Abstract
PUF family proteins are well-conserved regulators of cell proliferation in different developmental processes. They regulate target mRNAs by promoting degradation or by influencing translation through interaction with the translation initiation machinery. Here we show that Caenorhabditis elegans PUF-8 functions redundantly with the nuclear protein TCER-1 in the post-transcriptional maintenance of at least six germline mRNAs. The levels of spliced mRNAs in the puf-8(-) tcer-1(-) double mutant are only 10-30% of the wild type, whereas the unspliced forms increase by ∼2- to 3-fold compared with the wild type. These two proteins colocalise at the inner nuclear periphery, and their absence leads to reduced germ cell proliferation and to sterility. A yeast two-hybrid screen of 31 components of the nuclear pore complex and mRNA processing machineries identified seven proteins involved in mRNA export as potential partners of PUF-8. One of these, the nuclear cap-binding protein NCBP-2, colocalises with PUF-8 in the nucleus. A 50 amino acid N-terminal domain of PUF-8 is essential for interaction with NCBP-2 and for PUF-8 to function redundantly with TCER-1. These results reveal two important unexpected aspects of PUF proteins: that, in addition to the C-terminal PUF domain, the N-terminal domain is crucial for PUF function, and that PUF proteins have a novel role in mRNA maintenance. We propose that PUF proteins, in addition to their known cytoplasmic roles, participate in nuclear processing and/or export of mRNAs.
Keywords: PUF-8, TCER-1, C. elegans, Germ cells, mRNA processing, mRNA export
INTRODUCTION
Proteins of the PUF family are present in diverse eukaryotes including yeast, plants and metazoans ranging from invertebrates to mammals. These proteins regulate a wide variety of developmental processes, including embryonic patterning, stem cell maintenance and neuronal adaptation (Ariz et al., 2009; Crittenden et al., 2002; Forbes and Lehmann, 1998; Kaye et al., 2009; Schweers et al., 2002; Subramaniam and Seydoux, 2003; Tadauchi et al., 2001; Walser et al., 2006). Although many of these functions are species specific, regulation of stem cell proliferation appears to be the most ancient and well-conserved function of PUF proteins.
Evidence so far indicates that the PUF proteins function as post-transcriptional regulators. Results from large-scale experiments suggest that these proteins influence the expression of a large number of mRNAs (Galgano et al., 2008; Gerber et al., 2004; Kershner and Kimble, 2010; Mainpal et al., 2011; Morris et al., 2008). However, PUF-mediated regulation has been established only for a few of them. In the case of these well-demonstrated target mRNAs, the relevant PUF protein has been shown to function by direct binding to specific sequences within the 3′ UTR (Crittenden et al., 2002; Kadyrova et al., 2007; Murata and Wharton, 1995; Zhang et al., 1997). A few examples of positive regulation have been reported, but in most cases PUF proteins negatively regulate their target mRNAs (Kaye et al., 2009). The mechanism of positive regulation is not clear, whereas negative regulation is known to be accomplished either by promoting mRNA degradation or interfering with translation initiation (Deng et al., 2008; Goldstrohm et al., 2006; Goldstrohm et al., 2007). The common theme emerging from these studies is that PUF proteins are sequence-specific regulators of mRNAs in the cytoplasm.
A characteristic of PUF proteins is the presence of a conserved RNA-binding domain called the PUF domain, which contains eight imperfect repeats of ∼40 amino acids each, in their C-terminal region (Wickens et al., 2002). The entire PUF domain is essential for RNA binding and, in the case of the Drosophila PUF protein Pumilio, the pumilio mutant phenotypes are mostly rescued by the PUF domain alone (Wharton et al., 1998). Intriguingly, all PUF proteins contain a region of ∼200-1100 amino acids in their N-terminal region that is distinct from the PUF domain. The functional significance of this rather large N-terminal region is not known. Its amino acid sequence has not been conserved among the different PUF family members and does not share similarity with any other known protein motif.
Here we report the identification of an unexpected novel function for the Caenorhabditis elegans PUF protein PUF-8. In germ cells, in addition to being in the cytoplasm, we find that PUF-8 is also present in the nucleus, where it colocalises with at least two nuclear proteins involved in mRNA processing/export. PUF-8 functions redundantly with one of them, TCER-1, which is predicted to link transcription with splicing, to ensure normal levels of germ cell proliferation and fertility. Our results indicate that PUF-8 functions along with TCER-1 at a post-transcriptional step to maintain normal levels of certain germline mRNAs. Further, our yeast two-hybrid experiments suggest that PUF-8 interacts with some of the mRNA processing/export machinery components, for which a 50 amino acid region of its N-terminus is crucial. Importantly, the same 50 amino acid N-terminal region is essential to rescue the puf-8(-) tcer-1(-) double mutant.
MATERIALS AND METHODS
C. elegans strains
Strains used in this study are described in supplementary material Table S1. They were maintained as described (Brenner, 1974), except that all transgenic lines were grown at 25°C to avoid silencing of the germline-expressed transgenes (Strome et al., 2001). Generation of double-mutant and double-transgenic lines, as well as transgene rescue experiments, were performed using standard genetic techniques. RNAi experiments were carried out as described (Mainpal et al., 2011).
Construction of transgenes
The tcer-1 transgene construct pPK6 was prepared as follows: a 1813 bp genomic region upstream of the tcer-1 start codon and a part of the coding region (up to the BamHI site) was PCR amplified using primers KS2315 and KS2132 and cloned between SacII and BamHI sites of the pBluescript KS+ plasmid vector (see supplementary material Table S2 for primer sequences). The remainder of the coding region was amplified using primers KS2133 and KS2134 and cloned between the BamHI and SmaI sites of the plasmid resulting from the above cloning. In the resulting plasmid, GFP coding sequences were PCR amplified from pMP15 (Ariz et al., 2009) using primers KS2135 and KS2326 and inserted between the SmaI and SalI sites. To this, 1730 bp of tcer-1 downstream genomic sequences, which were amplified using primers KS2316 and KS2361, were added between SalI and ApaI sites. A 5596 bp genomic fragment containing the unc-119 gene was PCR amplified using primers KS1222 and KS1223 and introduced into the above plasmid between NgoMIV and SacII as the transgene selection marker. The GFP in pPK6 was replaced with mCherry coding sequences amplified using KS2944 and KS2945 to generate pPK20 (supplementary material Table S1).
The ncbp-2 transgene construct pPK46 was generated as follows: the histone H2B sequences in pPKS114 (Ariz et al., 2009) were replaced with ncbp-2 coding sequences amplified from C. elegans genomic DNA using primers KS3703 and KS3704. One copy of the cMyc epitope was included as part of the KS3704 primer. The GFP coding sequences were then replaced with the mCherry sequences amplified using KS3526, which contains another copy of cMyc, and KS2978. Three additional copies of the cMyc epitope were incorporated upstream of mCherry using primers KS3709 and KS3710. These two partially overlapping primers were annealed, converted into full double strands by primer extension and digested with BglII and BamHI. The resulting fragment was ligated at the BamHI site present upstream of the mCherry sequences. The emr-1 transgene construct pPK19 is similar to pPK46 except that it does not contain cMyc sequences.
The transgene constructs bearing puf-8 deletions were made as follows: the BamHI-NotI fragment of pMP15, containing part of puf-8 and gfp coding sequences, was replaced with the PCR fragment amplified from pMP15 using primers KS3022 and KS3175 to introduce the HA epitope at the C-terminus of GFP. This construct, termed pPK25, served as the parental plasmid for all puf-8 deletion constructs. pPK30, which does not contain amino acids 3-143, was generated in two steps. First, the upstream and coding sequences of puf-8 in pPK25 were removed by KpnI digestion. To this, a PCR fragment bearing the upstream puf-8 sequences up to the second amino acid, amplified using primers KS109 and KS3303, was added at the KpnI site. Second, the sequences coding PUF-8 amino acids 144-535 were amplified using primers KS3304 and KS3320 and inserted at the ApaI site introduced by primer KS3303. Other deletion constructs were generated by similar two-step clonings by substituting KS3303 and KS3304 with the appropriate primers: KS3303 and KS3675 for pPK45 (Δ3-49aa), KS3551 and KS3609 for pPK39 (Δ51-100aa), and KS3612 and KS3304 for pPK44 (Δ111-143aa).
All constructs were generated in duplicate - two parallel, independent PCR and cloning events - and introduced into the unc-119(-) strain by biolistic bombardment as described (Jadhav et al., 2008; Praitis et al., 2001). We routinely obtained 10-15 independent lines per construct; at least three independent transgenic lines with an identical expression pattern were used for the rescue of mutants. The three lines tested yielded similar results in the rescue experiments described in this study.
Fluorescence microscopy
Gonad dissection and DAPI staining were performed as described (Ariz et al., 2009). For leptomycin B (Sigma, L2913) treatment, ∼100 young adults were incubated for 2.5 hours at 25°C in 50 μl M9 buffer containing 1 ng/ml leptomycin B. The worms were then recovered and examined for GFP fluorescence. To detect NCBP-2 protein, dissected gonads were fixed in aldehyde fixative (3.5% formaldehyde, 0.25% glutaraldehyde, 0.1 M potassium phosphate, pH 7.2) for 15 minutes, followed by incubation in methanol for 5 minutes and acetone for 10 minutes, both at -20°C. Non-specific epitopes were blocked by incubating fixed gonads in PBT (0.1% BSA and 0.1% Tween 20 in phosphate-buffered saline) for 30 minutes. The gonads were then incubated overnight at 4°C with 1:500 dilution of anti-human CBP20 antibody (200 μg/ml rabbit polyclonal; Santa Cruz, sc-48793). After washing three times with PBT, the gonads were incubated with 1:50 dilution of FITC-conjugated goat anti-rabbit secondary antibody (1 mg/ml; Jackson ImmunoResearch, 115-095-068) overnight at 4°C, then washed three times with PBT and mounted in Vectashield (Vector Laboratories). These immunostained gonads, as well as worms expressing GFP and mCherry fluorescence reporters, were observed using a Zeiss Axioskop II mot plus microscope. Fluorescence images were acquired using an Axiocam HRm camera and deconvolved using Axiovision software (both Carl Zeiss).
Quantitation of mRNAs
The growth rate of puf-8(-) tcer-1(-) worms was slower than that of the wild type and both single mutants. Therefore, to compare RNA levels in age-matched populations, we first determined the time required by the different genotypes to initiate spermatogenesis. For this, we collected embryos from gravid hermaphrodites for a 2-hour period and allowed them to hatch and grow. We monitored germline development in these synchronous populations of larvae by DAPI staining at different time points. Compared with ∼58 hours at 20°C for the wild-type and tcer-1(-), the puf-8(-) worms took ∼64 hours and puf-8(-) tcer-1(-) took ∼94 hours to initiate spermatogenesis.
We isolated total RNA from 100 synchronously grown worms at the above time points using Tri Reagent (Sigma) following the manufacturer’s protocol. The RNA samples were treated with DNase I for 15 minutes, heat inactivated and used for reverse transcription (RT) using oligo(dT)18 as the primer. To reduce errors arising from pipetting smaller volumes, the cDNA was diluted to 500 μl and 10 μl aliquots were used as template for the PCR amplification. We used Southern hybridisation with appropriate radiolabelled probes to detect the amplified products. Hybridisation signals were detected and quantified using a phosphorimager (Bio-Rad). Use of Southern hybridisation enabled detection while product accumulation was still in the linear range, which was determined for each product by performing hybridisation at different cycle points. To further control variations, the whole process, from growth of worms to RT-PCR, was repeated at least eight times. To account for small variations caused by the slight reduction in the number of germ cells in the double mutant at the above mentioned time point, a set of least-varying mRNAs, which were ∼1.5-fold higher in the wild type than in the double mutant, were used for normalisation. In addition, we calculated the mRNA to pre-mRNA ratio, which is independent of the overall RNA levels. Although more laborious than the real-time PCR methods, the Southern hybridisation-based approach avoided the non-specific signals arising from primer dimers. To account for variations in the RNA and cDNA yields, and to further account for any variation in the germ cell numbers, we used the levels of germline hip-1 mRNA (http://nematode.lab.nig.ac.jp), which did not vary significantly in these experiments, for normalisation. Primers used for RNA quantitation are listed in supplementary material Table S3.
Yeast two-hybrid assay
The method described by James et al. (James et al., 1996) was followed. Briefly, PCR-amplified products were inserted into the vector pGBDU-C2 or pACT2 to serve as the DNA-binding (BD) or activation domain (AD) fusions, respectively. The BD and AD fusions tested are listed in supplementary material Tables S4 and S5, respectively. In the case of the PUF-8 deletions N16-BD and N18-BD, the two fragments flanking the deletion were amplified and ligated separately, and then reamplified using KS3022 and KS3024, which was then cut with BglII and cloned into the BamHI site of pGBDU-C2. All constructs were prepared in duplicate as described above. Yeast strain PJ69-4α (James et al., 1996) was transformed as described (Gietz and Woods, 2002) and colonies were selected on appropriate selection plates. To test interaction between BD and AD fusions, three colonies from each transformation were suspended in 50 μl aliquots of water and 10 μl aliquots were spotted on (1) agar plates lacking histidine but containing 2 mM 3-aminotriazole and (2) agar plates without adenine. Only the BD and AD combinations that grew robustly on both selection plates are reported in Fig. 4.
Fig. 4.
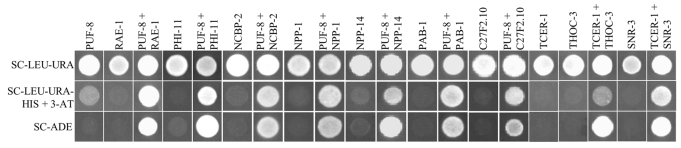
PUF-8 and TCER-1 interact with mRNA processing/export components in the yeast two-hybrid assay. Growth of yeast strains carrying the various constructs (labelled at top) in media lacking the indicated nutrients (left). In row 2, the minimal medium used contains 2 mM 3-aminotriazole (3-AT). PUF-8 was expressed as a Gal4BD fusion and all others were Gal4AD fusions. Panels marked with a single protein are negative controls expressing the indicated fusion protein along with either Gal4BD or Gal4AD alone. SC, synthetic complete medium; LEU, leucine; URA, uracil; HIS, histidine; ADE, adenine. Growth observed in rows 2 and 3 indicates activation of the HIS3 and ADE2 reporters, respectively. These two reporters are under the control of two different GAL4-responsive promoters; activation of both reporters provides strong support for the functional reconstitution of GAL4 by the interacting protein pairs (James et al., 1996). Therefore, to avoid potential false positives, only those fusion protein pairs showing robust growth on both selection plates (rows 2 and 3) were scored as interacting pairs.
RESULTS
PUF-8 functions redundantly with TCER-1, a component of the RNA processing machinery, to regulate germ cell proliferation
Depletion of PUF-8 results in several germ cell defects. These include reduced proliferation of germline stem cells, dedifferentiation of primary spermatocytes into germ cell tumours, failure of the sperm/oocyte switch and improper chromosome segregation during meiosis (Ariz et al., 2009; Bachorik and Kimble, 2005; Subramaniam and Seydoux, 2003). Surprisingly, these phenotypes are temperature sensitive: even strains homozygous for the null alleles of puf-8, although subfertile, can be maintained continuously when grown at 20°C (Subramaniam and Seydoux, 2003).
To identify gene(s) that potentially compensate for the lack of PUF-8 at 20°C, a synthetic genetic screen was carried out on the puf-8(+/-) background (Vaid et al., 2013). To map one of the synthetic mutants identified in that screen, we performed an RNAi screen of genes present in a limited genetic interval on chromosome II on the puf-8(zh17) genetic background, which is a strong loss-of-function allele. This RNAi screen identified tcer-1 as functionally redundant with puf-8. Consistent with the earlier results, 100% of the puf-8(zh17) worms were fertile at 20°C (n=150). Similarly, all of the tcer-1(RNAi) worms were fertile and did not show any observable defects (n=500). By contrast, the puf-8(zh17) tcer-1(RNAi) double-mutant worms were 100% sterile at 20°C (n=500). Subsequently, we confirmed the RNAi results by generating a puf-8(-) tcer-1(-) genetic double-mutant strain using tcer-1(tm1452), which has a 392 bp deletion that includes most of the second exon of tcer-1. Similar to the tcer-1(RNAi) worms, tcer-1(tm1452) worms were normal (n=500). In the double-mutant worms, the total number of germ cells was severely reduced and the morphology of the chromatin, as revealed by DAPI staining, was abnormal. Although a few of these worms produced sperm, they did not develop a normal oocyte (Fig. 1). We conclude that puf-8 and tcer-1 function redundantly to ensure normal levels of germ cell proliferation.
Fig. 1.
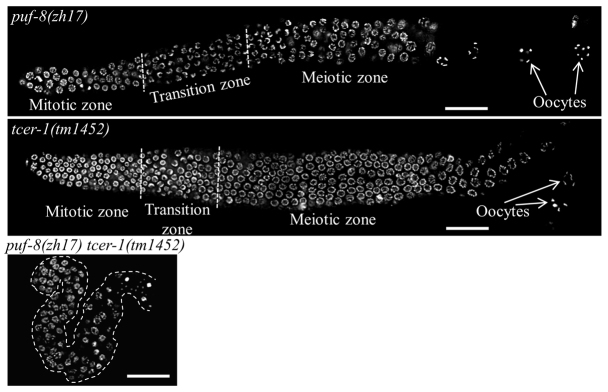
puf-8 functions redundantly with tcer-1. Dissected C. elegans adult gonads of the indicated genotype stained with DAPI. Left, distal; right, proximal. The three images are at identical magnification. The double-mutant gonad is significantly smaller than either single mutant and it contains fewer germ cells and no gametes. Scale bars: 25 μm.
The lack of any observable phenotype in tcer-1(tm1452) is most likely due to its functional redundancy with a partial duplication, ZK1127.6, present ∼1.4 kb upstream and on the opposite strand with respect to the tcer-1 locus [data obtained from WormBase (www.wormbase.org)]. Consistent with this notion, ZK1127.6(tm2782) tcer-1(RNAi) resulted in 40% embryonic lethality (n=100). Although a deletion allele, ZK1127.6(tm2782), is available, owing to the very close proximity of the two loci we were unable to generate the ZK1127.6(tm2782) tcer-1(tm1452) double mutant. However, puf-8(zh17) ZK1127.6(tm2782) worms were fertile (n=500), which indicates that this partial duplication does not function redundantly with puf-8. Further, a transgene expressing ZK1127.6::GFP under the control of the upstream and downstream sequences of ZK1127.6 did not show any expression in the germline (data not shown).
PUF-8 colocalises in the nucleus with TCER-1
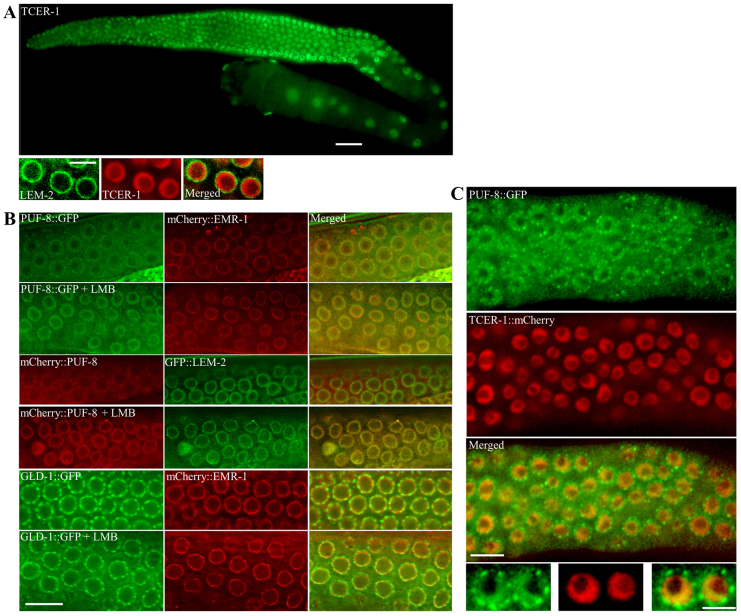
TCER-1 has been annotated as a transcription elongation regulator based on its similarity to the mammalian protein CA150 (TCERG1) and its yeast orthologue PRP40p (Ghazi et al., 2009). CA150 has been shown to be essential for mRNA processing and, based on its ability to interact with the C-terminal domain (CTD) of RNA polymerase II and the components of the spliceosome, CA150 has been implicated in coupling transcription and splicing (Lin et al., 2004; Pearson et al., 2008). To begin to address its functional redundancy with PUF-8, we determined the distribution pattern of TCER-1 in the germline using TCER-1::GFP and TCER-1::mCherry transgenes. Both of these transgenes were expressed using the promoter and downstream sequences of tcer-1, and were able to restore fertility in puf-8(-) tcer-1(-) worms. As shown in Fig. 2A, TCER-1::GFP was present in all germ cell nuclei, starting from the mitotic cells at the distal end to the oocytes at the proximal end of the gonad. TCER-1::mCherry showed an identical expression pattern (data not shown). Both of these transgenes were expressed in somatic nuclei as well (data not shown).
Fig. 2.
PUF-8 and TCER-1 colocalise at the inner nuclear periphery. (A) Expression pattern of tcer-1::gfp transgene in a wild-type background (top) and of gfp::lem-2, a nuclear envelope marker, and tcer-1::mCherry transgenes (bottom). Only a few germ cells from the distal part of the gonad are shown at higher magnification. Left, distal; right, proximal. (B) Row 1 (from top) shows expression patterns of PUF-8::GFP and mCherry::EMR-1, another nuclear envelope marker, in worms carrying both transgenes. Row 3, PUF-8 is fused to mCherry and the nuclear envelope is visualised using the GFP::LEM-2 marker. Row 5, expression patterns of GLD-1::GFP (shown here as a control) and mCherry::EMR-1. Rows 2, 4 and 6 are as rows 1, 3 and 5, respectively, but after treatment with leptomycin B (LMB). (C) Expression patterns of PUF-8::GFP and TCER-1::mCherry in worms carrying both transgenes. Beneath, two nuclei from each panel shown at a higher magnification. Except for A (top), images have been deconvolved using the Iterative Deconvolution module of Axiovision software to enhance the axial resolution of the fluorescence signal. (B,C) Only a part of the distal gonad, revealing a few germ cell nuclei, is shown. Scale bars: 25 μm in A (top); 10 μm in B and C (top); 5 μm in A (bottom) and C (bottom).
We confirmed the nuclear localisation of the TCER-1::mCherry fusion protein using a double-transgenic line carrying both TCER-1::mCherry and GFP::LEM-2 - a nuclear envelope marker (Gorjánácz et al., 2007) - transgenes (Fig. 2A). At higher magnification, we noticed that the TCER-1 reporter fusions were present at the inner periphery of the nucleus, often in a crescent-shaped fashion (Fig. 2A,C). However, we did not detect any fluorescence signal corresponding to either of the TCER-1 reporter fusions in the cytoplasm.
Since removal of PUF-8 did not affect TCER-1 expression (data not shown), we examined whether PUF-8 functions in the nucleus along with TCER-1. An earlier study from our laboratory reported that PUF-8 is present on P granules (Ariz et al., 2009). Since weak nuclear signals might have been missed, we re-examined these transgenic lines with the help of a nuclear envelope marker. For this, we generated a double-transgenic line that carries both PUF-8::GFP and mCherry::EMR-1 transgenes. The PUF-8::GFP transgene rescues the puf-8(-) mutant (Ariz et al., 2009), and EMR-1 serves as a nuclear envelope marker (Gorjánácz et al., 2007). As reported previously (Ariz et al., 2009), we observed a perinuclear distribution of PUF-8::GFP that is reminiscent of germ cell-specific P granules. Significantly, we noticed PUF-8::GFP in the nucleus as well, where it was distributed in a diffuse pattern at the inner periphery of the nucleus (Fig. 2B). In several nuclei, the PUF-8::GFP distribution was asymmetrical, with a crescent-shaped pattern that was strikingly similar to that of TCER-1.
To confirm the nuclear presence of PUF-8, we treated the worms with leptomycin B, a known blocker of nuclear export (Nakielny and Dreyfuss, 1999). As would be expected for a protein that shuttles between cytoplasm and nucleus, leptomycin B treatment led to the accumulation of PUF-8::GFP at the inner periphery of the nucleus (Fig. 2B). Nuclear localisation of PUF-8 revealed by PUF-8::GFP does not seem to be an artefact of GFP fusion: a similar fusion with GLD-1, which is another P granule-associated RNA-binding protein, was not present in the nucleus and did not accumulate in the nucleus following leptomycin B treatment (Fig. 2B).
To further confirm the presence of PUF-8 in the nucleus, we examined the expression pattern of PUF-8 using the mCherry reporter. Surprisingly, the nuclear presence of mCherry::PUF-8 was not obvious in the untreated worms. Since mCherry is known to have a weaker fluorescence intensity and less signal stability than GFP (Shaner et al., 2005), we suggest that the low levels of PUF-8 present at the inner nuclear periphery could not be detected with the mCherry fusion. However, previous studies have shown that leptomycin B treatment, by blocking nuclear export, can cause significant nuclear accumulation of shuttling proteins such that their nuclear presence can be readily detected (Fukuda et al., 1997; Zheng and Guan, 1994). Consistently, upon leptomycin B treatment, mCherry::PUF-8 became concentrated near the nuclear envelope, which was strikingly similar to the behaviour of PUF-8::GFP (Fig. 2B). We then generated double-transgenic lines expressing both PUF-8::GFP and TCER-1::mCherry fusions to test whether these proteins colocalise. As shown in Fig. 2C, these two proteins indeed show significant overlap in several germ cell nuclei. We conclude that PUF-8 is present in the same region of the germ cell nucleus as TCER-1.
PUF-8 functions redundantly with TCER-1 to maintain normal levels of mature mRNAs
Since the yeast and mammalian orthologues of TCER-1 have been implicated in mRNA processing (Abovich and Rosbash, 1997; Pearson et al., 2008), we investigated whether PUF-8 and TCER-1 have a role in mRNA processing. We measured the levels of unspliced and spliced versions of several mRNAs in puf-8(-), tcer-1(-) and puf-8(-) tcer-1(-) mutant worms and compared them with wild type. We selected 16 mRNAs that are known to be expressed in the germline (Kim et al., 2001) [data obtained from The Nematode Expression Pattern Database (http://nematode.lab.nig.ac.jp)]. Seven of these have recently been identified as potential targets of PUF-8 (Mainpal et al., 2011). We monitored RNA levels using quantitative RT-PCR in synchronously grown populations of worms (see Materials and methods). Selective amplification of pre-mRNAs was achieved by designing one PCR primer to match an intron and the other to match an exon. The mature mRNAs were amplified using oligonucleotides belonging to two exons separated by at least one intron as PCR primers. As the level of hip-1 mRNA, which encodes an HSP70-interacting protein, did not vary significantly among these samples, it was used to normalise variations arising from differences in the amount of RNA extracted from different samples.
As shown in Fig. 3, in the puf-8(-) tcer-1(-) double mutant, the levels of six of the 16 mRNAs that we measured, namely egg-5, pal-1, cyb-2.1, pos-1, spn-4 and oma-1 mRNAs, were only ∼10-30% of the wild type. By contrast, the pre-mRNA levels of all but one of them - the unspliced version of oma-1 mRNA could not be amplified - showed a ∼2- to 3-fold increase in the double mutant compared with the wild type. The levels of the unspliced and spliced forms of these mRNAs did not vary significantly between the wild type and single mutants. These results show that PUF-8 and TCER-1 function redundantly to maintain normal levels of the spliced form of these mRNAs.
Fig. 3.
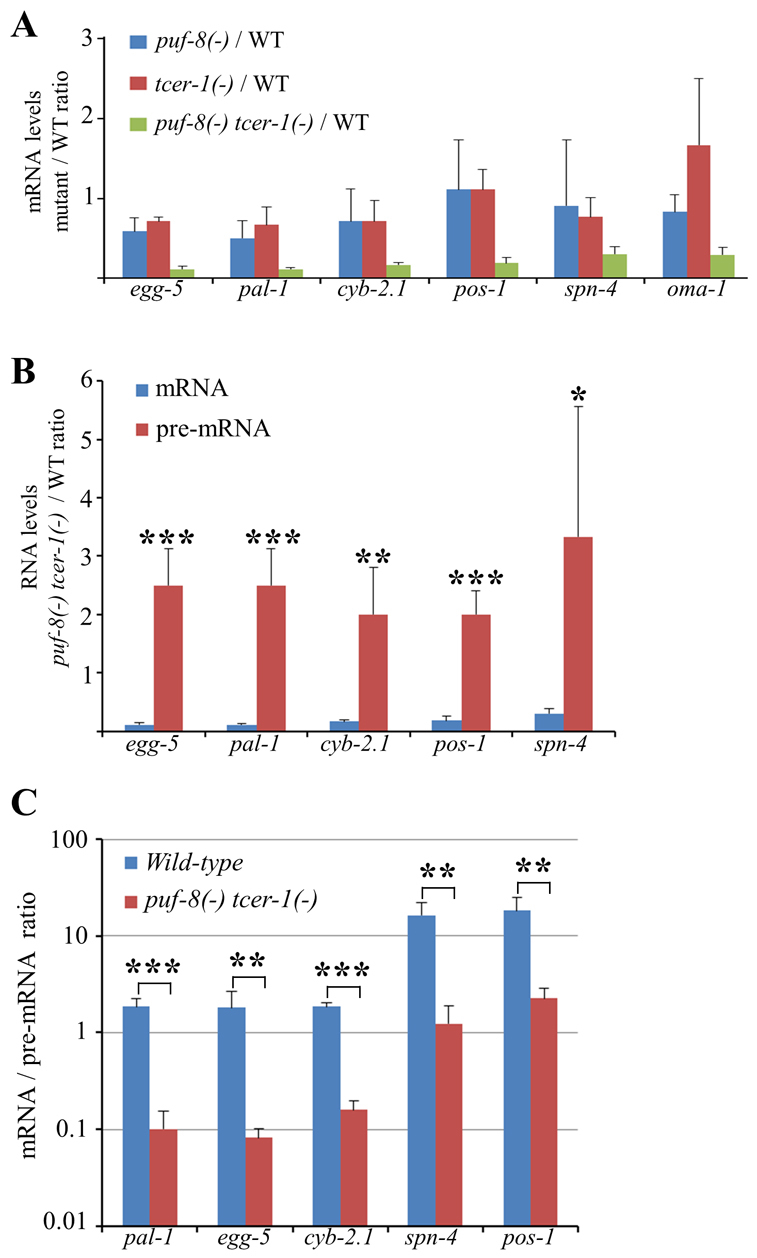
The levels of several germline mRNAs are dependent on PUF-8 and TCER-1. (A) Mutant to wild type ratios of the indicated mRNA levels quantitated by RT-PCR (see Materials and methods). (B) Comparison of the levels of the spliced (mRNA) and unspliced (pre-mRNA) versions of the indicated mRNAs in the puf-8(-) tcer-1(-) double mutant. ***P=0.0003; **P=0.0038; *P=0.0344; Student’s t-test, mRNA versus pre-mRNA levels. (C) Comparison of mRNA/pre-mRNA ratio between the wild type and the puf-8(-) tcer-1(-) double mutant. This ratio is independent of overall RNA levels, and thus eliminates potential inaccuracies arising from variations in the overall RNA levels between the wild type and double mutant. ***P=0.0001; **P=0.003; Student’s t-test. Error bars indicate s.d.
PUF-8 and TCER-1 interact with mRNA processing/export components in a yeast two-hybrid assay
The mammalian orthologue of TCER-1 has been shown to interact with the components of mRNA processing/export machineries (Deckert et al., 2006; Goldstrohm et al., 2001; Lin et al., 2004; Makarov et al., 2002; Rappsilber et al., 2002). Therefore, we selected the C. elegans orthologues of these components and tested them in yeast two-hybrid assays for their ability to interact with TCER-1. Of the 31 proteins that we were able to test (supplementary material Table S5), TCER-1 interacted with two of them, namely THOC-3 and SNR-3 (Fig. 4).
To gain insight into the nature of the functional redundancy between PUF-8 and TCER-1, we then tested whether PUF-8 similarly interacts with any of these 31 proteins. We found that PUF-8 interacted with the splicing factor PHI-11, the nuclear cap-binding protein NCBP-2, poly(A)-binding protein PAB-1, C27F2.10, which is the C. elegans orthologue of the yeast TREX 2 complex protein THP1p, and three nuclear pore complex (NPC) components, namely NPP-1, NPP-14 and RAE-1 (Fig. 4). These three NPC complex proteins are the C. elegans orthologues of the mammalian NPC components Nup54, Nup214 and Rae1, respectively (Galy et al., 2003). Their yeast orthologues are Nup57, Nup159 and Gle2p (Rodriguez et al., 2004; Strambio-De-Castillia et al., 2010). These three NPC components, the TREX 2 complex, the cap-binding complex and the poly(A)-binding protein play crucial roles in transcription-coupled mRNA processing and export (Brune et al., 2005; Iglesias and Stutz, 2008; Köhler and Hurt, 2007; Rodriguez et al., 2004). Significantly, out of the eight NPC components tested, PUF-8 showed interaction only with the three that have been directly implicated in mRNA export (Gorsch et al., 1995; Murphy et al., 1996; Terry and Wente, 2007).
In an initial attempt to establish whether PUF-8 interacts in vivo with any of the proteins identified by the yeast two-hybrid assay, we chose NCBP-2, for which specific antibodies were available, and examined whether it colocalised with PUF-8::GFP in the nucleus. We generated double-transgenic lines that express both of these proteins in the germline. We expressed NCBP-2 as a fusion protein tagged with the cMyc epitope and the mCherry reporter in the germline using the pie-1 promoter. In these lines, cMyc::NCBP-2::mCherry was present in all germ cell nuclei. We further validated this expression pattern using an antibody specific for the human nuclear cap-binding protein CBP20 (NCBP2), which is known to recognise the worm NCBP-2 in immunostaining experiments (Lall et al., 2005). Although the anti-CBP20 antibody showed intense staining near the inner nuclear periphery, it also showed some punctate staining in the cytoplasm, which is potentially due to cross-reactivity with a cytoplasmic protein that shares similarity with NCBP-2. We confirmed the nuclear localisation by generating double-transgenic lines expressing both cMyc::mCherry::NCBP-2 and the nuclear envelope marker GFP::LEM-2.
If NCBP-2 indeed interacts with PUF-8, then these proteins would colocalise. To test this, we generated double-transgenic lines expressing both cMyc::mCherry::NCBP-2 and PUF-8::GFP fusion proteins. As shown in Fig. 5, these proteins exhibited significant colocalisation in the nucleus. Unfortunately, owing to the low levels of PUF-8::GFP, we were unable to detect it in western blots and, as a consequence, we could not test the interaction between NCBP-2 and PUF-8 using the co-immunoprecipitation strategy.
Fig. 5.
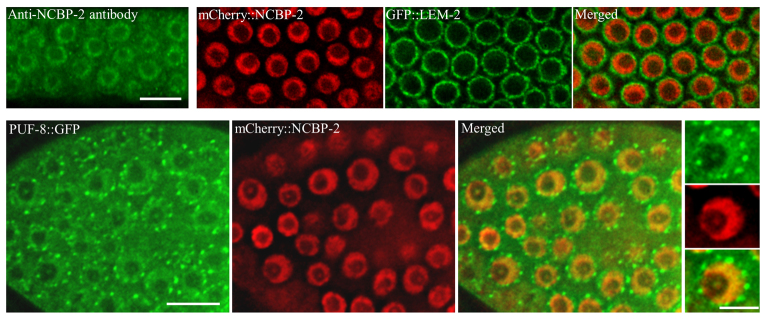
PUF-8 colocalises with the nuclear cap-binding protein NCBP-2. The expression pattern of NCBP-2 as revealed by immunostaining with anti-human CBP20 antibody, the human orthologue of NCBP-2, is shown top left. All other images show the expression patterns of the indicated transgene-expressed fusion proteins. In the bottom row, single nuclei from each panel are shown at higher magnification to the right. Scale bars: 10 μm (top and bottom left); 5 μm (bottom right).
The N-terminal domain of PUF-8 is essential for interaction with mRNA processing/export components in the yeast two-hybrid assay and for redundancy with TCER-1
To uncover the biological significance of the interactions revealed by the yeast two-hybrid assays, we sought to define the amino acids of PUF-8 that are crucial for these interactions, and then test whether these residues are crucial for PUF-8 function in vivo. We generated a series of truncations and deletions of PUF-8 and tested their ability to interact with three of the interactors described above, namely NCBP-2, RAE-1 and C27F2.10, in the yeast two-hybrid assay. This analysis revealed that the N-terminal, and not the PUF domain-containing C-terminal, region of PUF-8 is necessary and sufficient for the interactions. Further, amino acids 51-100 were found to be crucial for the ability of PUF-8 to interact with all three proteins (Fig. 6A).
Fig. 6.
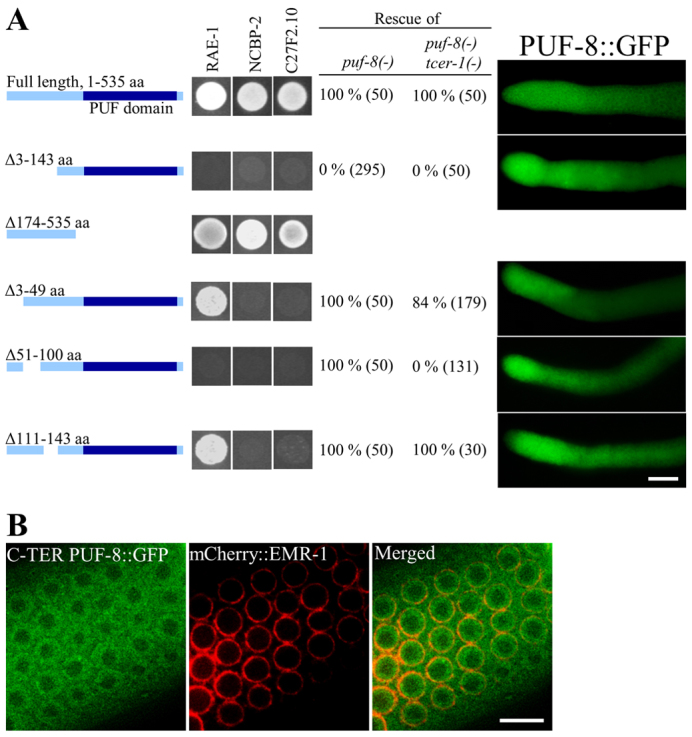
A 50 amino acid N-terminal domain of PUF-8 is crucial for both protein-protein interactions and functional redundancy with TCER-1. (A) (Left) Yeast two-hybrid assay as in Fig. 4 but using the BD fusions of various PUF-8 N-terminal deletions and the AD fusions indicated above each panel. The regions deleted are illustrated alongside. (Centre) Rescue of the indicated mutant strain by puf-8::gfp transgenes bearing the same deletions used in the yeast two-hybrid assay. The number of worms observed is given in parenthesis. (Right) Expression patterns of the corresponding transgenes in the distal gonad as visualised by GFP fluorescence. The level and pattern of expression were approximately the same for all the transgenes. Transgenic lines bearing deletion of amino acids 174-535 could not be generated. (B) Expression patterns of the PUF-8 C-terminal region (amino acids 144-535) fused to GFP and the nuclear envelope marker mCherry::EMR-1 in worms carrying both transgenes. Scale bars: 20 μm in A; 10 μm in B.
We then generated transgene constructs carrying similar deletions in PUF-8 and tested their ability to rescue the puf-8(-) and puf-8(-)tcer-1(-) mutant worms (Fig. 5A). These constructs express the PUF-8 open reading frame as a PUF-8::GFP fusion under the control of puf-8 upstream and downstream sequences. As reported above, the transgene with the full-length PUF-8 coding region restored fertility to puf-8(-) mutant worms. In addition, we found that this full-length construct could restore fertility to the puf-8(-) tcer-1(-) double mutant as well. By contrast, the transgene expressing only the C-terminal region of PUF-8 (amino acids 144-535) failed to rescue the puf-8(-) single mutant itself. Deletion of amino acids 3-49 or 111-143 did not significantly affect the ability of the transgene to rescue puf-8(-) or puf-8(-) tcer-1(-) worms. By contrast, deletion of amino acids 51-100, which were crucial for PUF-8 interaction with RAE-1, NCBP-2 and C27F2.10 in the yeast two-hybrid assay, failed to rescue the puf-8(-) tcer-1(-) double mutant. Strikingly, this deletion was able to rescue the puf-8(-) single mutant (Fig. 6A). To rule out the possibility that some of these constructs were not expressed in the germline, we monitored their expression pattern and level by GFP fluorescence (Fig. 6A).
These results indicate that the N-terminal domain of PUF-8 is crucial for its ability to function redundantly with TCER-1 and suggest that the interactions identified by the yeast two-hybrid experiments might indeed be relevant in vivo for PUF-8 function. As shown in Fig. 6B, the C-terminal half of PUF-8 itself localises to the nucleus, which indicates that the nuclear localisation signal is present within the PUF domain. As a consequence, we were unable to test whether nuclear localisation is essential for PUF-8 to function redundantly with TCER-1.
DISCUSSION
The results presented here uncover two unexpected aspects of PUF proteins. One is the presence of PUF-8 in the nucleus and the requirement of PUF-8 for normal levels of several germline mRNAs. This is surprising because PUF proteins have been thought to function in the cytoplasm and to reduce mRNA levels by recruiting the deadenylase complex (Goldstrohm et al., 2006; Goldstrohm et al., 2007). The second novel aspect of these findings is the importance of the PUF-8 N-terminal domain. Although all PUF proteins contain a large N-terminal region, ranging from 200 to 1000 amino acids, its sequence has not been well conserved and its function is not known. In the case of Drosophila Pumilio, the PUF domain seems to be sufficient for mRNA binding, interaction with other protein partners and to rescue the mutant phenotype (Wharton et al., 1998; Sonoda and Wharton, 1999). As a consequence, the N-terminal part of PUF proteins has been largely ignored. Contrary to this, the results of our deletion analysis reveal that this region is functionally important in the case of PUF-8. We predict that the N-terminal domain of other PUF proteins might have similar essential functions.
We find that the levels of six germline mRNAs are significantly reduced in puf-8(-) tcer-1(-) double-mutant worms. There are a number of possible reasons for this. First, the difference is due to there being fewer germ cells in the mutant. Second, the effect is at the transcriptional level. These two possibilities are, however, inconsistent with the 2- to 3-fold increase in unspliced mRNAs that we observed in the double mutant compared with the wild type. A third possibility is that the mRNAs are more rapidly degraded in the cytoplasm of the double mutant. Our current results do not completely rule out this possibility - it would have been more convincing had we seen mRNA accumulation in the nucleus by in situ hybridisation. Unfortunately, we were unable to detect in situ hybridisation signals in the nucleus, presumably owing to very low levels of nuclear RNA. Since the accumulation of aberrantly processed mRNAs will trigger the nuclear degradation machinery, nuclear accumulation might not be in proportion to the level of reduction of processed mRNAs. However, we do not favour the cytoplasmic mRNA degradation model for the following reasons. First, we did not see any expression of TCER-1 outside the nucleus, which is consistent with the known expression patterns of its yeast and mammalian orthologues. Thus, of the two proteins, only PUF-8 is present in the cytoplasm. If the observed reduction in mRNA levels were due to cytoplasmic degradation, then we should have seen the reduction in the puf-8(-) single mutant itself. Second, PUF proteins are known to promote the degradation, rather than stability, of mRNAs in the cytoplasm. Finally, as mentioned above, we do see an increase in the levels of unspliced mRNAs in the double mutant (Fig. 3), which points to defects in nuclear mRNA processing or export.
How do PUF-8 and TCER-1 function to maintain mRNA levels? We propose a model in which both proteins promote nuclear mRNA processing and/or export. We speculate that TCER-1, just like its mammalian counterpart, functions as part of the general transcription-coupled export machinery, whereas PUF-8, through its ability to bind specific mRNAs and to interact with proteins involved in mRNA export, such as NCBP-2, provides an alternative export pathway for some of the mRNAs. This model is consistent both with our current results and the existing literature. Of the eight NPC components tested in the yeast two-hybrid assay, PUF-8 interacted only with those whose yeast orthologues have been shown to directly function in mRNA export. Further, PUF-8 colocalises with NCBP-2, the yeast and mammalian orthologues of which function in mRNA export.
Importantly, we find that the N-terminal region crucial for these interactions is essential for the ability of PUF-8 to function redundantly with TCER-1. Similarly, five out of the six mRNAs with significantly reduced levels in the puf-8(-) tcer-1(-) double mutant have been shown previously to affinity purify with PUF-8 (Mainpal et al., 2011). It is important to note that the N-terminus is not essential for the nuclear localisation of PUF-8, as the C-terminal part alone readily localises to the nucleus (Fig. 6B). Thus, the interactions with mRNA export factors do not appear to be crucial for the ability of PUF-8 to enter the nucleus. Although this might be the first report of a nuclear role for a metazoan PUF protein, the nuclear localisation and involvement in RNA processing of PUF proteins have been documented in yeast and plants. For example, the yeast PUF protein Nop9p and the trypanosome PUF7 localise to the nucleolus and function in rRNA maturation (Droll et al., 2010; Thomson et al., 2007). Similar observations have been reported for the Arabidopsis PUF protein APUM23 (Abbasi et al., 2010).
As per our model, the presence of PUF-8 in the nucleus will be essential for its function in mRNA processing or export. We were unable to test this because the nuclear targeting signal seems to be within the conserved PUF domain. Our attempts to generate transgenic lines expressing the PUF domain of other worm PUF proteins, which would have established whether they too enable nuclear localisation, did not succeed. However, the PUF domain of human PUM1 has been shown to readily translocate into the nucleus (Kedde et al., 2010), suggesting that nuclear localisation is a conserved feature of the PUF domain. Specific mutations within the PUF domain will be required to identify the nuclear targeting signal, if any. Another prediction is that the mRNA binding is essential for the proposed nuclear function of PUF-8. We are currently testing this prediction using transgenic reporter fusions with the mRNAs identified in this study.
Supplementary Material
Acknowledgments
We thank Geraldine Seydoux for critical reading of the manuscript and for several useful suggestions; the anonymous reviewers for their valuable comments and suggestions; Alex Hajnal for puf-8(zh17); the Mitani laboratory for tcer-1(tm1452) and zk1127.6(tm2782); Susan Strome for SS747; and the C. elegans Genetics Consortium for XA3507 strains.
Footnotes
Funding
K.P. is a recipient of a National Doctoral Fellowship from the All India Council of Technical Education. This work was supported by the Wellcome Trust and the National Agricultural Innovation Project (NAIP) scheme of the Indian Council of Agricultural Research. Deposited in PMC for immeidate release.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.087833/-/DC1
References
- Abbasi N., Kim H. B., Park N. I., Kim H. S., Kim Y. K., Park Y. I., Choi S. B. (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64, 960–976 [DOI] [PubMed] [Google Scholar]
- Abovich N., Rosbash M. (1997). Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89, 403–412 [DOI] [PubMed] [Google Scholar]
- Ariz M., Mainpal R., Subramaniam K. (2009). C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germline stem cell mitosis. Dev. Biol. 326, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachorik J. L., Kimble J. (2005). Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc. Natl. Acad. Sci. USA 102, 10893–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune C., Munchel S. E., Fischer N., Podtelejnikov A. V., Weis K. (2005). Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA 11, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Bernstein D. S., Bachorik J. L., Thompson B. E., Gallegos M., Petcherski A. G., Moulder G., Barstead R., Wickens M., Kimble J. (2002). A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417, 660–663 [DOI] [PubMed] [Google Scholar]
- Deckert J., Hartmuth K., Boehringer D., Behzadnia N., Will C. L., Kastner B., Stark H., Urlaub H., Lührmann R. (2006). Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26, 5528–5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Singer R. H., Gu W. (2008). Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes Dev. 22, 1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droll D., Archer S., Fenn K., Delhi P., Matthews K., Clayton C. (2010). The trypanosome Pumilio-domain protein PUF7 associates with a nuclear cyclophilin and is involved in ribosomal RNA maturation. FEBS Lett. 584, 1156–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A., Lehmann R. (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679–690 [DOI] [PubMed] [Google Scholar]
- Fukuda M., Asano S., Nakamura T., Adachi M., Yoshida M., Yanagida M., Nishida E. (1997). CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308–311 [DOI] [PubMed] [Google Scholar]
- Galgano A., Forrer M., Jaskiewicz L., Kanitz A., Zavolan M., Gerber A. P. (2008). Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE 3, e3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Mattaj I. W., Askjaer P. (2003). Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol. Biol. Cell 14, 5104–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A. P., Herschlag D., Brown P. O. (2004). Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi A., Henis-Korenblit S., Kenyon C. (2009). A transcription elongation factor that links signals from the reproductive system to lifespan extension in Caenorhabditis elegans. PLoS Genet. 5, e1000639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- Goldstrohm A. C., Albrecht T. R., Suñé C., Bedford M. T., Garcia-Blanco M. A. (2001). The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21, 7617–7628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm A. C., Hook B. A., Seay D. J., Wickens M. (2006). PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13, 533–539 [DOI] [PubMed] [Google Scholar]
- Goldstrohm A. C., Seay D. J., Hook B. A., Wickens M. (2007). PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 282, 109–114 [DOI] [PubMed] [Google Scholar]
- Gorjánácz M., Jaedicke A., Mattaj I. W. (2007). What can Caenorhabditis elegans tell us about the nuclear envelope? FEBS Lett. 581, 2794–2801 [DOI] [PubMed] [Google Scholar]
- Gorsch L. C., Dockendorff T. C., Cole C. N. (1995). A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129, 939–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N., Stutz F. (2008). Regulation of mRNP dynamics along the export pathway. FEBS Lett. 582, 1987–1996 [DOI] [PubMed] [Google Scholar]
- Jadhav S., Rana M., Subramaniam K. (2008). Multiple maternal proteins coordinate to restrict the translation of C. elegans nanos-2 to primordial germ cells. Development 135, 1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova L. Y., Habara Y., Lee T. H., Wharton R. P. (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519–1527 [DOI] [PubMed] [Google Scholar]
- Kaye J. A., Rose N. C., Goldsworthy B., Goga A., L’Etoile N. D. (2009). A 3’UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron 61, 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J. A., Elkon R., Agami R. (2010). A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 12, 1014–1020 [DOI] [PubMed] [Google Scholar]
- Kershner A. M., Kimble J. (2010). Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. USA 107, 3936–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. K., Lund J., Kiraly M., Duke K., Jiang M., Stuart J. M., Eizinger A., Wylie B. N., Davidson G. S. (2001). A gene expression map for Caenorhabditis elegans. Science 293, 2087–2092 [DOI] [PubMed] [Google Scholar]
- Köhler A., Hurt E. (2007). Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8, 761–773 [DOI] [PubMed] [Google Scholar]
- Lall S., Piano F., Davis R. E. (2005). Caenorhabditis elegans decapping proteins: localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol. Biol. Cell 16, 5880–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K. T., Lu R. M., Tarn W. Y. (2004). The WW domain-containing proteins interact with the early spliceosome and participate in pre-mRNA splicing in vivo. Mol. Cell. Biol. 24, 9176–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainpal R., Priti A., Subramaniam K. (2011). PUF-8 suppresses the somatic transcription factor PAL-1 expression in C. elegans germline stem cells. Dev. Biol. 360, 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov E. M., Makarova O. V., Urlaub H., Gentzel M., Will C. L., Wilm M., Lührmann R. (2002). Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298, 2205–2208 [DOI] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G. (2008). 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18, 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A. R., Mukherjee N., Keene J. D. (2008). Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol. Cell. Biol. 28, 4093–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y., Wharton R. P. (1995). Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80, 747–756 [DOI] [PubMed] [Google Scholar]
- Murphy R., Watkins J. L., Wente S. R. (1996). GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell 7, 1921–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S., Dreyfuss G. (1999). Transport of proteins and RNAs in and out of the nucleus. Cell 99, 677–690 [DOI] [PubMed] [Google Scholar]
- Pearson J. L., Robinson T. J., Muñoz M. J., Kornblihtt A. R., Garcia-Blanco M. A. (2008). Identification of the cellular targets of the transcription factor TCERG1 reveals a prevalent role in mRNA processing. J. Biol. Chem. 283, 7949–7961 [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Ryder U., Lamond A. I., Mann M. (2002). Large-scale proteomic analysis of the human spliceosome. Genome Res. 12, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M. S., Dargemont C., Stutz F. (2004). Nuclear export of RNA. Biol. Cell 96, 639–655 [DOI] [PubMed] [Google Scholar]
- Schweers B. A., Walters K. J., Stern M. (2002). The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics 161, 1177–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Steinbach P. A., Tsien R. Y. (2005). A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909 [DOI] [PubMed] [Google Scholar]
- Sonoda J., Wharton R. P. (1999). Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M. P. (2010). The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11, 490–501 [DOI] [PubMed] [Google Scholar]
- Strome S., Powers J., Dunn M., Reese K., Malone C. J., White J., Seydoux G., Saxton W. (2001). Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol. Biol. Cell 12, 1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K., Seydoux G. (2003). Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Curr. Biol. 13, 134–139 [DOI] [PubMed] [Google Scholar]
- Tadauchi T., Matsumoto K., Herskowitz I., Irie K. (2001). Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20, 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Wente S. R. (2007). Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178, 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E., Rappsilber J., Tollervey D. (2007). Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA 13, 2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaid S., Ariz M., Chaturbedi A., Anil Kumar G., Subramaniam K. (2013). PUF-8 negatively regulates RAS/MAPK signalling to promote differentiation of C. elegans germ cells. Development (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser C. B., Battu G., Hoier E. F., Hajnal A. (2006). Distinct roles of the Pumilio and FBF translational repressors during C. elegans vulval development. Development 133, 3461–3471 [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Sonoda J., Lee T., Patterson M., Murata Y. (1998). The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1, 863–872 [DOI] [PubMed] [Google Scholar]
- Wickens M., Bernstein D. S., Kimble J., Parker R. (2002). A PUF family portrait: 3’UTR regulation as a way of life. Trends Genet. 18, 150–157 [DOI] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J., Wickens M. P. (1997). A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390, 477–484 [DOI] [PubMed] [Google Scholar]
- Zheng C.-F., Guan K.-L. (1994). Cytoplasmic localization of the mitogen-activated protein kinase activator MEK. J. Biol. Chem. 269, 19947–19952 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.