at the breaker stage in the transgenic overexpressor fruits (Fig. 5). There were also indications of enhanced ACO expression at breaker in some transgenic lines, which was consistent with elevated levels of ethylene production and higher levels of transcript for the cell wall-degrading enzyme polygalacturonase (PG). No consistent differences in the levels of NOR, PSY1, or ACS2 transcripts were observed between wild-type and transgenic lines at the early stages of ripening, although wild-type levels of all three genes were elevated in wild-type fruits at later stages, especially breaker + 7 d (Supplemental Fig. S3). ACO1 and ACS2 encode enzymes for the key steps in ethylene biosynthesis (Alexander and Grierson, 2002). These observations indicate that APRR2-Like overexpression stimulates the expression of RIN and enhances ripening-related ethylene biosynthesis. Both of these events are known to trigger the expression of downstream effector genes such as PG (Klee and Giovannoni, 2011).
Figure 5.
The tomato APPR2-Like gene impacts ripening-related gene expression. qPCR analysis of ACO (A), RIN (B), and PG (C) is shown in the wild type (WT) and transgenic APRR2-Like overexpression lines (229, 232, and 264) during development and ripening at the following stages: IMG, immature green; B, breaker; B+4, breaker + 4 d; B+7, breaker + 7 d. Error bars indicate se; n = 3 fruits from each line at each developmental stage.
The absence of changes in phytoene synthase transcript levels, a gene product that is a critical intermediate of carotenoid biosynthesis, is consistent with the increase in flux through the carotenoid pathway without elevation in PSY1 transcript levels, as found with the hp mutants (Enfissi et al., 2010). The expression of genes specifically involved in plastid metabolism, such as the tomato GLK1 and accD, which is involved in fatty acid biosynthesis, was unchanged (Supplemental Fig. S3). cv Micro-Tom is known to harbor the uniform mutation that causes a stop codon in the GLK2 gene; therefore, APRR2-Like cannot be mediating its effects by altering the levels of a functional GLK2 protein (Carvalho et al., 2011). Also, a list of genes showing differential expression in GLK2 overexpressors did not include APRR2-Like (see supplemental data in Powell et al., 2012). The effects of APRR2-Like overexpression on LeMADS-RIN, ACO, and PG transcript levels indicates that it plays a role in the ripening process above and beyond enhancing pigment levels. This is in contrast to observations with GLK2 overexpressors reported by Powell et al. (2012), where no obvious effect on the timing of ripening was reported. Our results suggest that the APRR2-Like gene product may form part of a ripening signal mechanism involving the plastid compartment (Barry et al., 2012). The mechanism of action clearly demands further investigation.
Preliminary data indicate that the elevated pigment phenotype of the APRR2-Like overexpression lines in the cv Micro-Tom tomato background can be transferred to globed tomato fruits, as indicated in the visual phenotypes of the F1 cv Micro-Tom × cv M82 hybrid line (Supplemental Fig. S4).
An Ortholog of the Tomato APRR2-LIKE Gene Is Associated with Increased Pigmentation in Sweet Pepper
Pepper (Capsicum annuum) genotypes exhibit a wide variation for color and color intensity at both the immature and mature stages of fruit development (Thorup et al., 2000). A pepper ortholog of the APRR2-Like gene would represent a strong candidate as a regulator of fruit color and color intensity. We have identified a likely APRR2-Like gene in sweet pepper by undertaking a BLAST search of a pepper bacterial artificial chromosome library using the tomato APRR2-Like sequence. This pepper gene (GenBank accession no. KC175445) maps to pepper chromosome 8 in a region that shows strong synteny with a region of tomato chromosome 8 containing the APRR2-Like locus (Solyc08g077230).
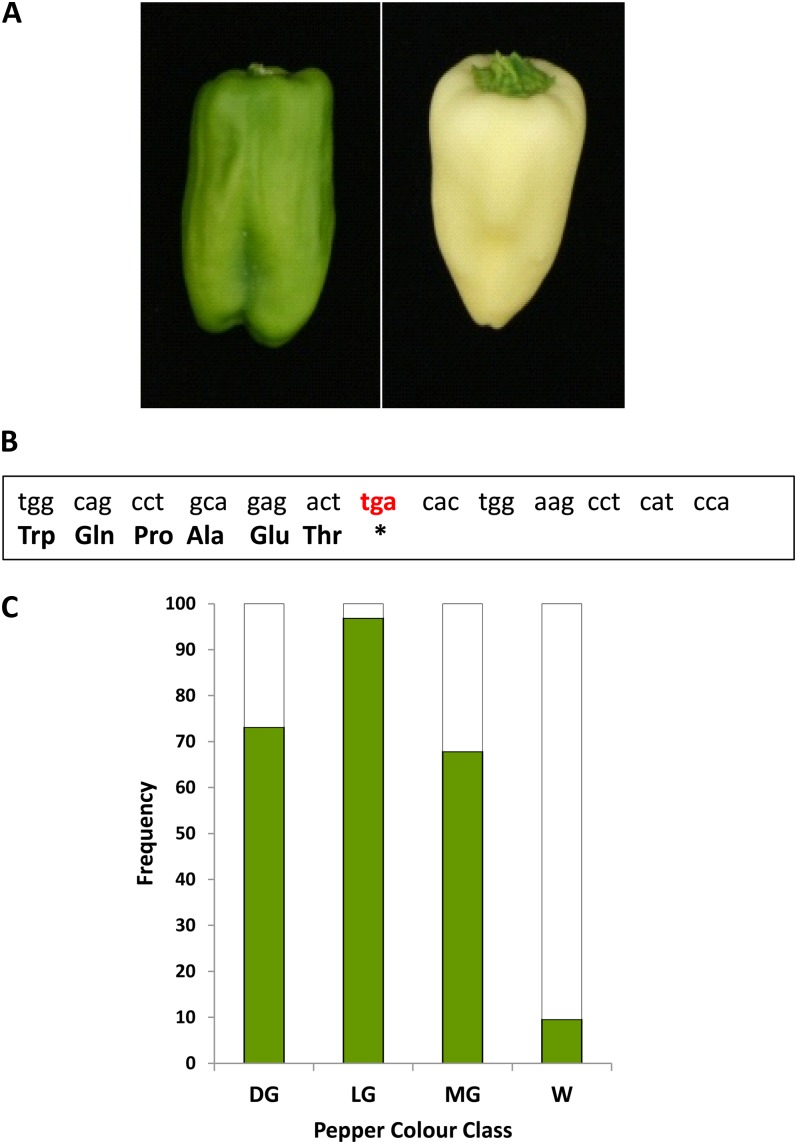
We have several lines of evidence that this APRR2-Like gene in pepper can influence color in these fruits. (1) Sequencing of the gene from wild-type and white-fruited parental lines from a mapping population revealed evidence for a polymorphism between these two sets of genotypes, and sequence alignment revealed a G-to-A substitution resulting in a stop codon in a white-fruited parent (Fig. 6; Supplemental Fig. S5). (2) Analysis of the pigment content of lines from this blocky pepper population, which segregated for fruit color, revealed a strong association between immature fruit color intensity and the single-nucleotide polymorphism identified in the gene (Fig. 6; for fruit chlorophyll content, see Supplemental Fig. S6). These effects are all consistent with the gene modulating pigmentation in immature pepper fruits. Recently, a major quantitative trait locus (QTL) explaining over 50% of the variation in chlorophyll content in a pepper × Capsicum chinense mapping population has been reported (Brand et al., 2012). The QTL was responsible for increasing plastid compartment size in the fruits. This QTL was located on pepper chromosome 8 and associated with marker T1341. This marker is close to C2_At1g32540, which is a COSII marker mapped on both the tomato and pepper genetic maps close to T1341 and a pepper APRR2-Like gene.
Figure 6.
Association between pepper color and the presence of a null pepper APPR2-Like gene. A, Dark green and white fruits. B, Fragment of the pepper APRR2-Like allele showing the stop codon. C, Association of an allele with a stop codon (white bar) with mature pepper color in mapping population color types as follows: DG, dark green; LG, light green; MG, medium green; W, white.
The mechanism of action of the tomato or pepper APRR2-Like genes is not known, and the function of APRR2 in Arabidopsis is also unclear. In a recent study (Rohrmann et al., 2011), a novel qRT-PCR platform was used to quantify the expression level of more than 1,000 tomato transcription factors during ripening in cv Ailsa Craig and hp1 mutant fruits. Interestingly, the authors observed that the expression of transcription factors of the “GLK2-like” family was considerably altered in the hp1 mutant. This included a gene with the same sequence as tomato APRR2-Like. These data indicate a link between the functional deficiency of the DDB1 signal transduction gene in the hp1 mutant and APRR2-Like gene expression. Evidence from Arabidopsis indicates that the APRR2 gene product interacts with the calmodulin-like protein CLM9, which is involved in tolerance to abiotic stress and the regulation of seed germination by ABA (Perochon et al., 2010). Additionally, APRR1, a gene related to APRR2, has been reported to encode a protein that interacts with the ABSCISIC ACID INSENSITIVE3 (ABI3) transcription factor (Kurup et al., 2000). ABI3 is known to be involved in promoting the expression of genes containing ABA-responsive elements. One possibility is that in tomato, the APRR2-Like genes interact with ABI3, in some way influencing ABA signaling related to plastid development and ripening. The mechanistic basis of the APRR2-Like gene effects are now the subject of further investigation in our laboratories.
Despite our limited understanding of the role of the APRR2-Like gene product in tomato ripening, it is an important target for breeding. Indeed, fruit color QTLs have been reported in the Solanum pennellii introgression lines harboring the region containing the APRR2-Like locus (Liu et al., 2003). In this study and elsewhere, microarray platforms (Lee et al., 2012), the recently published qRT-PCR platform for tomato transcription factors (Rohrmann et al., 2011), and the RNAseq data for all tomato genes (Tomato Genome Consortium, 2012) are all proving to be important resources for facilitating the discovery of novel ripening regulators, especially when combined with new network inference-based approaches.
MATERIALS AND METHODS
Plant Material
All experiments were performed using wild-type tomato (Solanum lycopersicum ‘Micro-Tom’ and ‘Ailsa Craig’) and cv Ailsa Craig near-isogenic lines containing the rin and Cnr nonripening mutations. cv Ailsa Craig, rin, and Cnr plants were grown in 24-cm-diameter pots in M3 compost (Levington Horticulture), and cv Micro-Tom plants were grown in 9-cm-diameter pots under standard glasshouse conditions. Plants were watered daily. Flowers were tagged at anthesis, and fruit development was recorded as days post anthesis. MG fruit were defined as 40 DPA and were characterized as being green and shiny with no obvious color change. Breaker fruit were defined as those showing the first signs of ripening-associated color change from green to orange. Mutants rin and Cnr were taken as 49-DPA equivalents to breaker wild-type fruits. Subsequent ripening stages were defined in days post breaker. All plant samples for the preparation of total RNA were taken at the same time each day, frozen in liquid nitrogen, and stored at −70°C until required. Three separate transgenic events were analyzed with three fruit per line taken at specified stages of development for the various analyses.
A blocky pepper (Capsicum annuum) double haploid mapping population was developed based on the interspecific cross between the white-fruited line [Whi]B∼B (New Mexico State University Chile Pepper Institute) and the red-fruited line [PimG]B (Syngenta breeding line). This population segregates for immature and mature fruit color and was grown under standard commercial practices in open fields in Naples, Florida, and in Gilroy, California, in the spring and summer of 2011. The population was planted in a randomized block design with three replicate plots per genotype in the trial. Each plot contained 20 plants per genotype.
Phylogenetic Comparison
Phylogenetic comparisons between the tomato APRR2-Like protein and other GLK and APRR2-related proteins were made using the Unweighted Pair Group Method with Arithmetic algorithm in MEGA5.0 (http://www.megasoftware.net/).
Plasmid Construction and Generation of Transgenic Plants
The transgenic construct (pGWB-APRR2-like) was designed to constitutively overexpress a functional APRR2 gene with the cauliflower mosaic virus 35S promoter. A 2,090-bp PCR fragment of the tomato APRR2-Like gene including the full coding sequence was ligated in the sense orientation between the cauliflower mosaic virus 35S promoter and terminator using the Gateway cloning method (Invitrogen). Plasmids containing the ligated fragment in the correct orientation were identified by restriction digest analysis and by sequencing. After transfer to Agrobacterium tumefaciens strain C58 by electroporation, the construct was used to transform tomato cotyledon explants (Sun et al., 2006). Transgenic plants that rooted on 75 mg L−1 kanamycin were transferred to compost and grown as described above. Primers for gene cloning were as follows: APRR2-Like-F, 5′-GACCCACCTAACTATAATCA-3′; APRR2-Like-R, 5′-AATGGAGCACACTCATCGGC-3′.
qRT-PCR Analysis
qRT-PCR was performed with mRNA isolated from different tissues. Primers were designed by using Primer 3 software (http://primer3.sourceforge.net/). The probe was duel labeled by fluorescein at the 5′ end and TAMARA at the 3′ end. qRT-PCR, with an annealing temperature of 60°C, was performed in triplicate on a 384-well plate in the LightCycler 480 Real-Time PCR Detection System (Roche) using LightCycler Taqman Master according to the manufacturer’s recommended protocol. The elongation factor was used as a control to normalize the qRT-PCR values across different samples (GenBank accession no. X14449). Primers are listed in Supplemental Table S1.
Plastid Number and Size Determination
Immediately after excision with a sterile razor blade, MG and ripening tomato fruit (pericarp only, 1-mm2 sections) cells were fixed in 3.5% glutaraldehyde solution for 1 h in darkness. Fruit tissue was disrupted as described previously (Forth and Pyke, 2006) and as follows: heat treated at 65°C in a solution of disodium EDTA (EDTA-Na2; 0.1 m, pH 9.0) for 20 min followed by maceration with clean forceps on glass microscope slides. Ripe-fruit pericarp was disrupted in a solution of EDTA-Na2 (0.1 m, pH 9.0) at room temperature. Tissue was stored at 4°C in EDTA-Na2 solution for up to 6 months. Samples were imaged using a Leica CTR5000 microscope and a Leica DFC 420C digital camera. Images were acquired using the Leica Application Suite (version 3) software, and cell plan area measurement was taken using Image-Pro Plus (version 5.1).
Metabolite Analysis
Total chlorophyll and carotenoid measurements were taken for both inner and outer pericarp following the method of Forth and Pyke (2006). A section of the pericarp was cut from MG and ripe fruit, and inner and outer pericarp were separated, weighed, and ground in liquid nitrogen. Twenty milliliters of 60%:40% hexane:acetone was added to the melted samples. The hexane:acetone was removed and stored in a glass bottle wrapped in foil and replaced repeatedly with fresh hexane:acetone until no longer discolored by grinding of the fruit. The absorbance of the samples was immediately measured in a Phillips PU 8720 scanning spectrophotometer, and the chlorophyll and carotenoid contents were calculated with the following equations: total chlorophyll (mg g−1) = [8.02 × A663 + 20.20 × A645] × V/(1,000 × W); total carotenoid (mg g−1) = (OD450)/0.25 × V/(1,000 × W), where V = volume of the extract (mL) and W = weight of fresh leaves (g).
The carotenoid pigments and tocopherols were extracted from freeze-dried tomato material that had been homogenized and ground to a fine powder using a standard protocol (Fraser et al., 2000). Typically, 10 mg of material was extracted with chloroform (1 mL), and a partition was created with a methanol:water (1:1) mixture. The lower chloroform layer was removed, and the remaining aqueous phase was reextracted with chloroform (1 mL). The pooled chloroform extracts were dried under a stream of nitrogen gas and stored at −20°C until further use. Prior to separation by HPLC, the residue was resuspended in ethyl acetate (HPLC grade) and centrifuged at 12,000g for 5 min to remove debris. The HPLC separation of carotenoids and tocopherols was performed with a Waters Alliance 2600S system. Separations were performed on a reverse-phase C30, 5-µm column (250 × 4.6 mm i.d.) with a C30 guard column (20 × 4.6 mm; YMC), maintained at 25°C as described by Fraser et al. (2000). The separation method used consisted of the mobile phase used for routine analysis composed of methanol (A), methanol:water (80:20, v/v) containing 0.2% (w/v) ammonium acetate (B), and tert-butyl methyl ether (C). Elution from the column was carried out from 95% A and 5% B for 12 min, then a step to 80% A, 5% B, and 15% C, followed by a linear gradient to 30% A, 5% B, and 65% C at 30 min. The column was then returned to the initial conditions and equilibrated over 30 min. A flow rate of 1 mL min−1 was employed. Pigments were detected using an online photo diode array (200–600 nm) with the elution being monitored continuously. Identification was performed by the comparison of spectral properties and cochromatography with authentic standards. Quantification was carried out using dose-response curves prepared from authentic standards previously purified by HPLC and/or thin-layer chromatography.
Color Measurements in Tomato
Color was determined on each fruit in both epidermis and pulp using a Minolta CR200 model colorimeter. Following the recording of the Hunter color scale values of L, a, and b, tomato color index (TCI) was calculated: TCI = (2,000a)/(L × (a2 + b2)0.5) (Langley et al., 1994). Results were means ± se of three determinations for each fruit of the three replicates.
Analysis of Pigment Variant in Pepper
The association between immature pepper fruit color and the presence of the normal or white allele of the APRR2-Like gene was determined on a double haploid pepper population segregating from white to dark green for immature fruit color. Sequence polymorphism in the APRR2-Like gene from 149 individuals was analyzed and correlated with observed immature fruit color.
Statistical Analysis
The mean values of qRT-PCR, chlorophyll and carotenoid contents, and color measurement were taken from the measurements of three biological replicates, and se was calculated. Data were analyzed by Statistical Analysis System, version 14, software (GenStat) by Student’s t test to assess significant differences among the means.
Determination of Chloroplast Size in Tomato
Fresh glasshouse-grown tomato fruit were shipped to Syngenta’s Jealott’s Hill International Research Station from Nottingham University. A 1-cm3-thick hand-cut section was removed from the midpoint of each fruit, and each section was mounted in water on a glass microscope slide and covered with a glass coverslip. The chloroplasts were visualized within each section using a Bio-Rad Radiance 2100 laser scanning confocal microscope using the following settings: 488-nm argon ion laser line, 50% power, 40× objective, 1,024 × 1,024 resolution, no zoom, 50 lines per second scan speed. Three random fields of view were imaged within each section. The images were then exported as tagged image files, and the size of the chloroplasts was analyzed in ImageJ.
Network Inference
ANNs have previously shown strong modeling capabilities when correctly applied to the analysis of complex biological data across a range of domains and applications (Lancashire et al., 2010). In this investigation, we used multilayer feed-forward back-propagation ANNs in an attempt to elucidate gene-gene relationships from a large-scale transcriptomic data set. We initially looked for novel fruit-ripening genes using an unseeded ANN method, in which the 551 transcription factors present on the tomato Affymetrix GeneChip were considered as potential regulators of ripening. ANNs do not model well in data sets with extremely high dimensionals, so effective step-wise parameterization approaches (Lancashire et al., 2008) were employed to reduce the number of markers in the data set to 100. This approach consists of two phases and has been previously applied with success in several studies (Lancashire et al., 2005, 2010; Matharoo-Ball et al., 2007).
Phase 1. Screening for Contextual Probes
The algorithm first considers each of the n transcription factors as single inputs to the algorithm, thus creating n one-input models. A cross-validation strategy, combined with an early-stopping method, was applied to avoid over fitting of the model on the training set of data. At each one-input submodel, Monte Carlo cross validation was applied by randomly assigning the samples of the data set to a training, test, or validation set, with 20%, 20%, or 60% of the samples, respectively. This was repeated 50 times, for which a complete random reshuffling of the samples in the three subsets was undertaken. Training was repeated from the beginning over the 50 times and was constantly monitored by applying each of the models undergoing training to the test subset. Each model was then assessed for its predictive performance on this set. The training step was performed over 3,000 epochs, with an early stopping after 1,000 epochs when there was no observed improvement of the performance for the model on the test subset.
Once all of the genes had been tested through this method, they were ranked according to their mean square error at predicting the target phenotype, in this case breaker versus red ripe. The top 100 ranked transcription factors were selected for an ANN-based network inference.
Phase 2. Network Inference Modeling
The ANN algorithm described above was used in such a way that 99 transcription factors were the input to predict the output for the remaining transcription factor. The models were then parameterized to provide a weight and sign (i.e. activation or repression) to the relationship between each of the 99 transcription factors and the output transcription factor. This process was repeated with each of the remaining 99 transcription factors used as an output transcription factor. All 100 interaction models were then integrated to produce a network. If all interactions were considered, then 9,900 potential interactions would be present in the top 100 genes. Thus, for ease of interpretation, only the top 100 (1%) of interactions were mapped, indicating the strongest influences. The interaction maps depicted potential gene-gene interactions that may be important in fruit development pathways.
In Silico Network Verification
In silico verification of the ANN model was based on the top 100 transcription factors identified by phase 1 above being run through the network inference phase in a larger data set that considered all time points in the ripening process (n = 8, time points = MG to breaker + 7 d). In this analysis the APRR2-Like gene showed a high level of centrality determined by examination of a hierarchical network derived from the weightings generated in the phase 2 process described above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KC147634 and KC175445.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ANN with two developmental points.
Supplemental Figure S2. Chlorophyll levels in leaves from the wild type and APPR2-Like transgenic lines.
Supplemental Figure S3. qPCR data for additional ripening-related genes.
Supplemental Figure S4. Phenotypes of representative cv M82 × APRR2 F1 fruits.
Supplemental Figure S5. Pepper APRR2-Like complementary DNA (A) and predicted amino acid sequences from parent lines with green and white immature fruits (B).
Supplemental Figure S6. Chlorophyll content of individual immature white (W) and green (G) pepper lines.
Supplemental Table S1. qPCR primers for expression analysis.
Supplemental Data Set S1. GeneChip data set linked to Supplemental Figure S1.
Acknowledgments
We thank Peter G. Walley for the RNA extractions for the GeneChip experiments and Sarah Perfect for expert help with the analysis of chloroplast area determinations in the tomato samples. Malcolm Bennett and Michael Wilson provided useful discussions during the preparation of the manuscript. Christopher Gerrish is thanked for technical help and Laura Perez for the cv M82 × APRR2 fruit images in Supplemental Figure S4.
Glossary
- ABA
abscisic acid
- ANN
artificial neural network
- qRT
quantitative real-time
- qPCR
quantitative PCR
- MG
mature green
- QTL
quantitative trait locus
- EDTA-Na2
disodium EDTA
References
- Alexander L, Grierson D. (2002) Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot 53: 2039–2055 [DOI] [PubMed] [Google Scholar]
- Barry CS, Aldridge GM, Herzog G, Ma Q, McQuinn RP, Hirschberg J, Giovannoni JJ. (2012) Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiol 159: 1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A, Borovsky Y, Meir S, Rogachev I, Aharoni A, Paran I. (2012) pc8.1, a major QTL for pigment content in pepper fruit, is associated with variation in plastid compartment size. Planta 235: 579–588 [DOI] [PubMed] [Google Scholar]
- Carvalho RF, Campos ML, Pino LE, Crestana SL, Zsögön A, Lima JE, Benedito VA, Peres LE. (2011) Convergence of developmental mutants into a single tomato model system: ‘Micro-Tom’ as an effective toolkit for plant development research. Plant Methods 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Mustilli AC, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, et al (2004) Manipulation of DET1 expression in tomato results in photomorphogenic phenotypes caused by post-transcriptional gene silencing. Plant J 40: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfissi EMA, Barneche F, Ahmed I, Lichtlé C, Gerrish C, McQuinn RP, Giovannoni JJ, Lopez-Juez E, Bowler C, Bramley PM, et al (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Forth D, Pyke KA. (2006) The suffulta mutation in tomato reveals a novel method of plastid replication during fruit ripening. J Exp Bot 57: 1971–1979 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM. (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43: 228–265 [DOI] [PubMed] [Google Scholar]
- Fraser PD, Pinto MES, Holloway DE, Bramley PM. (2000) Technical advance: application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J 24: 551–558 [DOI] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J. (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53: 717–730 [DOI] [PubMed] [Google Scholar]
- Goff SA, Klee HJ. (2006) Plant volatile compounds: sensory cues for health and nutritional value? Science 311: 815–819 [DOI] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. (2009) TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kurup S, Jones HD, Holdsworth MJ. (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J 21: 143–155 [DOI] [PubMed] [Google Scholar]
- Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. (1994) The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancashire L, Ugurel S, Creaser C, Schadendorf D, Rees R, Ball G (2005) Utilizing artificial neural networks to elucidate serum biomarker patterns which discriminate between clinical stages in melanoma. In Proceedings of the 2005 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology. pp 455–460 [Google Scholar]
- Lancashire LJ, Powe DG, Reis-Filho JS, Rakha E, Lemetre C, Weigelt B, Abdel-Fatah TM, Green AR, Mukta R, Blamey R, et al. (2010) A validated gene expression profile for detecting clinical outcome in breast cancer using artificial neural networks. Breast Cancer Res Treat 120: 83–93 [DOI] [PubMed] [Google Scholar]
- Lancashire LJ, Rees RC, Ball GR. (2008) Identification of gene transcript signatures predictive for estrogen receptor and lymph node status using a stepwise forward selection artificial neural network modelling approach. Artif Intell Med 43: 99–111 [DOI] [PubMed] [Google Scholar]
- Langley KR, Martin A, Stenning R, Murray AJ, Hobson GE, Schuch WW, Bird CR. (1994) Mechanical and optical assessment of the ripening of tomato fruit with reduced polygalacturonase activity. J Sci Food Agric 66: 547–554 [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei ZJ, Tieman D, Klee H, Giovannoni J. (2012) Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Lieberman M, Segev O, Gilboa N, Lalazar A, Levin I. (2004) The tomato homolog of the gene encoding UV-damaged DNA binding protein 1 (DDB1) underlined as the gene that causes the high pigment-1 mutant phenotype. Theor Appl Genet 108: 1574–1581 [DOI] [PubMed] [Google Scholar]
- Lin ZF, Hong YG, Yin MG, Li CY, Zhang K, Grierson D. (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Semel Y, Zamir D. (2007) An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr Opin Genet Dev 17: 545–552 [DOI] [PubMed] [Google Scholar]
- Liu YS, Gur A, Ronen G, Causse M, Damidaux R, Buret M, Hirschberg J, Zamir D. (2003) There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol J 1: 195–207 [DOI] [PubMed] [Google Scholar]
- Liu YS, Roof S, Ye ZB, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J. (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Matharoo-Ball B, Hughes C, Lancashire L, Tooth D, Ball G, Creaser C, Elgasim M, Rees R, Layfield R, Atiomo W. (2007) Characterization of biomarkers in polycystic ovary syndrome (PCOS) using multiple distinct proteomic platforms. J Proteome Res 6: 3321–3328 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C. (1999) Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11: 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perochon A, Dieterle S, Pouzet C, Aldon D, Galaud JP, Ranty B. (2010) Interaction of a plant pseudo-response regulator with a calmodulin-like protein. Biochem Biophys Res Commun 398: 747–751 [DOI] [PubMed] [Google Scholar]
- Powell ALT, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Rohrmann J, Tohge T, Alba R, Osorio S, Caldana C, McQuinn R, Arvidsson S, van der Merwe MJ, Riano-Pachon DM, Mueller-Roeber B, et al (2011) Combined transcription factor profiling, microarray analysis and metabolite profiling reveals the transcriptional control of metabolic shifts occurring during tomato fruit development. Plant J 68: 999–1013 [DOI] [PubMed] [Google Scholar]
- Sun HJ, Uchii S, Watanabe S, Ezura H. (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47: 426–431 [DOI] [PubMed] [Google Scholar]
- Thorup TA, Tanyolac B, Livingstone KD, Popovsky S, Paran I, Jahn M. (2000) Candidate gene analysis of organ pigmentation loci in the Solanaceae. Proc Natl Acad Sci USA 97: 11192–11197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, McQuinn R, Chung M, Poole M, Rose JKC, Seymour G, Grandillo S, Giovannoni J, et al (2009) Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Wang SH, Liu JK, Feng YY, Niu XL, Giovannoni J, Liu YS. (2008) Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. Plant J 55: 89–103 [DOI] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA. (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56: 432–444 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]