A simple genetic manipulation triggers enhanced nitrogen use efficiency in lettuce.
Abstract
Plant nitrate (NO3−) acquisition depends on the combined activities of root high- and low-affinity NO3− transporters and the proton gradient generated by the plasma membrane H+-ATPase. These processes are coordinated with photosynthesis and the carbon status of the plant. Here, we present the characterization of romaine lettuce (Lactuca sativa ‘Conquistador’) plants engineered to overexpress an intragenic gain-of-function allele of the type I proton translocating pyrophosphatase (H+-PPase) of Arabidopsis (Arabidopsis thaliana). The proton-pumping and inorganic pyrophosphate hydrolytic activities of these plants are augmented compared with control plants. Immunohistochemical data show a conspicuous increase in H+-PPase protein abundance at the vasculature of the transgenic plants. Transgenic plants displayed an enhanced rhizosphere acidification capacity consistent with the augmented plasma membrane H+-ATPase proton transport values, and ATP hydrolytic capacities evaluated in vitro. These transgenic lines outperform control plants when challenged with NO3− limitations in laboratory, greenhouse, and field scenarios. Furthermore, we report the characterization of a lettuce LsNRT2.1 gene that is constitutive up-regulated in the transgenic plants. Of note, the expression of the LsNRT2.1 gene in control plants is regulated by NO3− and sugars. Enhanced accumulation of 15N-labeled fertilizer by transgenic lettuce compared with control plants was observed in greenhouse experiments. A negative correlation between the level of root soluble sugars and biomass is consistent with the strong root growth that characterizes these transgenic plants.
The survival of plants depends on the ability of root systems to establish themselves in locations where water and nutrients are available for uptake and translocation (Hawes et al., 2003). As a response to limiting mineral elements, plants have the ability to allocate a greater proportion of their biomass to the root system (Hermans et al., 2006). Evidence overwhelmingly shows that carbon and nitrogen (N) metabolisms are interrelated for the sustained growth and development of plants (Zheng, 2009). Physiological and biochemical studies have shown that when plants are deficient in N, their photosynthetic output is negatively affected. Interestingly, an increase in carbon supply promotes N uptake and assimilation (Coruzzi and Bush, 2001). Of note, the expression of two root high-affinity nitrate (NO3−) transporter genes (AtNRT1.1 and AtNRT2.1) in Arabidopsis (Arabidopsis thaliana) has been shown to be regulated by N status, photosynthesis, or via direct supply of Suc to the roots (Lejay et al., 1999). Active transport across the plasma membrane (PM) of root epidermal and cortical cells is the first step in NO3− acquisition. This transport is coupled to a H+ electrochemical gradient generated by the PM H+-ATPases (Miller and Smith, 1996; Forde, 2000).
NO3− is the major form of plant-available N in agricultural soils. Application of large quantities of N fertilizers is the norm for the production of high-yielding domesticated crops. This practice has intrinsic energetic and ecological costs (Bi et al., 2007), since single-year crop recoveries average below 50% (Mosier et al., 2004). Lettuce (Lactuca sativa) is a crop that receives N rates typically exceeding 150 kg N ha−1 (Kerns et al., 1999), and crop N recoveries are often low depending on climate, soil, and management conditions (Jackson et al., 1994; Sanchez, 2000; Karam et al., 2002). N not taken up by the crops is often lost to the environment, potentially having adverse ecological impacts (Mosier et al., 2004). Approximately 76 MJ (Nagy, 1999) in the form of natural gas is required to produce 1 kg of N fertilizer. It has been estimated that energy associated with fertilizer and water can exceed 50% of the total utilized in irrigated crop production (Alexandrou et al., 2009). A number of studies have sought to improve the N use efficiency of vegetable crops through fertilizer management schemes (Sanchez and Doerge, 1999), but far fewer have attempted modification of the plant to better exploit soil N (Jackson, 1995; Johnson et al., 2000).
The partitioning of photoassimilates between their sites of production (source tissues) and their sites of utilization in harvestable regions is a major determinant of crop yield (Giaquinta, 1983). The potential of regulating the translocation of assimilates promises substantial opportunities for yield increases in crops. A recent study provided immunohistochemical and immunogold-labeling data consistent with a PM localization of the type I proton translocating pyrophosphatase (H+-PPase) Arabidopsis Vacuolar Pyrophosphatase1 (AVP1) in sieve element and companion cell complexes of Arabidopsis source leaves (Paez-Valencia et al., 2011). Furthermore, the binding of the phloem-specific AtVOZ1 transcription factor to the cis-acting region of the AVP1 promoter (Mitsuda et al., 2004), together with the conspicuously localized expression of the AVP1p::GUS reporter under control or phosphate limitations in Arabidopsis, are consistent with a relevant role for the H+-PPase in Arabidopsis vascular tissue (Yang et al., 2007). Up-regulation of either the Arabidopsis or Thellungiella halophila type I H+-PPases triggers enhanced growth/biomass and photosynthetic capacity in a variety of agriculturally important crops (Gaxiola et al., 2001; Li et al., 2005, 2008, 2010; Park et al., 2005; Yang et al., 2007; Bao et al., 2008; Lv et al., 2008, 2009; Pasapula et al., 2011) grown under normal or stressful conditions, such as water scarcity, salinity, and nutrient limitation. Interestingly, it was recently shown that the metabolite profile in Arabidopsis plants engineered with an AVP1 overexpression cassette (AtAVP1-OX) is shifted toward N metabolism and changes in carbon signaling (Gonzalez et al., 2010). Furthermore, changes in the expression of AVP1 correlate with apoplastic pH alterations and rhizosphere acidification (Li et al., 2005; Yang et al., 2007). A recent report on the proton-pumping activity of the PM H+-ATPase from roots of AtAVP1-OX plants grown under nonlimiting nutrient conditions showed that the initial velocity and steady state of the proton gradients generated by the PM H+-ATPase are significantly greater (50%–70%; α = 0.05) than in the wild type (Undurraga et al., 2012). Rhizosphere acidification is a central mechanism for plant mineral nutrition, since it contributes to nutrient solubility and the PM proton motive force (Palmgren, 2001; Marschner, 2002). Accordingly, it has been shown that AVP1 transgenic Arabidopsis, tomato (Solanum lycopersicum), and rice (Oryza sativa) plants outperform controls when grown under phosphate limitation and accumulate higher contents of potassium under all conditions tested (Yang et al., 2007). Interestingly, immunohistochemical data on AVP1-overexpressing transgenic Arabidopsis (Gaxiola et al., 2001) and cotton (Gossypium hirsutum) plants (Pasapula et al., 2011) are consistent with an enhanced H+-PPase abundance at the vasculature of leaves.
In this study, we evaluate the potential for improved NO3− use efficiency in transgenic romaine lettuce ‘Conquistador’ plants with enhanced H+-PPase abundance and activity. In addition, a complementary DNA (cDNA) clone encoding a putative high-affinity NO3− transporter, called LsNRT2.1, was characterized with regard to its tissue expression patterns and responses to NO3− and sugars in control and transgenic lettuce plants. N status in aboveground tissues, and the levels of soluble sugars and total carbon and N in roots, were also determined in control and transgenic plants.
RESULTS
Molecular and Biochemical Characterization of Lettuce Plants Engineered with a 35S:AVP1D Expression Cassette
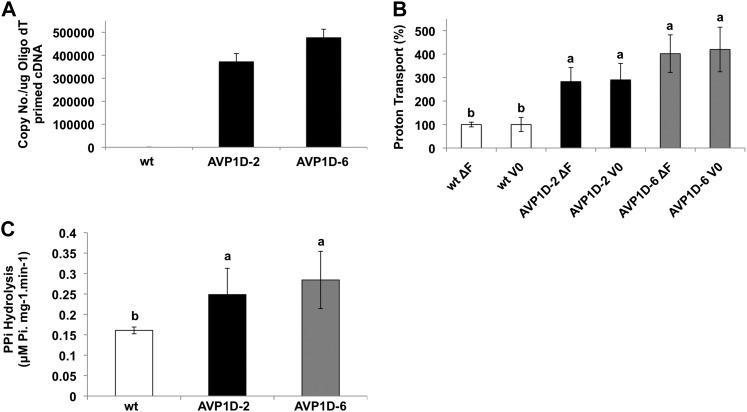
Lettuce ‘Conquistador’ was engineered with a 35S:AVP1D expression cassette. The AVP1D gene is an intragenic E229D gain-of-function allele of the Arabidopsis type I H+-PPase AVP1 gene that has been shown to have an in vitro-coordinated increase of both inorganic pyrophosphate (PPi) hydrolytic activity and PPi-dependent H+-translocation (Zhen et al., 1997). Interestingly, tomato plants engineered with this 35S:AVP1D expression cassette have been shown to develop robust root systems that enable them to outperform controls under water deprivation (Park et al., 2005) and limiting phosphate nutrition (Yang et al., 2007). Here, we present the characterization of two T4 independent transgenic lettuce lines, AVP1D-2 and AVP1D-6. AVP1D-specific primers (Supplemental Table S2) were used to determine the expression levels of the transgene in the AVP1D-2 and AVP1D-6 lines. The expression of AVP1D ranges from about 38,000 to 48,000 copies µg−1 cDNA for the AVP1D-2 and AVP1D-6 lines, respectively (Fig. 1A). Immunohistochemical analysis was used to monitor the expression patterns of the H+-PPase in the main rib of source leaves (L1) of control and transgenic lines. An H+-PPase immunospecific signal was detected mainly in companion cells and sieve elements of control and AVP1D transgenic lettuce. It should be noted that staining for both transgenic lines appeared significantly stronger compared with control leaf sections (Fig. 2). The antibody used was raised against a peptide corresponding to the PPi-binding site of the Arabidopsis H+-PPase that is conserved among all of the reported plant H+-PPases (Kim et al., 1994; Park et al., 2005). Biochemical experiments were set to examine whether the enhanced H+-PPase mRNA and protein levels documented in these AVP1D transgenic lettuce plants had an effect on the pump activities. PPi-dependent proton-pumping activities from root microsomal fractions of control and transgenic lettuce plants showed that both the initial velocity and the steady state were 2- and 3-fold enhanced in the AVP1D-2 and AVP1D-6 lines compared with controls, respectively (Fig. 1B). Furthermore, PPi hydrolytic activities were about 2-fold higher in AVP1D-2 and AVP1D-6 than in controls (Fig. 1C).
Figure 1.
Molecular and biochemical characterization of AVP1D expression in transgenic lettuce. A, Quantification of AVP1 transcript levels in leaves of wild-type cv Conquistador (wt) and transgenic (AVP1D-2 and AVP1D-6) lettuce plants was performed by real-time PCR. Transcript levels are expressed as the number of copies μg−1 cDNA (see “Materials and Methods”). The data shown are means from triplicates per run of three independent mRNA extractions ± sd. B, H+-PPase proton-pumping activity. Data of initial velocity (V0) and steady state (ΔF) of H+-PPase-dependent H+ gradients across tonoplast vesicles from root microsomal fractions of control (wt) and transgenic lettuce (as indicated) were monitored by fluorescence quenching of the pH-sensitive dye 9-amino-6-chloro-2-methoxyacridine (see “Materials and Methods”). Tukey’s HSD test α = 0.05. C, H+-PPase hydrolytic activities from the same root microsomal fractions monitored as phosphate release (see “Materials and Methods”). The PPi hydrolysis was determined at pH 7 with or without K+. Tukey’s HSD test α = 0.05.
Figure 2.
Immunohistochemical localization of H+-PPase protein in source leaves of cv Conquistador and AVP1D-expressing lettuce plants. Representative micrographs are shown for cross sections of source leaves from cv Conquistador (A), AVP1D-2 (B), and AVP1D-6 (C) lettuce stained with Fast Green. A′, B′, and C′ show cross sections from the same leaves as above incubated with an antiserum raised against the H+-PPase (see “Materials and Methods”). The signal (dark brown precipitate) is mainly detected in companion cells (CC) and in sieve elements (SE) of control and AVP1D transgenic lettuce. X, Xylem. [See online article for color version of this figure.]
Higher H+-PPase Activity in Transgenic Lettuce Increases Plant Biomass and Root Acidification Capacity
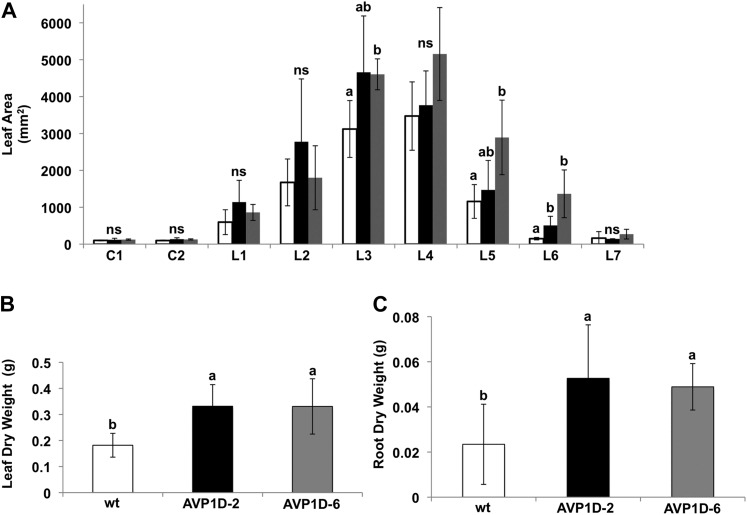
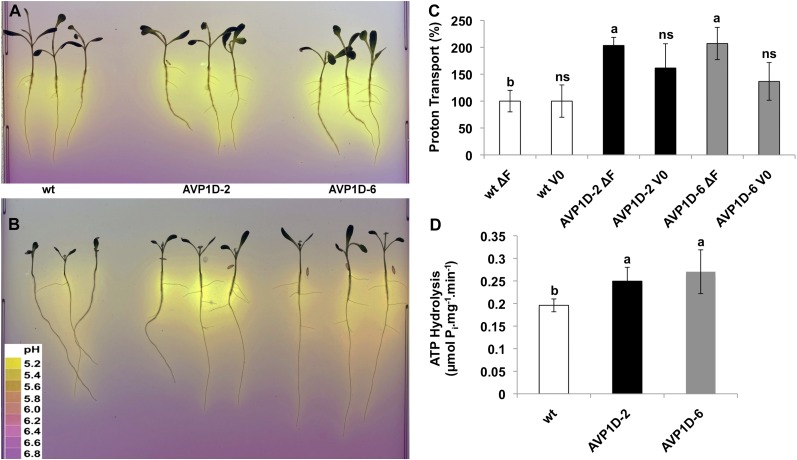
It has been shown that overexpression of type I H+-PPases under the control of either the 35S or Ubiquitin promoter increases root and shoot proliferation (Li et al., 2005, 2008, 2010; Bao et al., 2008; Lv et al., 2008, 2009). Here, we monitored shoot and root parameters of control and AVP1D transgenic lettuce plants grown in a complete hydroponic nutrient solution (Fig. 3). Area data of most leaves and cotyledons did not show statistically significant differences among control and AVP1D transgenic lines (Fig. 3A). However, both leaf and root dry weight data showed a tendency toward higher weights in both AVP1D transgenic lettuces compared with the controls (Fig. 3, B and C). To further characterize these AVP1D transgenic plants, we tested their root acidification capacity. Seeds of control, AVP1D-2, and AVP1D-6 plants were germinated in solid medium supplemented with complete nutrient solution and a pH indicator dye. As shown in Figure 4A, the acidification zone generated by both AVP1D-2 and AVP1D-6 seedlings appears more intense compared with the controls. In order to have quantitative data of this enhanced acidification capacity displayed by the AVP1D transgenic lettuce plants, we evaluated the PM H+-ATPase proton-pumping (Fig. 4C) and hydrolytic (Fig. 4D) activities from root microsomal fractions. Interestingly, there is no statistically significant difference in the initial velocities of control and AVP1D lettuce plants; however, their steady-state transport values are about 2-fold higher than in controls (Fig. 4C). Furthermore, the ATP hydrolytic capacities of AVP1D-2 and AVP1D-6 are 25% higher compared with the controls (Fig. 4D).
Figure 3.
Growth parameters of cv Conquistador and AVP1D-expressing lettuce lines grown in normal medium. A, cv Conquistador and AVP1D transgenic lettuce were grown under hydroponic control conditions for 14 d, and their leaf areas were determined as described in “Materials and Methods.” The average areas of cotyledons (C1 and C2) and leaves (L1–L7) of cv Conquistador (white bars), AVP1D-2 (black bars), and AVP1D-6 (gray bars) were determined (n = 9 per line). B and C, Average dry weight data of leaves (B) and roots (C) of the same plants described in A. Tukey’s HSD test α = 0.05; ns = not significantly different; wt, wild type.
Figure 4.
Root acidification and PM H+-ATPase activity of cv Conquistador and AVP1D-expressing lettuce. A and B, Seedlings of wild-type cv Conquistador (wt) and AVP1D-2 and AVP1D-6 transgenic lettuce were germinated in the presence of normal (A) or low (B; 0.5 mm) NO3−-containing solid medium with the pH indicator bromocresol purple. The pH change was visualized via changes in medium color. The inset in B shows a color bar generated by documenting the color change of bromocresol purple in the same medium at specific pH values. The photographs show plants grown for 20 d (A) and 25 d (B). C, PM H+-ATPase proton-pumping activity. Data of initial velocity (V0) and steady state (ΔF) of H+-ATPase-dependent H+ gradients across PM vesicles from root microsomal fractions of control (wt) and transgenic lettuce (as indicated) were monitored by fluorescence quenching of the pH-sensitive dye 9-amino-6-chloro-2-methoxyacridine (see “Materials and Methods”). D, PM H+-ATPase hydrolytic activities from the same root microsomal fractions monitored as phosphate release (see “Materials and Methods”). The vanadate-sensitive ATP hydrolysis was measured at pH 6.5. Tukey’s HSD test α = 0.05; ns = not significantly different. [See online article for color version of this figure.]
AVP1D Transgenic Lettuce Plants Outperform Controls When Grown under Limiting NO3− Conditions
Overexpression of type I H+-PPases has been associated with increased photosynthesis rates and root development (Lv et al., 2008). The enhanced root acidification capacity and biomass displayed by AVP1D transgenic lettuce plants (Figs. 3 and 4) are indicative of the augmented availability of photosynthates. Coordinating the activity of root ion-uptake systems with photosynthesis in shoots has been proposed to be the way in which plants balance their nutrient homeostasis (Lejay et al., 2003). NO3− uptake has been correlated with the availability of photosynthates in roots (Delhon et al., 1996). Furthermore, root acidification that depends on photosynthate supply is a universal strategy for nutrient uptake in plants (Marschner, 2002). In order to test if the robust and active root systems of the AVP1D-2 and AVP1D-6 plants confer a growth advantage under limiting NO3− conditions, we germinated seeds in low-NO3− (0.5 mm) solid medium supplemented with a pH indicator dye. Interestingly, the seedlings of both transgenic lines displayed an enhanced acidification capacity compared with controls (Fig. 4B). Furthermore, visual inspection of the seedlings showed that both shoots and roots of the AVP1D lines are larger and that lateral root development is more robust compared with the controls (Fig. 4B).
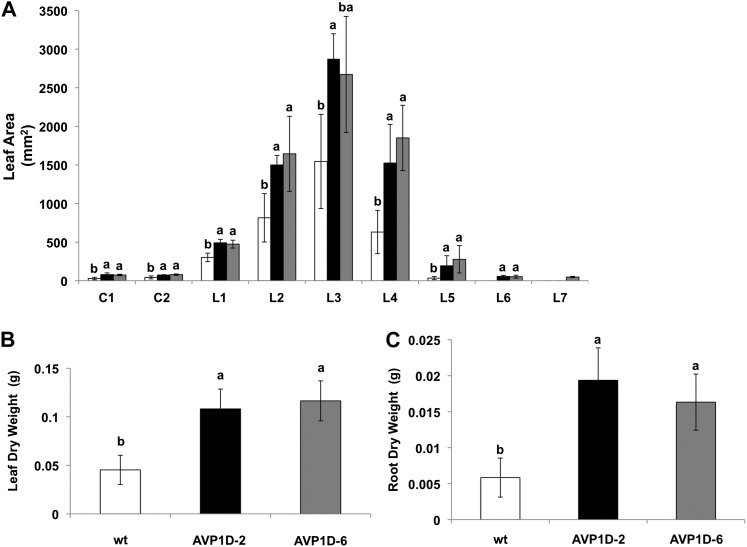
To further document this H+-PPase-triggered growth advantage under limiting NO3−, we monitored shoot and root parameters from control, AVP1D-2, and AVP1D-6 plants grown in a limiting NO3− (0.5 mm) hydroponic nutrient solution (Fig. 5). Cotyledon and leaf data of both AVP1D-2 and AVP1D-6 plants showed a statistically significant increase in area compared with controls (Fig. 5A). Consistently, leaf and root dry weight data showed that AVP1D-expressing lettuce lines developed at least 2-fold more leaf biomass and about 3-fold more root biomass than controls under limiting NO3− (Fig. 5, B and C).
Figure 5.
Growth parameters of cv Conquistador and AVP1D-expressing lettuce grown in low-NO3− medium. A, Wild-type cv Conquistador (wt) and AVP1D transgenic lettuce were grown hydroponically under low-NO3− (0.5 mm) conditions for 20 d, and their leaf areas were determined as described in “Materials and Methods.” Average areas of cotyledons (C1 and C2) and leaves (L1–L7) of cv Conquistador (white bars), AVP1D-2 (black bars), and AVP1D-6 (gray bars) were determined (n = 9 per line). B and C, Average dry weight data of leaves (B) and roots (C) of the same plants described in A. Tukey’s HSD test α = 0.05.
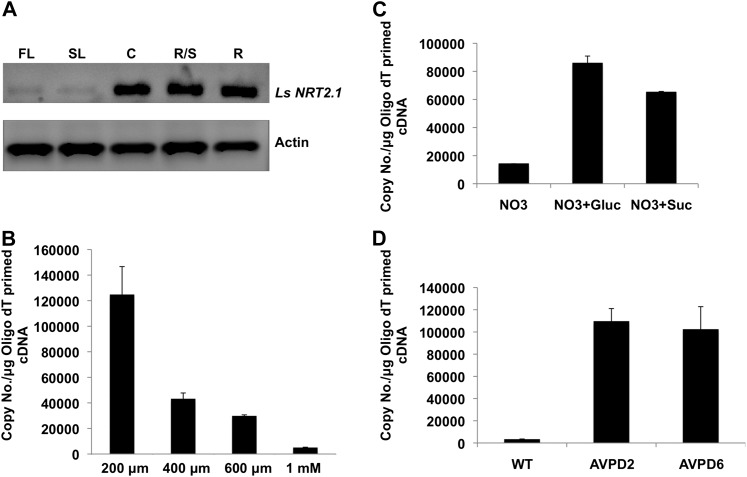
Expression Analysis of LsNRT2.1 in Response to NO3− Concentration and Sugars
As shown above (Fig. 5), overexpression of the AVP1D gene in lettuce plants enhances shoot, and more significantly root, development when plants are grown under limiting NO3− conditions as compared with controls. NO3− deficiency has been shown to affect photosynthesis and the partitioning of reduced carbon between source and sink tissues (Hermans et al., 2006). Interestingly, the regulation of the high-affinity NO3− transporter AtNRT2.1 in Arabidopsis roots is coordinated with photosynthesis and carbon allocation (Lejay et al., 2003). In order to determine if the enhanced carbon allocation hinted by the robust root systems and the acidification capacity of the AVP1D expression lines correlates with an increased expression of a lettuce ortholog of AtNRT2.1, we examined the levels of expression of the lettuce NRT2.1 NO3− transporter gene (LsNRT2.1; Supplemental Figures S1 and S2S2; Supplemental Tables S1 and S2; Supplemental Text S1) in leaves, cotyledons, root-shoot junctions, and roots using semiquantitative reverse transcription (RT)-PCR. Expression was detected in cotyledons, root-shoot junctions, and roots of control lettuce seedlings that were N starved for 5 d and then incubated in the presence of 1 mm NO3−-containing solution (Fig. 6A). In order to further characterize this LsNRT2.1 gene, we monitored its expression in roots of control seedlings in response to different concentrations of NO3− in the growth medium. The peak of expression was detected when NO3−-starved seedlings were transferred for 6 h to a 200 µm NO3−-containing growth medium (Fig. 6B). This expression diminished about 60% when the NO3− concentration was 400 µm, reaching the lowest expression at 1 mm NO3− (Fig. 6B). To study the response of LsNRT2.1 expression to sugars, control seedlings were grown in 1 mm NO3−-containing medium and treated with Glc and Suc (100 µm). After 6 h of root exposure to Glc or Suc, the expression levels of LsNRT2.1 were measured by real-time PCR. Exogenous Glc and Suc were able to up-regulate the basal expression of the LsNRT2.1 gene about 5- and 3.6-fold, respectively (Fig. 6C). The more robust root systems and enhanced acidification capacity displayed by the AVP1D lettuce plants exposed to limiting NO3− conditions (Figs. 4 and 5) suggest that these root systems should have higher availability of photosynthates. We tested the behavior of LsNRT2.1 expression in roots of control, AVP1D-2, and AVP1D-6 plants grown in the presence of 1 mm NO3− medium. The expression of LsNRT2.1 in AVP1D-2 and AVP1D-6 was 7-fold higher compared with control plants (Fig. 6D).
Figure 6.
Characterization of a lettuce NRT2.1 gene (LsNRT2.1). A, Ethidium bromide staining of semiquantitative RT-PCR expression analysis of LsNRT2.1 in first and second leaves (FL and SL), cotyledons (C), root-shoot junction (R/S), and roots (R) of control plants N starved for 5 d and then incubated in the presence of 1 mm NO3−-containing solution. LsNRT2.1 expression levels in different tissues are relative to actin. B, Analysis of LsNRT2.1 expression in roots of control seedlings starved for NO3− and incubated for 6 h under different NO3− concentrations as indicated (for details, see “Materials and Methods”). C, Response of LsNRT2.1 expression in roots of control plants incubated in the presence of 1 mm KNO3, 100 mm Glc + 1 mm KNO3, or 100 mm Suc + 1 mm KNO3 as indicated (for details, see “Materials and Methods”). D, Expression levels of LsNRT2.1 in roots of control (wild type [WT]), AVP1D-2, and AVP1D-6 plants grown in the presence of 1 mm NO3−. Quantification was performed by real-time PCR, and the transcript levels in B, C, and D are expressed as the number of copies μg−1 cDNA (see “Materials and Methods”). All real-time PCR experiments were done in triplicate from three independent RNA extractions.
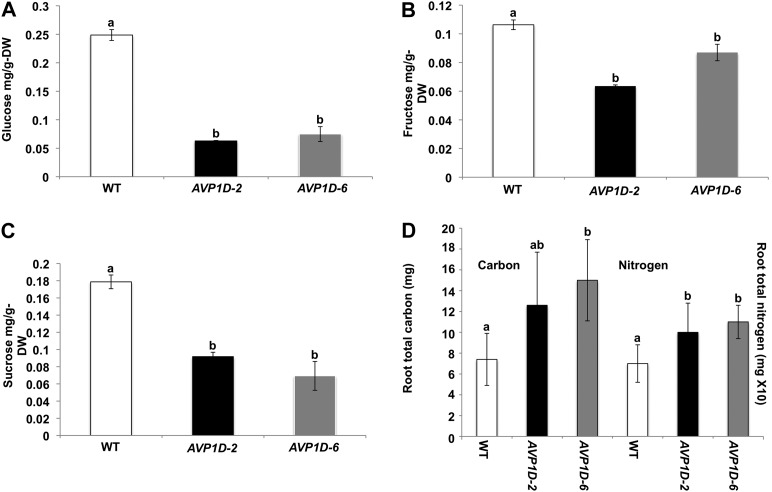
Evaluation of Soluble Sugar Levels and Total Carbon and N in Roots of AVP1D Transgenic Lettuce and Controls
We measured the levels of Glc, Fru, and Suc in roots to test if carbon allocation was enhanced in the AVP1D-2 and AVP1D-6 lines compared with the controls (Fig. 7, A–C). Plants were grown in the presence of 1 mm NO3− medium. The levels of all these soluble sugars were reduced in the AVP1D-2 and AVP1D-6 lines. The levels of Glc and Suc were about 5- and 2-fold lower than in controls in the AVP1D-2 and AVP1D-6 lines, respectively (Fig. 7, A and C). By comparison, the level of Fru was only slightly reduced in the roots of the transgenic lettuce (Fig. 7B). However, root total carbon in the AVP1D-2 and AVP1D-6 lines was about 2-fold higher than in controls (Fig. 7D). These data show a distinct negative correlation between soluble sugars and biomass. Interestingly, root total N from the aforementioned plants was about 50% higher than in controls (Fig. 7D).
Figure 7.
Quantification of root soluble sugars and total carbon. A to C, Roots of wild-type cv Conquistador (WT) and AVP1D-2 and AVP1D-6 transgenic lettuce plants were collected at midday and treated as described (see “Materials and Methods”). Glc (A), Fru (B), and Suc (C) contents (mg g−1 dry weight [DW]) were determined. Results are averages of three replicates ± se. D, Total carbon per root was determined as described (see “Materials and Methods”). Results are averages of five plants per line grown under identical conditions as the plants used for A to C. Tukey’s HSD test α = 0.05.
Greenhouse and Field Experiments
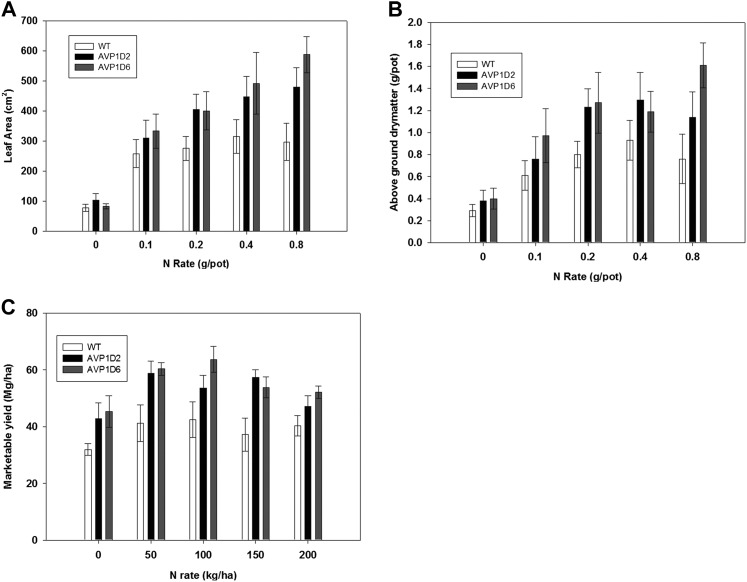
All the above experiments were performed in artificial medium; in order to explore the behavior of these AVP1D transgenic lettuce plants under real soil conditions, we performed greenhouse and field experiments. Lettuce leaf area and aboveground dry matter were significantly (P < 0.01) increased by N rate (Fig. 8, A and B). Furthermore, cultivar effects were also statistically significant, where AVP1D-6 outperformed AVP1D-2, which in turn outperformed the control plants.
Figure 8.
Greenhouse and field experiments. A and B, Leaf area (A) and aboveground dry matter (B) responses to N rate and cultivar in a greenhouse experiment. Responses showed a significant (P > 0.01) linear and quadratic response to N and significant differences among cultivars (as indicated). C, Marketable yield of lettuce to N rate and cultivar. Significant (P < 0.05) quadratic response to N and significant (P < 0.01) differences among cultivars were seen, where both AVP1D-2 and AVP1D-6 produced greater marketable yield compared with unmodified wild-type cv Conquistador (WT).
Leaching of N in this greenhouse experiment was high due to the frequent watering required to maintain plant vigor in the warm greenhouse. Aboveground N accumulation from soil and fertilizer N (the latter measured by 15N) increased with N rate and was statistically (P < 0.05) higher in AVP1D transgenic lettuce plants compared with the controls. On average, over 90% of the N accumulated in the plant was derived from the applied 15N-labeled fertilizer, and the engineered cultivars recovered more fertilizer N (from 10% to 50% more) than the unmodified controls across all N rates (Table I).
Table I. Total aboveground N and fertilizer N to N rate and cultivar in the greenhouse.
Significant linear (L) and quadratic (Q) responses to N rate at P < 0.01 were seen. The cultivar effect was significant at P < 0.05. Fertilizer N was calculated from the 15N label (see “Materials and Methods”). **P < 0.01.
| N Rate | Total Aboveground N
|
Total Aboveground Fertilizer N |
||||
|---|---|---|---|---|---|---|
| Conquistador | AVP1D-2 | AVP1D-6 | Conquistador | AVP1D-2 | AVP1D-6 | |
| mg per pot | ||||||
| 0 g per pot | 3.7 | 5.4 | 5.4 | – | – | – |
| 0.1 g per pot | 22.5 | 25.1 | 28.1 | 19.7 | 22.0 | 24.5 |
| 0.2 g per pot | 33.1 | 44.7 | 49.2 | 30.9 | 40.8 | 44.9 |
| 0.4 g per pot | 47.5 | 59.7 | 51.7 | 41.4 | 55.6 | 48.4 |
| 0.8 g per pot | 33.5 | 46.1 | 67.7 | 31.4 | 43.3 | 63.8 |
| Statistics | L**Q** | L**Q** | L**Q** | L**Q** | L**Q** | L**Q** |
Romaine lettuce showed a significant (P < 0.05) quadratic response, where marketable yields were essentially maximized to the first N rate (50 kg N ha−1) and N rates beyond 100 kg N ha−1 reduced yields (Fig. 8C). Cultivar response was highly significant (P < 0.01), where both AVP1D-2 and AVP1D-6 produced more marketable yields across all N rates compared with control plants. The AVP1D6 cultivar produced 40% more marketable product per unit of N at the optimal N rate of 50 kg N ha−1 than the controls.
DISCUSSION
In this study, two transgenic lettuce lines (AVPD1-2 and AVPD1-6) with enhanced H+-PPase abundance and activity (Figs. 1 and 2) were used to evaluate the potential of this genetic manipulation for improving NO3− uptake efficiency in lettuce. The experiments described in this work demonstrate that this technology was instrumental in improving lettuce N use efficiency under control and limiting NO3− regimens in laboratory, greenhouse, and field scenarios (Figs. 3, 5, and 8; Table I). In all instances, more production (including marketable yield) was obtained per unit of N input for AVP1D-engineered lettuce versus controls.
The type I H+-PPase from Arabidopsis (AVP1) has been identified as a yield-enhancing gene (Gonzalez et al., 2009, 2010). There is a large body of evidence showing that overexpression of type I H+-PPases under the control of either the 35S or Ubiquitin promoter in different crops increases root and shoot proliferation (Li et al., 2005, 2008, 2010; Park et al., 2005; Yang et al., 2007; Bao et al., 2008; Lv et al., 2008, 2009; Pasapula et al., 2011). However, only the work by Yang and collaborators has shown that these H+-PPase-enhanced root systems represent an advantage when plants are challenged with limiting phosphate nutrition (Yang et al., 2007; Gaxiola et al., 2011). Furthermore, it was in this work that an enhanced root acidification capacity was correlated with the up-regulation of the H+-PPase (Yang et al., 2007). This enhanced rhizosphere acidification capacity was shown to be sensitive to vanadate, suggesting that up-regulation of the H+-PPase has a positive effect on the activity of the PM H+-ATPase (Yang et al., 2007). A recent report on the proton-pumping activity of the PM H+-ATPase from roots of transgenic Arabidopsis plants engineered with a 35S:AVP1 expression cassette showed that the initial velocity and steady state of the proton gradients generated by the PM H+-ATPase are significantly greater (50%–70%; α = 0.05) than in the wild type (Undurraga et al., 2012). Rhizosphere acidification is a central mechanism for plant mineral nutrition, since it contributes to nutrient solubility and the PM proton motive force (Palmgren, 1998; Marschner, 2002). AVP1D-2 and AVP1D-6 transgenic lettuce plants display an increased rhizosphere acidification capacity under both normal and limiting NO3− conditions (Fig. 4). Furthermore, direct evidence shows that the augmented root acidification capacity displayed by AVP1D lettuce plants correlates with enhanced PM H+-ATPase pumping and hydrolytic activities (Fig. 4). The enhanced biomass (shoot and root) and rhizosphere acidification capacities displayed by the AVP1D-engineered lettuce under normal or limiting NO3− are indicative of the augmented availability of photosynthates in these plants. Suc produced by photosynthesis is the cornerstone of higher plant metabolism. It is the main substrate for respiration and biosynthesis. NO3− uptake has been correlated with the availability of photosynthates in roots (Delhon et al., 1996). Carbon and N metabolisms are interrelated for the sustained growth and development of plants (Zheng, 2009). For example, amino acid biosynthesis requires the coordination of N and carbon metabolism, since the N assimilation demands carbon skeletons in the form of 2-oxoglutarate, ATP, and reductant (Lancien et al., 2000). Interestingly, it has been shown that the metabolite profile in AVP1-overexpressing transgenic Arabidopsis is shifted toward N metabolism and changes in carbon signaling (Gonzalez et al., 2010).
Plant NO3− acquisition depends on the combined activities of root high- and low-affinity NO3− transporters and the proton gradient generated by the PM H+-ATPase (Forde, 2000). Plant NRT2 genes studied so far are expressed preferentially in roots (Miller et al., 2007). In Arabidopsis, the regulation of AtNRT2.1 transcript levels in roots by light, Suc supply to the roots, and N limitation has been extensively documented (Lejay et al., 1999). The response to NO3− limitation (Fig. 6B) as well as Suc and Glc treatments of the LsNRT2.1 gene reported here (Fig. 6C) is consistent with the behavior of the Arabidopsis and rice NRT2.1 transporter genes (Feng et al., 2011). Of note, it has been shown that Suc-mediated diurnal regulation is common to a wide range of nutrient-uptake genes in Arabidopsis roots, including ammonium transporters and high-affinity sulfate, phosphate, and potassium transporters (Lejay et al., 2003). These results are consistent with an integration of photosynthesis in the shoots with nutrient uptake in the roots. The constitutive up-regulation of the LsNRT2.1 gene in both AVP1D-2 and AVP1D-6 transgenic lettuce (Fig. 6D), and the conspicuous increase in H+-PPase abundance at the vasculature of leaves shown here (Fig. 2) and elsewhere (Gaxiola et al., 2001; Pasapula et al., 2011), further suggest that the enhanced expression of the H+-PPase could favor the transport of photosynthates from source to sink tissues, as hypothesized previously (Paez-Valencia et al., 2011; Gaxiola et al., 2012). In keeping with this, the enhanced root biomass (Figs. 3 and 5) is consistent with an augmented transport of reduced carbon to the roots of these transgenic lettuce plants. Furthermore, an increased root acidification capacity (Fig. 4) depends on an ATP supply generated by the respiration of reduced carbon molecules. The data presented here showing a reduction in the level of soluble sugars (Fig. 7, A–C) and an increase in total carbon (Fig. 7D) in the roots of the AVP1D-2 and AVP1D-6 lines suggest that the reduction of the pool sizes of these soluble sugars is due to a high metabolic activity. A similar negative correlation between the level of soluble sugars and strong growth has been described previously in Arabidopsis (Meyer et al., 2007). Of note, total root N also increased in both transgenic lettuce lines, but the carbon-N ratios were similar to controls (Fig. 7D). The data from the greenhouse study show greater aboveground N accumulation from total and fertilizer N for the AVP1D-2 and AVP1D-6 lines compared with controls in all N rates tested (Table I). These data are consistent with the enhanced root acidification capacity (Fig. 4) and the observed up-regulation of the LsNRT2.1 gene in both AVP1D-2 and AVP1D-6 transgenic lettuce compared with controls (Fig. 6D). Furthermore, the enhanced biomass and marketable yield of the AVP1D transgenic lettuce under control and limiting N suggest that this genetic modification results in an integral N and carbon metabolism.
In summary, this simple genetic manipulation can trigger agriculturally relevant phenotypes that will likely allow more efficient utilization of N and facilitate high production at a fraction of the economic and environmental cost.
MATERIALS AND METHODS
Plant Material, Transformation, and Growth Conditions
Lettuce (Lactuca sativa ‘Conquistador’) transformation was performed by means of the Agrobacterium tumefaciens-mediated transformation method described elsewhere (Michelmore et al., 1987). A. tumefaciens strain GV3101 carrying the pGR395 (CaMV35S::AVP1D) vector was used for this study. The pRG395 plasmid was generated by cloning the AVP1D gene downstream of a tandem repeat of the 35S promoter of Cauliflower mosaic virus as described (Zhen et al., 1997; Gaxiola et al., 2001). T1 seeds were screened in 100 mg L−1 kanamycin-containing medium, and 15 positive lines were selected and then transferred to soil. Segregation analysis of T2 seeds from self-pollinated T1 plants was carried out on kanamycin selection medium, and homozygous T2 AVP1D lines were selected. From these 15 lines, two T4 homozygous lines (AVP1D-2 and AVP1D-6) were chosen for use in all the experiments reported in this study. For a typical experiment, lettuce seeds (the wild type and transgenic lines AVP1D-2 and AVP1D-6) were germinated in soil (SunGro Redi-Earth) in chambers at 21°C with 70% relative humidity and a 16-h-light photoperiod (150 μmol m−2 s−1). After germination, seedlings were carefully removed and transferred to 11-L tanks containing modified Hoagland nutrient solution [1.5 mm Ca(SO4), 0.75 mm MgSO4, 0.50 mm KH2PO4, 72 µm Fe-EDTA, and 1 mL L−1 1,000× trace element solution]. The only source of N was KNO3; two concentrations of KNO3 were used for most of the experiments, 6.25 mm (sufficient) and 0.5 mm (low). Potassium chloride (KCl) was used to keep the potassium concentration at 1 mm.
Greenhouse Experiment
Surface soil mapped as Casa Grande (fine-loamy, mixed, hyperthermic, Typic Natriargid [reclaimed]) was collected at the Maricopa Agricultural Center, sieved, and 1.6 kg was weighed into 15-cm-diameter pots. The preplant NO3−-N test for this soil was 50 mg kg−1. All pots received 0.34 g of phosphorus as monocalcium phosphate so that phosphorus would not be limiting. The treatments included cv Conquistador, AVP1D-2, and AVP1D-6 romaine lettuce and N rates. The total seasonal rates of N were 0, 0.1, 0.2, 0.4, and 0.8 g per pot. The N was applied as a potassium nitrate solution in eight split applications to achieve these seasonal total rates. The potassium nitrate solution was labeled with 10 atom % 15N. The experimental design was a randomized complete block with four replications.
Lettuce seedlings (one-leaf stage) were transplanted into the prewatered pots on September 15, 2011. The first N fertilization occurred on September 20 and continued twice weekly through harvest. These plants were grown to the eight-leaf stage (October 14) and harvested by cutting the aboveground plant at the soil surface. Total leaf area was measured using a LiChor area meter (LI 3100 C), and fresh and dry weights were determined as described above. Total N and 15N percentage were determined by combustion mass spectroscopy at the Colorado Plateau Stable Isotope Laboratory at Northern Arizona University. N derived from fertilizer and N fertilizer recovery were determined using methods described elsewhere (Sanchez and Blackmer, 1988). Statistical analysis was performed using SAS (SAS Institute), where responses to N were evaluated by trend analysis and differences among cultivars were evaluated by lsd.
Field Experiment
This study was performed in the field where we collected soil for the greenhouse experiment. The entire plot area received 125 kg phosphorus ha−1 as monoammonium phosphate, which is the common practice for low-phosphorus testing soils. Thus, the entire plot area also received 54 kg N ha−1 with the preplant phosphate fertilizer. The treatment design was a split plot with cultivar subplots (cv Conquistador, AVP1D-2, and AVP1D-6) within N main plots. The N rate treatments of 0, 50, 100, 150, and 200 kg N ha−1 (not including that applied with the preplant phosphate fertilizer) were applied preplant as a polymer-coated urea controlled-release N product (ESN, distributed by Agrium Advanced Technologies). All N and phosphorus fertilizers were roto-mulched into the beds. Individual main plots were 25 m2, and the experimental design was a randomized complete block with four replications.
Lettuce cultivars were seeded in elevated double-row beds on 1-m centers with a hand planter (Jang JP1 Clean Seeder) and thinned by hoe at the four-leaf stage to approximately 71,000 plants ha−1. The planting date was November 14, 2011. The stands were established by sprinkler irrigation. After establishment, all required irrigations were applied by level (no-slope) furrows.
Lettuce was harvested at maturity (March 16, 2012) by cutting and weighing all plants from 3 m of double-row beds. Marketable yield was determined after grading using standard practices. Statistical analysis was performed using SAS (SAS Institute), where responses to N were evaluated by trend analysis and differences among cultivars were evaluated by lsd.
AVP1D and LsNRT2.1 Quantitative Real-Time PCR
Total cellular RNA was purified with Trizol reagent following the manufacturer’s instructions. RT was carried out using the RETROscript kit (Ambion) according to the manufacturer’s instructions. The resulting cDNAs were used as templates for quantitative RT-PCR. Plasmid controls were constructed for the AVP1 and LsNRT2.1 genes by cloning specific amplicons into pBluescript SK+ and pGEM T-Easy vectors, respectively. Purified plasmid clones were quantified spectrophotometrically, and the copy number was calculated as described previously (Whelan et al., 2003). The copy number and concentration of the plasmid DNA were used to determine the precise number of molecules added to the real-time PCR in order to provide a standard for specific cDNA quantification. Real-time quantitative analyses were performed on the My iQ Single Color Real Time PCR Detection System (Bio-Rad). A standard curve was drawn plotting the natural log of the threshold cycle against the natural log of the number of molecules. The threshold cycle was calculated under default settings for the real-time sequence detection software (Bio-Rad). The linear equation fitted to the data points was used to calculate the precise number of cDNA molecules present per microgram of total oligo(dT) primered cDNA on the same reaction plate as the standard. Primer sequences are listed in Supplemental Table S1. Real-time PCR was performed on optical 96-well plates (Bio-Rad) under universal cycling conditions (10 min at 95°C, 35 cycles of 15 s at 95°C and 60 s at 60°C) followed by the generation of a dissociation curve to check for amplification specificity. The real-time PCRs contained 10 µL of SYBR Green Master Mix (Bio-Rad), 500 nm of specific forward and reverse primers, 1 µL of cDNA, and the appropriate amount of water for a final volume of 20 µL per reaction. Expression levels for each sample were calculated on three technical replicates.
Immunohistochemistry
Lettuce source leaves were fixed in 3.7% formaldehyde, 50% ethanol, and 5% acetic acid for 3 h at room temperature. Fixed material was dehydrated in a graded ethanol series (50%, 60%, 70%, 80%, 90%, and 100%), and absolute ethanol was replaced by histological clearing agent (xylene; Sigma-Aldrich). Tissues were embedded in Paraplast at 60°C. The embedded tissue was sliced into 10-μm sections and placed onto poly-Lys-coated slides. Sections on positively charged slides were deparaffinized in xylene, hydrated in graded alcohol (100%, 90%, 80%, 70%, 60%, 50%, and 30% distilled, deionized water), and pretreated with Antigen Retrieval Solution (BioGenex) under a steamer at 98°C for 10 min. The slides were treated with Power Universal Reagent (BioGenex) to reduce nonspecific binding and incubated with polyclonal anti-H+-PPase antibody (Park et al., 2005) or preimmune serum at a 1:1,000 dilution in phosphate-buffered saline. Three rinses with phosphate-buffered saline-Tween (0.1%, v/v) were followed by incubation with the DAKO EnVision system following the manufacturer’s instructions. After several washes, the immunohistochemical preparations were contrasted with Fast Green (Sigma).
H+-ATPase and H+-PPase H+-Pumping and Hydrolytic Activities
Microsomal fractions containing PM vesicles and tonoplast were isolated from roots by differential centrifugation, as described elsewhere (Giannini and Briskin, 1988) with some modifications. Plant root material was homogenized in buffered medium (pH 8.0) containing 250 mm Suc, 10% glycerol, 100 mm MOPS-1,3-bis(tris[hydroxymethyl]methylamino) propane (BTP), 2 mm EDTA, 2 mm dithiothreitol, 2 mm phenylmethylsulfonyl fluoride, 0.4% polyvinylpyrrolidone-40T, 0.3% bovine serum albumin, and 100 mm KCl. The homogenate was filtered through a membrane Miracloth (Calbiochem) and subjected to subsequent centrifugation to obtain the microsomal fraction. The microsomal fraction was solubilized in a medium containing 15% glycerol, 20 mm Tris-HCl (pH 7.6), 1 mm EGTA, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride.
The H+-ATPase and H+-PPase activities were determined colorimetrically by the release of phosphate during ATP or PPi hydrolysis using the method described elsewhere (Fiske and Subbarow, 1925) with modifications proposed by Façanha and De Méis (1998). The reactions were performed using the reaction medium composed of 20 mm MOPS-BTP (pH 6.5) for ATPase and MOPS-Tris (pH 7.0) for H+-PPase, 100 mm KCl, 5 mm MgSO4, 0.2 mm Na2MoO4, 0.2 mm Na2VO4, and 1 mm ATP or 0.1 mm PPi.
The proton gradient was measured by monitoring the rate of decrease in fluorescence of the metachromatic fluorescent probe 9-amino-6-chloro-2-metoxiacridina using a Fluoroskan Ascent spectrofluorimeter (model FL; Thermo Labsystems). The reaction medium was composed of 10 mm MOPS-BTP (pH 6.5) for ATPase, 10 mm MOPS-BTP (pH 7.0) for H+-PPase, 100 mm KCl, 5 mm MgSO4, 250 mm Suc, 2 mm 9-amino-6-chloro-2-metoxiacridina, and 30 µg of protein, using 1 mm ATP or 0.1 mm PPi as substrate, respectively. From the data, the initial velocity and the variation of maximum fluorescence were determined. The H+ pump specific activities were calculated by measuring H+ transport in the presence or absence of 0.2 mmol L−1 vanadate and 5 nmol L−1 concanamycin, specific inhibitors of P-type ATPase and V-ATPase, respectively. The reaction was triggered by the addition of 1 mmol L−1 ATP or 0.2 mmol L−1 PPi, and 3 mmol L−1 mesoxalonitrile 4-trifluoromethoxyphenylhydrazone was used to dissipate the H+ gradient.
Medium for Root Acidification
Seeds were germinated on plates containing modified Hoagland nutrient solution with normal or low KNO3 (see above), 0.8% agar, 1 mm MES (pH 6.8), and 0.04% (w/v) of the pH indicator bromocresol purple as reported previously (Yang et al., 2007). Two weeks after germination, images were captured using a Nikon 5100 camera.
Leaf Area Determination
Control, AVP1D-2, and AVP1D-6 plants were grown for 15 d in tanks containing modified Hoagland nutrient solution supplemented with normal or low NO3−. Leaves were carefully excised with a scalpel blade and scanned on an HP Scanjet G4010. The leaf areas were determined using the ImageJ 1.4.1 program (http://rsb.info.nih.gov/download.html).
Root and Shoot Fresh and Dry Weight Measurements
Control, AVP1D-2, and AVP1D-6 plants were grown for 15 d in tanks containing modified Hoagland nutrient solution supplemented with low or high N. Roots and shoots were separated with a scalpel blade, and their fresh weights were determined. The excess of water from roots was removed with tissue paper. Then, all plant material was oven dried at 70°C for 72 h in paper bags, and the dry weight was determined.
Relative Quantitative RT-PCR
Relative quantitative RT-PCR was used to determine the expression of LsNRT2.1 in vegetative tissue of wild-type lettuce plants. Total RNAs were isolated from first and second leaves, cotyledon, root-shoot junction, and roots using Trizol reagent according to the manufacturer’s instructions. RT was carried out using the RETROscript kit (Ambion). Gene-specific primers were designed according to the predicted mRNA to obtain the amplified product of 123 bp. Actin forward and reverse primers (Supplemental Table S1) were used as internal controls in RT-PCR under the following cycling conditions: denaturing step at 95°C for 30 s, amplification step for 35 cycles (98°C for 10 s, 58°C for 1 min, and 72°C for 45 s), and a final elongation step at 72°C for 10 min. The PCR products were visualized on ethidium bromide-stained 1% agarose gels.
LsNRT2.1 Gene Expression Experiments
In order to test the expression of the LsNRT2.1 gene in roots of control plants, a group of 20 seedlings were germinated in soil, transferred to control hydroponic medium, and allowed to grow for 3 weeks. Twelve plants of similar size were then transferred to deionized water for 5 d. Finally, groups of three plants were transferred at the beginning of the photoperiod to medium supplemented with different KNO3 concentrations (200 µm, 400 µm, 600 µm, and 1 mm). Roots of each group of plants were collected after 6 h of incubation.
A similar procedure was followed in order to analyze the response of the LsNRT2.1 gene to sugar added to the root solution. Briefly, plants were grown under control conditions for 3 weeks, transferred to deionized water for 5 d, and then transferred to medium containing 100 mm Suc plus 1 mm KNO3, 100 mm Glc plus 1 mm KNO3, or 1 mm KNO3 only.
For all experiments, plants of the same age were cultivated under similar conditions and harvested at the same time at hour 8 of the light period. The plants were grown in a controlled-environment chamber with a 16-h/8-h day/night cycle (150 μmol m−2 s−1) at a temperature of 21°C with 70% relative humidity. Tissues used for gene expression analysis were collected at hour 8 of the illuminated period and immediately frozen in liquid N for subsequent RNA extraction.
Determination of Root Levels of Soluble Sugars and Total Carbon and N Contents
Control, AVP1D-2, and AVP1D-6 plants were grown through the eight-leaf stage in tanks containing modified Hoagland nutrient solution supplemented with 1 mm KNO3. Roots of six plants per line were harvested at midday and rinsed in cold distilled water for 5 s. This material was dried with tissue paper to determine fresh weight (dry weight was determined from a fraction of this material) and frozen in liquid N. The frozen material was extracted once with 80% (v/v) ethanol and twice with 50% (v/v) ethanol. The supernatants were mixed and used for the determination of soluble sugars. Fru, Glc, and Suc were determined by using the Suc/d-Glc/d-Fru assay kit (Roche).
In a follow-up experiment, control, AVP1D-2, and AVP1D-6 plants were grown in a modified Hoagland nutrient solution supplemented with 1 mm KNO3 through the eight-leaf stage. These plants were harvested, separated into shoots and roots, dried at 60°C for 48 h, and ground, and total N and carbon were determined by combustion analysis using the Carlo Erba CN analyzer.
Statistical Analysis
Statistical analysis (unless specified otherwise) was performed with the JMP 6 software package (SAS Institute). In all cases, the “Fit Y by X” command was used to perform one-way analysis to compare the effect of genotype (X factor) and quantitative measurement (Y factor). Multiple comparisons were performed by using, alternatively, Student’s t test and Tukey’s honestly significant difference (HSD) test, which are the most suitable ways to compare numerous means because they correct for multiple comparisons. The significance value (α) was 0.05 for all analyses.
Sequence data from this article can be found in the GenBank data libraries under the following accession numbers: JI578969.1, NP_172288, ABQ57241, AAS93686, AAC09320, AAC49531, CAC35729, ACN22072, CAD89798, NP_001045658, BAD04063, XP_002523687, AAK19519, and AAV73917.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Nucleotide sequence of a putative lettuce high-affinity NO3− transporter, LsNRT2.1.
Supplemental Figure S2. Unrooted, bootstrapped tree of high-affinity NO3− transporters.
Supplemental Table S1. Cloned sequences of known or putative high-affinity NO3− transporters used to generate Supplemental Figure S2.
Supplemental Table S2. Primers used for quantitative RT-PCR analysis.
Supplemental Text S1. Characterization of a NRT2.1 gene from lettuce.
Acknowledgments
We thank R. Michelmore (University of California, Davis) for the sequence of LsNRT2.1.
Glossary
- NO3–
nitrate
- N
nitrogen
- PM
plasma membrane
- PPi
inorganic pyrophosphate
- RT
reverse transcription
- KCl
potassium chloride
- BTP
1,3-bis(tris[hydroxymethyl]methylamino) propane
- HSD
honestly significant difference
- cDNA
complementary DNA
- H+-PPase
proton translocating pyrophosphatase
References
- Alexandrou A, Vyrlas P, Adhikari D, Goorahoo D. (2009) Energy inputs for cantaloupe production in San Joaquin Valley, California. Agric Eng Intern 9: 1–15 [Google Scholar]
- Bao A-K, Wang S-M, Wu G-Q, Xi J-J, Zhang J-L, Wang C-M. (2008) Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci 176: 232–240 [Google Scholar]
- Bi Y-M, Wang R-L, Zhu T, Rothstein SJ. (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics 8: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR. (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol 125: 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L. (1996) Diurnal regulation of NO3− uptake in soybean plants. IV. Dependence on current photosynthesis and sugar availability to the roots. J Exp Bot 47: 893–900 [Google Scholar]
- Façanha AR, De Méis L. (1998) Reversibility of H+-ATPase and H+-pyrophosphatase in tonoplast vesicles from maize coleoptiles and seeds. Plant Physiol 116: 1487–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G. (2011) Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot 62: 2319–2332 [DOI] [PubMed] [Google Scholar]
- Fiske CH, Subbarow Y. (1925) The colorimetric determination of phosphorus. J Biol Chem 66: 375–400 [Google Scholar]
- Forde BG. (2000) Nitrate transporters in plants: structure, function and regulation. Biochim Biophys Acta 1465: 219–235 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Edwards M, Elser JJ. (2011) A transgenic approach to enhance phosphorus use efficiency in crops as part of a comprehensive strategy for sustainable agriculture. Chemosphere 84: 840–845 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR. (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98: 11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Sanchez CA, Paez-Valencia J, Ayre BG, Elser JJ. (2012) Genetic manipulation of a “vacuolar” H+-PPase: from salt tolerance to yield enhancement under phosphorus-deficient soils. Plant Physiol 159: 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini JL, Briskin DP. (1988) Pyridine nucleotide oxidation by a plasma membrane fraction from red beet (Beta vulgaris L.) storage tissue. Arch Biochem Biophys 260: 653–660 [DOI] [PubMed] [Google Scholar]
- Giaquinta RT. (1983) Phloem loading of sucrose. Annu Rev Plant Physiol 34: 347–387 [Google Scholar]
- Gonzalez N, Beemster GT, Inzé D. (2009) David and Goliath: what can the tiny weed Arabidopsis teach us to improve biomass production in crops? Curr Opin Plant Biol 12: 157–164 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y, et al. (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes MC, Bengough G, Cassab G, Ponce G. (2003) Root caps and rhizosphere. J Plant Growth Regul 21: 352–367 [Google Scholar]
- Hermans C, Hammond JP, White PJ, Verbruggen N. (2006) How do plants respond to nutrient shortage by biomass allocation? Trends Plant Sci 11: 610–617 [DOI] [PubMed] [Google Scholar]
- Jackson LE. (1995) Root architecture in cultivated and wild lettuce (Lactuca spp.). Plant Cell Environ 18: 885–894 [Google Scholar]
- Jackson LE, Stivers LJ, Warren BT, Tanji KK. (1994) Crop nitrogen utilization and soil nitrate loss in a lettuce field. Fertilizer Research 37: 93–105 [Google Scholar]
- Johnson WC, Jackson LE, Ochoa O, van Wijk R, Peleman J, St. Clair DA, Michelmore RW. (2000) Lettuce, a shallow-rooted crop, and Lactuca serriola its wild progenitor, differ at QTL determining root architecture and deep soil water exploration. Theor Appl Genet 101: 1066–1073 [Google Scholar]
- Karam F, Mounzer O, Sarkis F, Lahoud R. (2002) Yield and nitrogen recovery of lettuce under different irrigation regimes. J Appl Hortic 4: 70–76 [Google Scholar]
- Kerns DL, Matheron MA, Palumbo JC, Sanchez CA, Still DW, Tickes BR, Umeda K, Wilcox MA (1999) Guidelines for Head Lettuce Production in Arizona. IPM series number 12. Cooperative Extension, College of Agriculture and Life Sciences, University of Arizona, Tucson [Google Scholar]
- Kim Y, Kim EJ, Rea PA. (1994) Isolation and characterization of cDNAs encoding the vacuolar H+-pyrophosphatase of Beta vulgaris. Plant Physiol 106: 375–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancien M, Gadal P, Hodges M. (2000) Enzyme redundancy and the importance of 2-oxoglutarate in higher plant ammonium assimilation. Plant Physiol 123: 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Müller CG, Krapp A, von Wirén N, Daniel-Vedele F, Gojon A. (2003) Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell 15: 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A. (1999) Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509–519 [DOI] [PubMed] [Google Scholar]
- Li B, Wei A, Song C, Li N, Zhang JR. (2008) Heterologous expression of the TsVP gene improves the drought resistance of maize. Plant Biotechnol J 6: 146–159 [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, Richter G, Blakeslee J, Bandyopadhyay A, Titapiwantakun B, Undurraga S, Khodakovskaya M, Richards EL, et al. (2005) Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science 310: 121–125 [DOI] [PubMed] [Google Scholar]
- Li Z, Baldwin CM, Hu Q, Liu H, Luo H. (2010) Heterologous expression of Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic creeping bentgrass (Agrostis stolonifera L.). Plant Cell Environ 33: 272–289 [DOI] [PubMed] [Google Scholar]
- Lv S, Zhang K, Gao Q, Lian L, Song Y, Zhang JR. (2008) Overexpression of an H+-PPase gene from Thellungiella halophila in cotton enhances salt tolerance and improves growth and photosynthetic performance. Plant Cell Physiol 49: 1150–1164 [DOI] [PubMed] [Google Scholar]
- Lv S-L, Lian L-J, Tao P-L, Li Z-X, Zhang K-W, Zhang J-R. (2009) Overexpression of Thellungiella halophila H+-PPase (TsVP) in cotton enhances drought stress resistance of plants. Planta 229: 899–910 [DOI] [PubMed] [Google Scholar]
- Marschner H (2002) Mineral Nutrition in Higher Plants, Ed 2. Academic Press, London [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt A, Willmitzer L, Selbig J, et al. (2007) The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R, Marsh E, Seely S, Landry B. (1987) Transformation of lettuce (Lactuca sativa) mediated by Agrobacterium tumefaciens. Plant Cell Rep 6: 439–442 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Miller AJ, Smith SJ. (1996) Nitrate transport and compartmentation in cereal root cells. J Exp Bot 300: 843–854 [Google Scholar]
- Mitsuda N, Hisabori T, Takeyasu K, Sato MH. (2004) VOZ: isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol 45: 845–854 [DOI] [PubMed] [Google Scholar]
- Mosier AR, Syers JK, Freney JR (2004) Nitrogen fertilizer: an essential component of increased food, feed, and fiber production. In AR Mosier, JR Freney, eds, SCOPE 65: Agriculture and the Nitrogen Cycle. Assessing the Impacts of Fertilizer Use on Food Production and the Environment. Island Press, Washington, DC, pp 3–18 [Google Scholar]
- Nagy C (1999) Coefficients for Agricultural Inputs in Western Canada. Canadian Agricultural Energy End-Use Data Analysis Centre. http://www.csale.usask.ca/PDFDocuments/energyCoefficientsAg.pdf (November 15, 2011)
- Paez-Valencia J, Patron-Soberano A, Rodriguez-Leviz A, Sanchez-Lares J, Sanchez-Gomez C, Valencia-Mayoral P, Diaz-Rosas G, Gaxiola R. (2011) Plasma membrane localization of the type I H(+)-PPase AVP1 in sieve element-companion cell complexes from Arabidopsis thaliana. Plant Sci 181: 23–30 [DOI] [PubMed] [Google Scholar]
- Palmgren MG (1998) Proton gradients and plant growth: role of the plasma membrane H+-ATPase. Adv Bot Res 28: 1–70 [Google Scholar]
- Palmgren MG. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52: 817–845 [DOI] [PubMed] [Google Scholar]
- Park S, Li J, Pittman JK, Berkowitz GA, Yang H, Undurraga S, Morris J, Hirschi KD, Gaxiola RA. (2005) Up-regulation of a H+-pyrophosphatase (H+-PPase) as a strategy to engineer drought-resistant crop plants. Proc Natl Acad Sci USA 102: 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapula V, Shen G, Kuppu S, Paez-Valencia J, Mendoza M, Hou P, Chen J, Qiu X, Zhu L, Zhang X, et al. (2011) Expression of an Arabidopsis vacuolar H+-pyrophosphatase gene (AVP1) in cotton improves drought and salt tolerance and increases fibre yield in the field conditions. Plant Biotechnol J 9: 88–99 [DOI] [PubMed] [Google Scholar]
- Sanchez CA. (2000) Response of lettuce to water and N on sand and the potential for leaching of nitrate-N. HortScience 35: 73–77 [Google Scholar]
- Sanchez CA, Blackmer AM. (1988) Recovery of anhydrous ammonia derived nitrogen-15 during three years of corn production in Iowa. Agron J 80: 102–108 [Google Scholar]
- Sanchez CA, Doerge TA. (1999) Using nutrient uptake patterns to develop efficient nitrogen management strategies for vegetables. Horttechnology 9: 601–606 [Google Scholar]
- Undurraga SF, Santos MP, Paez-Valencia J, Yang H, Hepler PK, Facanha AR, Hirschi KD, Gaxiola RA. (2012) Arabidopsis sodium dependent and independent phenotypes triggered by H+-PPase up-regulation are SOS1 dependent. Plant Sci 183: 96–105 [DOI] [PubMed] [Google Scholar]
- Whelan JA, Russell NB, Whelan MA. (2003) A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods 278: 261–269 [DOI] [PubMed] [Google Scholar]
- Yang H, Knapp J, Koirala P, Rajagopal D, Peer WA, Silbart LK, Murphy A, Gaxiola RA. (2007) Enhanced phosphorus nutrition in monocots and dicots over-expressing a phosphorus-responsive type I H+-pyrophosphatase. Plant Biotechnol J 5: 735–745 [DOI] [PubMed] [Google Scholar]
- Zhen RG, Kim EJ, Rea PA. (1997) Acidic residues necessary for pyrophosphate-energized pumping and inhibition of the vacuolar H+-pyrophosphatase by N,N′-dicyclohexylcarbodiimide. J Biol Chem 272: 22340–22348 [DOI] [PubMed] [Google Scholar]
- Zheng Z-L. (2009) Carbon and nitrogen nutrient balance signaling in plants. Plant Signal Behav 4: 584–591 [DOI] [PMC free article] [PubMed] [Google Scholar]