Abstract
Exon sizes in vertebrate genes are, with a few exceptions, limited to less than 300 bases. It has been proposed that this limitation may derive from the exon definition model of splice site recognition. In this model, a downstream donor site enhances splicing at the upstream acceptor site of the same exon. This enhancement may require contact between factors bound to each end of the exon; an exon size limitation would promote such contact. To test the idea that proximity was required for exon definition, we inserted random DNA fragments from Escherichia coli into a central exon in a three-exon dihydrofolate reductase minigene and tested whether the expanded exons were efficiently spliced. DNA from a plasmid library of expanded minigenes was used to transfect a CHO cell deletion mutant lacking the dhfr locus. PCR analysis of DNA isolated from the pooled stable cotransfectant populations displayed a range of DNA insert sizes from 50 to 1,500 nucleotides. A parallel analysis of the RNA from this population by reverse transcription followed by PCR showed a similar size distribution. Central exons as large as 1,400 bases could be spliced into mRNA. We also tested individual plasmid clones containing exon inserts of defined sizes. The largest exon included in mRNA was 1,200 bases in length, well above the 300-base limit implied by the survey of naturally occurring exons. We conclude that a limitation in exon size is not part of the exon definition mechanism.
Full text
PDF
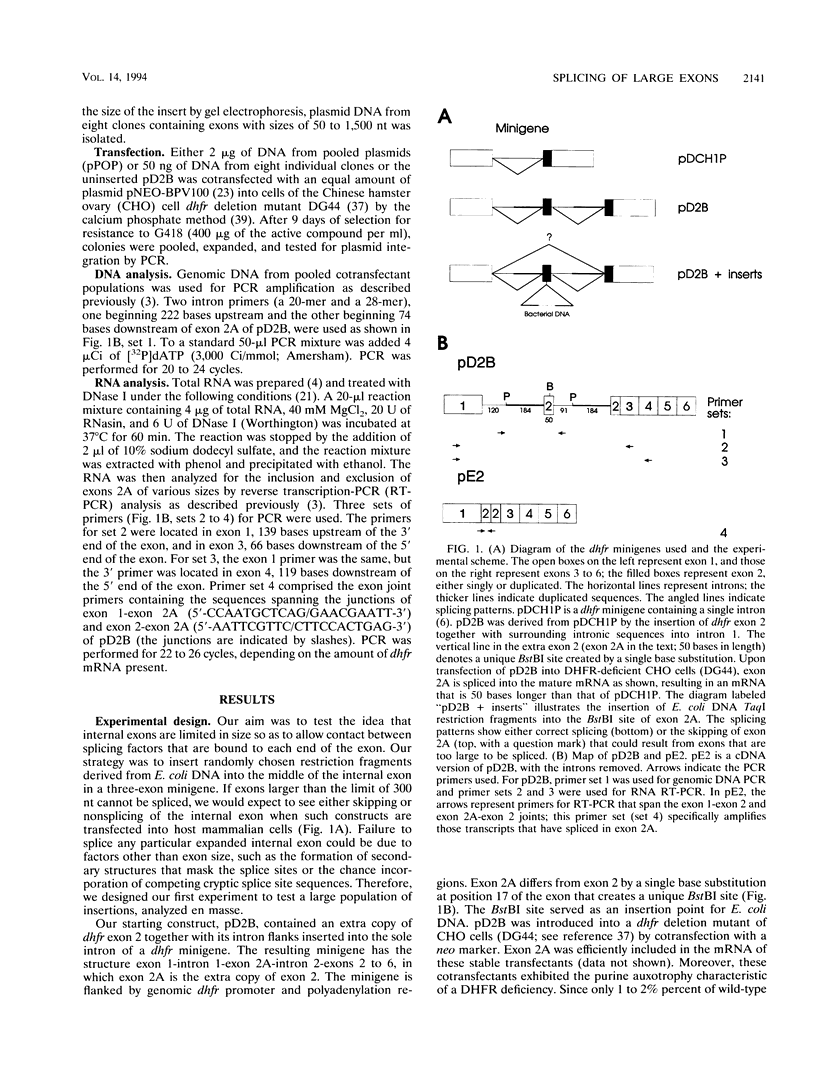



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carothers A. M., Urlaub G., Grunberger D., Chasin L. A. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1993 Aug;13(8):5085–5098. doi: 10.1128/mcb.13.8.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Mucha J., Grunberger D., Chasin L. A. Point mutation analysis in a mammalian gene: rapid preparation of total RNA, PCR amplification of cDNA, and Taq sequencing by a novel method. Biotechniques. 1989 May;7(5):494-6, 498-9. [PubMed] [Google Scholar]
- Chen I. T., Chasin L. A. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol Cell Biol. 1993 Jan;13(1):289–300. doi: 10.1128/mcb.13.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chu E., Voeller D., Koeller D. M., Drake J. C., Takimoto C. H., Maley G. F., Maley F., Allegra C. J. Identification of an RNA binding site for human thymidylate synthase. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):517–521. doi: 10.1073/pnas.90.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciudad C. J., Urlaub G., Chasin L. A. Deletion analysis of the Chinese hamster dihydrofolate reductase gene promoter. J Biol Chem. 1988 Nov 5;263(31):16274–16282. [PubMed] [Google Scholar]
- Crouse G. F., McEwan R. N., Pearson M. L. Expression and amplification of engineered mouse dihydrofolate reductase minigenes. Mol Cell Biol. 1983 Feb;3(2):257–266. doi: 10.1128/mcb.3.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. E., Jr Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science. 1978 Dec 22;202(4374):1257–1260. doi: 10.1126/science.364651. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Furdon P. J., Kole R. The length of the downstream exon and the substitution of specific sequences affect pre-mRNA splicing in vitro. Mol Cell Biol. 1988 Feb;8(2):860–866. doi: 10.1128/mcb.8.2.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. Genes-in-pieces revisited. Science. 1985 May 17;228(4701):823–824. doi: 10.1126/science.4001923. [DOI] [PubMed] [Google Scholar]
- Goossen B., Hentze M. W. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol Cell Biol. 1992 May;12(5):1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski P. J., Nasim F. U., Kuo H. C., Burch R. Combinatorial splicing of exon pairs by two-site binding of U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1991 Dec;11(12):5919–5928. doi: 10.1128/mcb.11.12.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg J., Maguire S., Belluscio L. A simple method for the preparation of plasmid and chromosomal E. coli DNA. Nucleic Acids Res. 1989 Nov 11;17(21):8893–8893. doi: 10.1093/nar/17.21.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler O., Jiang Y., Chasin L. A. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol Cell Biol. 1993 Oct;13(10):6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M., Reiss J., Cooper D. N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992 Sep-Oct;90(1-2):41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Kreivi J. P., Zerivitz K., Zefrivitz K., Akusjärvi G. A U1 snRNA binding site improves the efficiency of in vitro pre-mRNA splicing. Nucleic Acids Res. 1991 Dec 25;19(24):6956–6956. doi: 10.1093/nar/19.24.6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K. M., Spritz R. A. In vitro splicing pathways of pre-mRNAs containing multiple intervening sequences? Mol Cell Biol. 1987 Oct;7(10):3428–3437. doi: 10.1128/mcb.7.10.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980 Sep 19;209(4463):1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Rastinejad F., Blau H. M. Genetic complementation reveals a novel regulatory role for 3' untranslated regions in growth and differentiation. Cell. 1993 Mar 26;72(6):903–917. doi: 10.1016/0092-8674(93)90579-f. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Smith M. W. Structure of vertebrate genes: a statistical analysis implicating selection. J Mol Evol. 1988;27(1):45–55. doi: 10.1007/BF02099729. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Steingrimsdottir H., Rowley G., Dorado G., Cole J., Lehmann A. R. Mutations which alter splicing in the human hypoxanthine-guanine phosphoribosyltransferase gene. Nucleic Acids Res. 1992 Mar 25;20(6):1201–1208. doi: 10.1093/nar/20.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut T. W. Do exons code for structural or functional units in proteins? Proc Natl Acad Sci U S A. 1988 May;85(9):2944–2948. doi: 10.1073/pnas.85.9.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M. J., Ting A. C., Nordstrom J. L., Zimmer W., O'Malley B. W. Processing of high molecular weight ovalbumin and ovomucoid precursor RNAs to messenger RNA. Cell. 1980 Nov;22(1 Pt 1):219–230. doi: 10.1016/0092-8674(80)90170-1. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Kas E., Chasin L. A., Funanage V. L., Myoda T. T., Hamlin J. Effect of gamma rays at the dihydrofolate reductase locus: deletions and inversions. Somat Cell Mol Genet. 1986 Nov;12(6):555–566. doi: 10.1007/BF01671941. [DOI] [PubMed] [Google Scholar]
- Weil D., Brosset S., Dautry F. RNA processing is a limiting step for murine tumor necrosis factor beta expression in response to interleukin-2. Mol Cell Biol. 1990 Nov;10(11):5865–5875. doi: 10.1128/mcb.10.11.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R., Urlaub G., Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]