Abstract
In December 2012, scientists from around the world gathered in Waikoloa, Hawaii for ‘Auxin 2012’, a meeting organized by Paula McSteen (University of Missouri, USA), Ben Scheres (Utrecht University, The Netherlands) and Yunde Zhao (University of California, San Diego, USA). At the meeting, participants discussed the latest advances in auxin biosynthesis, transport and signaling research, in addition to providing context for how these pathways intersect with other aspects of plant physiology and development. Fittingly, the meeting began with a traditional Hawaiian ceremony that recognized the centrality of the harvest of plant life (’mea ho’oulu’ in Hawaiian) for continued human survival.
Keywords: Auxin, Biosynthesis, Signaling
Introduction
Auxin is a small molecule hormone that influences nearly every aspect of a plant’s life. Auxin response is modulated at multiple levels, including biosynthesis, conjugation, facilitated transport, and interaction with other hormones. The molecular mechanisms driving these processes have been elucidated over the last decades, and important new findings in all of these areas were presented at the ‘Auxin 2012’ meeting, which was held in Waikoloa, Hawaii in December 2012. Despite this remarkable progress, a central question was posed by many speakers: how does auxin orchestrate so many diverse processes with such precision? Excitingly, new technologies and new approaches to modeling that were presented at the meeting are now beginning to provide some answers to this question.
Auxin biosynthesis
A recent breakthrough in the field of auxin biosynthesis has been the discovery that the major source of auxin (or indole-3-acetic acid, IAA) is generated by a remarkably straightforward two-step pathway starting with the amino acid tryptophan (Trp) and relying on the serial action of two enzyme families: the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1-TRYPTOPHAN AMINOTRANSFERASE-RELATED (TAA1-TAR) family (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009) and the YUCCAs (Mashiguchi et al., 2011; Stepanova et al., 2011; Won et al., 2011). However, exactly how these enzymes function has been unclear. Yunde Zhao’s laboratory (University of California, San Diego, USA) has recently overcome technical difficulties to produce YUCCA6 in sufficient quantities and purity to allow them to determine the precise biochemical mechanism of YUCCA action, which involves flavin adenine dinucleotide (FAD) reduction and reaction with oxygen to produce IAA from indole pyruvic acid (IPyA) (Dai et al., 2013). New chemical tools, such as the TAA1 and TAR inhibitor l-kynurenine (He et al., 2011), also came up in many talks and are being used across the globe to pursue new questions about auxin biosynthesis and function. However, Joe Noel (The Salk Institute, USA) sounded a cautionary note about metabolic chaos and redundancy - enzymes tend to be promiscuous and enzymes in phylogenetic families are not always functionally redundant. He predicts that not all members of the TAA1-TAR family, which converts Trp to IPyA (Stepanova et al., 2008; Tao et al., 2008; Yamada et al., 2009), will act in auxin biosynthesis.
One answer to the paradox of the many roles of auxin might be that not all auxin is the same. The auxin precursor indole-3-butyric acid (IBA) is converted to active IAA by peroxisomal β-oxidation (reviewed by Strader and Bartel, 2011), and the contribution of IBA to the active auxin pool has recently been shown to be greater than previously anticipated (Strader et al., 2011). Lucia Strader (Washington University in St Louis, USA) presented evidence that IBA-derived auxin contributing to different developmental processes may be synthesized locally. Hiroyuki Kasahara (RIKEN, Japan) presented evidence that phenylacetic acid (PAA) acts as a natural auxin that is not transported in a polar fashion. PAA is detected in diverse plants, often at substantially higher levels than IAA, and can bind to auxin receptors. Like IAA, PAA is made by TAA1 and YUCCAs and can be found as amino acid conjugates. Higher order yucca mutants are only partially rescued by PAA treatment, suggesting potentially distinct roles for IAA and PAA. Wendy Peer (University of Maryland, USA) also described potential roles for oxindole-3-acetic acid (oxIAA), a likely auxin degradation product, in auxin and redox homeostasis. Furthermore, Chang-Hsien Yang (National Chung Hsing University, Taiwan) described how transcriptional repression of IAMT1, which encodes an IAA carboxyl methyltransferase that converts IAA to methyl-IAA (MeIAA), by the NAM, ATAF1/2, CUC2 (NAC)-like AUXIN SUPPRESSING FACTOR results in an array of developmental phenotypes. These alternative active and inactive forms of auxin raise the possibility that the regulation of auxin homeostasis is even more complex than we currently realize.
Auxin transcriptional responses
In the field’s formative years, genetic dissection of the auxin transcriptional response led to fundamental insights into ubiquitin-mediated protein degradation. This trend continued in several presentations. Auxin triggers the degradation of Auxin/INDOLE-3-ACETIC ACID (Aux/IAA) repressor proteins by promoting their interaction with TRANSPORT INHIBITOR RESPONSE1 (TIR1)-AUXIN SIGNALING F-BOX (AFB) F-box subunits of the E3 ubiquitin ligases (SCFTIR1/AFB) (Fig. 1) (reviewed by Calderón Villalobos et al., 2010). The ubiquitylation and subsequent degradation of Aux/IAA repressors leads to activation of AUXIN RESPONSE FACTOR (ARF) transcription factors. At the meeting, Mark Estelle (University of California, San Diego, USA) showed how a small number of mutations in TIR1 can substantially increase its binding affinity for Aux/IAAs and confer auxin hypersensitivity. Working in collaboration with Jennifer Nemhauser (University of Washington, USA), he demonstrated that these TIR1 variants also accelerate Aux/IAA degradation. Variation in this region among the TIR1-AFB family members might explain some of the observed differences in properties of co-receptor pairs (Calderón Villalobos et al., 2012; Havens et al., 2012). Additional TIR1 variants with mutations in the F-box region were also presented. These mutations led to increased TIR1 stability, probably by disrupting interaction with other components of the E3 ligase complex.
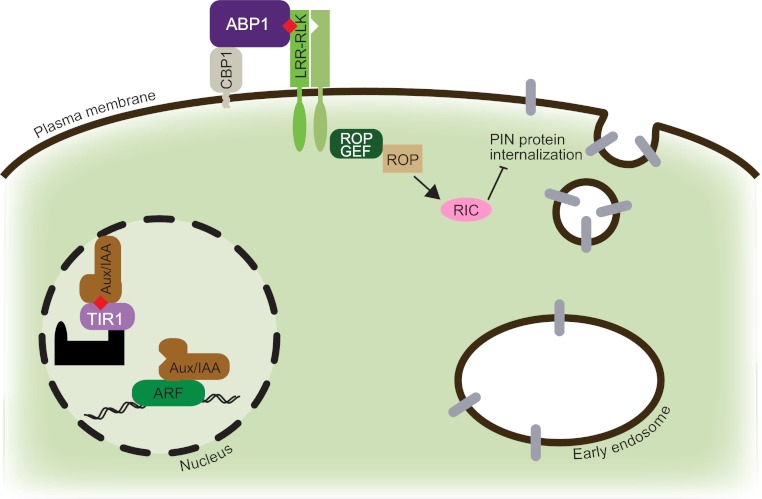
Fig. 1.
Model of auxin responses. Auxin (represented by a red diamond) generates both transcriptional and non-transcriptional responses. Transcriptional responses (occurring in the nucleus) are controlled primarily through the activities of ARF transcription factors (green) that promote auxin-induced gene expression, Aux/IAA transcriptional repressors (brown) that inhibit auxin-induced gene transcription, and the SCFTIR1/AFB ubiquitin ligase complex (black/lilac). Repression of ARF transcriptional activity is relieved by the auxin-dependent interaction of Aux/IAA proteins with TIR1/AFB F-box proteins. SCFTIR1/AFB promotes the polyubiquitylation and subsequent degradation of Aux/IAA proteins. Non-transcriptional auxin responses are controlled primarily through the activity of AUXIN BINDING PROTEIN1 (ABP1; purple), which is tethered to the plasma membrane by the glycosylphosphatidylinositol (GPI)-anchored protein CBP1 (pale gray). ABP1 binds auxin, possibly with an LRR-RLK co-receptor (green), and promotes the activation of ROP (beige), which then activates RIC (pink) to enhance F-actin bundling resulting in inhibition of clathrin-dependent internalization of the PIN family (gray) of auxin efflux carriers. See text for more details.
Another area in which studying auxin may uncover more widespread molecular mechanisms is transcriptional repression. Masao Tasaka (Nara Institute of Science and Technology, Japan) identified a mutant called macchi-bou2 (mab2) as a strong enhancer of the auxin transport mutant pinoid. MAB2 encodes a subunit of the MEDIATOR-CDK8 complex, which is implicated in repression (Ito et al., 2011). mab2 can partially suppress bodenlos mutants carrying a mutation that stabilizes one of the Aux/IAA repressors. Additional evidence for interactions between ARF proteins and MEDIATOR subunits was presented by Dolf Weijers (Wageningen University, The Netherlands); co-immunoprecipitation followed by mass spectrometry assays using different ARF proteins identified distinct and overlapping sets of MEDIATOR complex members.
A better understanding of ARFs is crucial for breaking the ‘auxin code’. This effort is being helped by work from several groups. The Weijers laboratory is leading the charge, by assembling profiles of ARF-specific protein complexes in various developmental stages and tissues, and by examining the atomic structure of ARF DNA-binding domains. Protein-binding microarray studies, also performed in the Weijers group, highlighted little specificity in DNA-binding site preference among ARFs, suggesting that spacing may play a larger role than previously suspected. Experimentally validated modeling of auxin response modules will be aided by higher resolution auxin-responsive transcript datasets (such as those presented by Bastiaan Bargmann, University of California, San Diego, USA), synthetic recreation of auxin transcriptional responses in yeast (such as those studied by Jennifer Nemhauser) and improved visualization tools [such as those developed by Teva Vernoux, Le Centre National de la Recherche Scientifique (CNRS), Lyon, France].
Non-transcriptional auxin responses
Although SCFTIR1/AFB-mediated signaling regulates many aspects of auxin-dependent plant growth and development, a number of cellular auxin responses do not require this pathway or precede changes in transcription. For example, many tropic responses are initiated by rapid re-distribution of auxin by the PIN-FORMED (PIN) auxin efflux carriers. Jae-Yean Kim (Gyeongsang National University, Korea) posed an intriguing question about how such auxin gradients can be formed and maintained, given that many plant cells are symplastically connected by channels called plasmodesmata. He provided evidence that reducing traffic through plasmodesmata by rapidly depositing callose plugs might play an essential role in PIN-driven auxin gradients. Kristoffer Jonsson from Rishi Bhalerao’s laboratory (Umeå Plant Science Centre, Sweden) presented evidence that the trans-Golgi network-localized protein ECHIDNA regulates secretion of the auxin transporter AUX1 to the plasma membrane, providing an additional layer of regulation to auxin transport rates.
Recently, much attention has been focused on identification of the core molecular mechanism responsible for non-transcriptional auxin perception and response (Chen et al., 2012; Lin et al., 2012; Robert et al., 2010; Shi and Yang, 2011; Xu et al., 2010). Jiří Friml (Institute of Science and Technology, Austria) and Zhenbiao Yang (University of California, Riverside, USA) presented strong evidence that AUXIN BINDING PROTEIN1 (ABP1) regulates local endocytosis of PIN carriers and, thereby, auxin transport. The Yang group has found that auxin rapidly activates RHO-RELATED PROTEIN FROM PLANTS (ROP2) and ROP6 on the cell surface in an ABP1-dependent manner (Xu et al., 2010). An ABP1-related protein was also implicated in auxin-regulated development of floral nectaries (as discussed by Clay Carter, University of Minnesota, Duluth, USA).
Several studies support the notion that the secreted form of ABP1 senses auxin in order to regulate non-transcriptional auxin responses (Shi and Yang, 2011). However, although ABP1 binds auxin, the dissociation constant (Kd) for auxin binding is in the micromolar range, whereas the physiological concentration of IAA required for ABP1-mediated auxin responses is in the nanomolar range. These observations led Zhenbiao Yang’s group to speculate that ABP1 requires a transmembrane partner to effectively bind auxin and transmit the auxin signal to cytoplasmic pathways, and they found evidence that members of a leucine-rich repeat receptor-like protein kinase (LRR-RLK) family can act as ABP1 functional partners. Interestingly, Elena Shpak (University of Tennessee, USA) described how the ERECTA family, also LRR-RLKs, regulate PIN1 levels at the shoot apical meristem to set the rate of new leaf initiation. In addition, Reidunn Aalen (University of Oslo, Norway) described the interaction between auxin and the small peptide INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), which acts as a ligand for the LRR-RLKs HAESA and HAESA-LIKE2, during lateral root emergence. Finally, Zachary Nimchuk (California Institute of Technology, USA) described several mechanisms by which another famous LRR-RLK, CLAVATA1, maintains the balance between ‘stemness’ and differentiation through manipulation of both auxin transport and auxin responses. On a related note, Klaus Palme (University of Freiburg, Germany) initiated a lively debate about the natural conditions in which auxin may regulate endocytosis, and presented experimental support for pathogen-induced auxin accumulation inhibiting endocytosis of the LRR-RLK FLAGELLIN SENSITIVE2 (FLS2). Together, these studies make a clear case for rapid, non-genomic auxin responses and their role in regulating many distinct aspects of plant growth and morphogenesis.
Interaction of auxin with other pathways
One session was devoted to auxin interactions with other pathways, but this idea was a recurrent theme throughout the meeting. For example, auxin was shown to integrate information about core metabolism; Karin Ljung (Swedish University of Agricultural Sciences, Sweden) and Jennifer Nemhauser presented evidence that auxin biosynthesis is strongly affected by levels of fixed carbon (Lilley et al., 2012; Sairanen et al., 2012). Ottoline Leyser (Sainsbury Laboratory Cambridge University, UK) described an experimentally validated model in which nitrogen can act through cytokinin to counterbalance auxin-mediated repression of bud activation.
It is now well established that auxin levels and responses are highly connected with other hormones. Joe Noel, for example, presented recent work on an enzyme called VAS1, which coordinates levels of auxin and ethylene synthesis. Auxin-mediated antagonism of cytokinin was also shown to be crucial for several developmental responses. Teva Vernoux (CNRS, Lyon, France) showed that sequential initiation of leaf primordia at the shoot apical meristem requires auxin-mediated induction of an inhibitor of cytokinin signaling called AHP6 in early primordia. Yka Helariutta (University of Helsinki, Finland) described a mutually inhibitory mechanism for vascular patterning that relied on auxin-dependent induction of AHP6-mediated repression of cytokinin signaling (Bishopp et al., 2011). Focusing downstream of the TARGET OF MONOPTEROS 5 (TMO5) family of direct ARF5 targets, Bert De Rybel (Wageningen University, The Netherlands) showed that auxin-dependent local production of cytokinin is crucial for proper root development (De Rybel et al., 2013). Cytokinin was also shown by Eva Benková (Institute of Science and Technology, Austria) to interfere with auxin-responsive development of lateral roots in two ways: by decreasing the expression of a subset of PIN genes and by inducing internalization of PIN1. Sabrina Sabatini (University of Rome, Italy) similarly presented a multi-level scheme whereby antagonism of auxin by cytokinin leads to proper balance between cell division and differentiation. Ive De Smet (University of Nottingham, UK) identified an additional antagonist of auxin signaling, a small signaling peptide that appears to interfere directly with auxin receptor-co-receptor interactions. In summary, the recurring mention of crosstalk between auxin and multiple other pathways suggests that this will continue to be a central theme in auxin biology for many years.
Auxin and development
How auxin shapes cell fate and growth patterns during development continues to be an area of active research, as evidenced by two sessions devoted to the topic. One area of convergence was the role of auxin in the gravitropic set-point angle (GSA), a mechanism that allows lateral organs to be maintained at a specific angle with respect to gravity (Digby and Firn, 1995). Jürgen Kleine-Vehn (University of Natural Resources and Life Sciences, Austria) suggested that a PIN-dependent auxin transport mechanism limits gravitropic responses in lateral roots, and experiments from Stefan Kepinski’s group (University of Leeds, UK) led to a model in which different non-vertical GSAs were achieved through varying levels of antagonism of the gravitropic response. Yusaku Uga (National Institute of Agrobiological Sciences, Japan) discussed results from a clever quantitative trait screen for deeper roots in rice. By growing plants in plastic colanders and measuring the number of roots that protruded through the basket, he was able to find a gene that contributes to a large percentage of variation in rooting depth. This gene, Dro1, is repressed by ARFs and affects the strength of the gravitropic response. Tests are currently underway to quantify the effect of introgression of this allele on rice yields. Joseph Dubrovsky [Universidad Nacional Autónoma de México (UNAM), Mexico] presented data on the dual action of auxin in both promoting (Dubrovsky et al., 2011; Dubrovsky et al., 2008) and inhibiting (Ivanchenko et al., 2010) lateral root initiation. Carlos Galvan-Ampudia (University of Amsterdam, The Netherlands) gave an interesting presentation about salt-induced negative root tropism. As with other tropic responses, salt triggers changes in auxin transport leading to differential cell elongation.
The role of auxin in organogenesis was also a frequent topic at the meeting. The PLETHORA (PLT) family of transcription factors are master regulators of root development and a focus of the Scheres Laboratory. PLTs were shown (by Dongping Bao, Wageningen University, The Netherlands) to directly regulate auxin biosynthesis, while Pankaj Dhonukshe (Utrecht University, The Netherlands) discussed how PLTs regulate auxin responses, leading to correct orientation of cell division planes (Dhonukshe et al., 2012). Interestingly, wound-induced jasmonates were shown (by Chuanyou Li, Institute of Genetics and Developmental Biology, China) to antagonize auxin-dependent induction of PLTs, and Stephan Pollmann [Centro de Biotecnología y Genómica de Plantas (UPM-INIA), Spain] demonstrated that they promote expression of some YUCCA genes. Marta Laskowski (Oberlin College, USA) used higher order plt mutants to illustrate the crucial role of the PLT family in the spacing of lateral primordia in both shoot and roots, leading her to propose a role for PLTs in the most primitive branching events in land plant evolution. Jan Traas (École Normale Supérieure de Lyon, France) demonstrated that the control of microtubule anisotropy regulates organ outgrowth, possibly through the activity of katanin. Lars Østergaard (John Innes Centre, UK) presented a model for gynoecium patterning, whereby transcription factor complexes containing INDEHISCENT (IND) and SPATULA (SPT) regulate not only auxin transport (Girin et al., 2011) but also auxin response through interaction with ARF3. The molecular mechanism by which fine-scale differences in auxin levels might contribute to cell fate determination remains a largely unanswered question, but several groups announced ambitious plans for applying new technologies to this problem. This is clearly an area to watch.
Technical advances and new tools
Several new tools for visualizing auxin were presented in the context of a wide array of biological questions. The DII-VENUS reporter developed by Teva Vernoux (Brunoud et al., 2012), which takes advantage of auxin-induced Aux/IAA degradation for a more accurate reflection of auxin levels, has been widely adopted. Ken Hayashi (Okayama University of Science, Japan) has developed a long-awaited alkoxy-auxin fluorescent compound that is transported like auxin, but is unable to activate an SCFTIR1/AFB-based auxin response, allowing visualization of auxin transport patterns without interfering with genomic auxin responses. In addition, Benjamin Babst (Brookhaven National Laboratory, USA) presented the development of [11C]-IAA (Reid et al., 2011), which, combined with in vivo PET imaging, can be used to study auxin transport and metabolism.
Modeling auxin transport has allowed a number of labs to generate new hypotheses. For example, Przemek Prusinkiewicz (University of Calgary, Canada) extended his previous work on leaf serration (Bilsborough et al., 2011) to lobed and compound leaves, and has shown that their diversity can be captured using only a few parameters. Ross Sozzani (University of Pavia, Italy) explained how new analytical methods can be combined with standard confocal microscopy to test protein-protein interactions and to quantify rates of intracellular protein transport and many other key parameters needed for testing developmental models.
Natural variation in auxin-related pathways is providing needed context. A wide sampling of Arabidopsis thaliana accessions were used by Christian Hardtke (University of Lausanne, Switzerland) who described a vertical view of systems biology: extending the study of one gene from cellular mechanism to ecological function. His gene of choice is BREVIS RADIX (BRX), which regulates root length through modulation of root apical meristem size. He showed that H+ pumps are hyperactive in brx roots and that this allows more robust root growth on acid soil. This predicted role is supported by isolation of brx mutations in accessions collected on acid soils (Gujas et al., 2012). A large collaborative Auxin Evo-Devo project is being spearheaded by Paula McSteen (University of Missouri, USA) and aims to take a comparative approach to all things auxin all across the flowering plant world. Andrea Gallavotti (Rutgers University, USA), a member of this consortium, showed how auxin transport and response are crucial to reproductive organ morphogenesis in maize. Additionally, David Pacheco-Villalobos (University of Lausanne, Switzerland) described how auxin biosynthesis affects root architecture in Brachypodium. Eva Sundberg (Swedish University of Agricultural Sciences, Sweden) reported evidence that auxin is crucial for the development of reproductive structures in moss and requires SHORT-INTERNODES/STYLISH (SHI/STY)-type transcription factors, suggesting perhaps an ancient role. Christine Beveridge (University of Queensland, Australia) working with pea showed that auxin transport might not be fast enough to regulate bud outgrowth following decapitation (Renton et al., 2012).
Conclusion
The Auxin 2012 meeting showcased a number of recent insights into the molecular mechanisms driving auxin biosynthesis, signaling and transport, and illustrates our need to understand the integration of each of these events to control plant growth and development. This is thus an exciting time for researchers interested in auxin biology. We anticipate that our growing understanding of auxin biology will ensure that future Auxin meetings will be as exciting as the one in Waikoloa.
Acknowledgments
We thank Paula McSteen, Ben Scheres and Yunde Zhao for excellent meeting organization and for putting together a stimulating scientific program; individuals mentioned in the review for permission to discuss unpublished data; and all meeting attendees for stimulating discussion. We apologize that we were unable to cover many of the interesting presentations in detail. ‘Mea ho’oulu’ is Hawaiian for harvest.
Footnotes
Funding
Our work on auxin is supported by the National Institutes of Health [L.C.S.]; the National Science Foundation [J.L.N.]; and the Paul G. Allen Family Foundation [J.L.N.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
References
- Bilsborough G. D., Runions A., Barkoulas M., Jenkins H. W., Hasson A., Galinha C., Laufs P., Hay A., Prusinkiewicz P., Tsiantis M. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 108, 3424–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishopp A., Help H., El-Showk S., Weijers D., Scheres B., Friml J., Benková E., Mähönen A. P., Helariutta Y. (2011). A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 21, 917–926 [DOI] [PubMed] [Google Scholar]
- Brunoud G., Wells D. M., Oliva M., Larrieu A., Mirabet V., Burrow A. H., Beeckman T., Kepinski S., Traas J., Bennett M. J., et al. (2012). A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106 [DOI] [PubMed] [Google Scholar]
- Calderón-Villalobos L. I., Tan X., Zheng N., Estelle M. (2010). Auxin perception - structural insights. Cold Spring Harb. Perspect. Biol. 2, a005546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón Villalobos L. I., Lee S., De Oliveira C., Ivetac A., Brandt W., Armitage L., Sheard L. B., Tan X., Parry G., Mao H., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Naramoto S., Robert S., Tejos R., Löfke C., Lin D., Yang Z., Friml J. (2012). ABP1 and ROP6 GTPase signaling regulate clathrin-mediated endocytosis in Arabidopsis roots. Curr. Biol. 22, 1326–1332 [DOI] [PubMed] [Google Scholar]
- Dai X., Mashiguchi K., Chen Q., Kasahara H., Kamiya Y., Ojha S., Dubois J., Ballou D., Zhao Y. (2013). The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 288, 1448–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., Möller B., Yoshida S., Grabowicz I., Barbier de Reuille P., Boeren S., Smith R. S., Borst J. W., Weijers D. (2013). A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell. (in press). [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., Weits D. A., Cruz-Ramirez A., Deinum E. E., Tindemans S. H., Kakar K., Prasad K., Mähönen A. P., Ambrose C., Sasabe M., et al. (2012). A PLETHORA-auxin transcription module controls cell division plane rotation through MAP65 and CLASP. Cell 149, 383–396 [DOI] [PubMed] [Google Scholar]
- Digby J., Firn R. D. (1995). The gravitropic set-point angle (GSA): the identification of an important developmentally controlled variable governing plant architecture. Plant Cell Environ. 18, 1434–1440 [DOI] [PubMed] [Google Scholar]
- Dubrovsky J. G., Sauer M., Napsucialy-Mendivil S., Ivanchenko M. G., Friml J., Shishkova S., Celenza J., Benková E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105, 8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky J. G., Napsucialy-Mendivil S., Duclercq J., Cheng Y., Shishkova S., Ivanchenko M. G., Friml J., Murphy A. S., Benková E. (2011). Auxin minimum defines a developmental window for lateral root initiation. New Phytol. 191, 970–983 [DOI] [PubMed] [Google Scholar]
- Girin T., Paicu T., Stephenson P., Fuentes S., Körner E., O’Brien M., Sorefan K., Wood T. A., Balanzá V., Ferrándiz C., et al. (2011). INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23, 3641–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujas B., Alonso-Blanco C., Hardtke C. S. (2012). Natural Arabidopsis brx loss-of-function alleles confer root adaptation to acidic soil. Curr. Biol. 22, 1962–1968 [DOI] [PubMed] [Google Scholar]
- Havens K. A., Guseman J. M., Jang S. S., Pierre-Jerome E., Bolten N., Klavins E., Nemhauser J. L. (2012). A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 160, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Brumos J., Li H., Ji Y., Ke M., Gong X., Zeng Q., Li W., Zhang X., An F., et al. (2011). A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23, 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Sono T., Tasaka M., Furutani M. (2011). MACCHI-BOU 2 is required for early embryo patterning and cotyledon organogenesis in Arabidopsis. Plant Cell Physiol. 52, 539–552 [DOI] [PubMed] [Google Scholar]
- Ivanchenko M. G., Napsucialy-Mendivil S., Dubrovsky J. G. (2010). Auxin-induced inhibition of lateral root initiation contributes to root system shaping in Arabidopsis thaliana. Plant J. 64, 740–752 [DOI] [PubMed] [Google Scholar]
- Lilley J. L., Gee C. W., Sairanen I., Ljung K., Nemhauser J. L. (2012). An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol. 160, 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Nagawa S., Chen J., Cao L., Chen X., Xu T., Li H., Dhonukshe P., Yamamuro C., Friml J., et al. (2012). A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22, 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., Hanada A., Yaeno T., Shirasu K., Yao H., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A. E., Kim S. W., Seiner B., Fowler F. W., Hooker J., Ferrieri R., Babst B. A., Fowler J. S. (2011). Radiosynthesis of C-11 labeled auxin (3-indolyl[1-11C]acetic acid) and its derivatives from gramine. J. Labelled Comp. Radiopharm. 54, 433–437 [Google Scholar]
- Renton M., Hanan J., Ferguson B. J., Beveridge C. A. (2012). Models of long-distance transport: how is carrier-dependent auxin transport regulated in the stem? New Phytol. 194, 704–715 [DOI] [PubMed] [Google Scholar]
- Robert S., Kleine-Vehn J., Barbez E., Sauer M., Paciorek T., Baster P., Vanneste S., Zhang J., Simon S., Čovanová M., et al. (2010). ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143, 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen I., Novák O., Pencík A., Ikeda Y., Jones B., Sandberg G., Ljung K. (2012). Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24, 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. H., Yang Z. B. (2011). Is ABP1 an auxin receptor yet? Mol. Plant 4, 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A. N., Robertson-Hoyt J., Yun J., Benavente L. M., Xie D. Y., Dolezal K., Schlereth A., Jürgens G., Alonso J. M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova A. N., Yun J., Robles L. M., Novak O., He W., Guo H., Ljung K., Alonso J. M. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Bartel B. (2011). Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 4, 477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Wheeler D. L., Christensen S. E., Berens J. C., Cohen J. D., Rampey R. A., Bartel B. (2011). Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23, 984–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Ferrer J. L., Ljung K., Pojer F., Hong F., Long J. A., Li L., Moreno J. E., Bowman M. E., Ivans L. J., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAS in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wen M., Nagawa S., Fu Y., Chen J. G., Wu M. J., Perrot-Rechenmann C., Friml J., Jones A. M., Yang Z. (2010). Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Greenham K., Prigge M. J., Jensen P. J., Estelle M. (2009). The Transport Inhibitor Response2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 151, 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]