Abstract
The majority of available cardiomyocyte markers are intercellular proteins, limiting our ability to enrich live cardiomyocytes from heterogeneous cell preparations in the absence of genetic labeling. Here, we describe enrichment of live cardiomyocytes from the hearts of adult mice in a label-free microfluidic approach. The separation device consisted of a vertical column (15 mm long, 700 μm diameter), placed between permanent magnets resulting in a field strength of 1.23 T. To concentrate the field at the column wall, the column was wrapped with 69 μm diameter nickel wire. Before passing the cells through the column, the cardiomyocytes in the cell suspension had been rendered paramagnetic by treatment of the adult mouse heart cell preparation with sodium nitrite (2.5 mM) for 20 min on ice. The cell suspension was loaded into the vertical column from the top and upon settling, the non-myocytes were removed by the upward flow from the column. The cardiomyocytes were then collected from the column by applying a higher flow rate (144 μl/min). We found that by applying a separation flow rate of 4.2 μl/min in the first step, we can enrich live adult cardiomyocytes to 93% ± 2% in a label-free manner. The cardiomyocytes maintained viability immediately after separation and upon 24 h in culture.
INTRODUCTION
Chronic cardiovascular disease such as heart failure is increasing to epidemic levels, affecting 1 in 5 persons. The beating heart muscle has no significant ability to regenerate and the viable tissue remaining after an injury, such as myocardial infarction, is often insufficient to maintain adequate cardiac output.1 Heart transplant is often not an available or appropriate option. Thus, there is a pressing need for alternative interventions2, 3 through innovative therapeutic solutions enabled by tissue engineering. Since cardiomyocytes (CM), the beating cells of the heart, are terminally differentiated, they cannot be propagated from the heart biopsies of adult patients. Recent advances in the stem cell field enable derivation of CM from embryonic stem cells (ESCs)4 or induced pluripotent stem cells (iPSCs).5 However, engineering advances are required to enable label-free separation of these cells from heterogeneous populations in a cost-effective manner.
To our knowledge, there are no label-free separation techniques to isolate CM at high purity and yield. Prior to the recent discovery of a signal regulatory protein α (SIRPA) as a CM surface marker6 all of the available cardiac markers were intracellular proteins, thus antibody staining or genetic labeling had to be used for cell isolation. The antibody staining of intracellular markers such as contractile proteins requires cell permeabilization, which unfortunately renders the cells non-viable and useless for cardiac cell therapy or tissue engineering. Genetic labeling of cells for clinical applications cannot be performed in humans due to ethical concerns. A mitochondrial die, Tetramethylrhodamine (TMRM), was reported effective in labeling and enrichment of CM;7 however, cells labeled with fluorescent probes cannot be used for clinical applications due to the unknown long-term effects of these organic molecules in humans. Although the newly identified SIRPA6 acts as a marker of CM derived from human pluripotent stem cells, the wide applicability of the SIRPA antibody is yet to be determined. In addition, separation of cells for clinical application using mouse derived antibodies may cause sensitization in patients and development of anti-mouse antibodies.8 Other separation approaches, such as dielectrophoresis,9 are yet to be extensively studied with CM due to the fact that electrical fields can affect these electrically excitable cells. Using engineering principles to achieve high efficiency label-free separation of CM will have a significant impact even when the CM surface markers are identified, as expensive antibodies will not be required.
Isolation methods that rely on inherent physical properties of cells have been used for label-free separation of cells. For example, Murthy et al. used a sieve-like microfluidic device to10 demonstrate the feasibility of enriching fibroblasts from CM on the basis of size. The high purity enrichment of CM from other cell types such as fibroblasts remains challenging because of the dimensional similarity of the suspended CM to other cell types including large fibroblasts present in native heart isolates.11 Another set of techniques relies of differential adhesion properties. In native heart isolates, CM are a cell type that takes the longest time to attach to the surfaces of tissue culture plates. Non-myocytes, such as fibroblasts, take significantly less time. These characteristics form a basis of an enrichment technique called pre-plating. The native heart isolate is incubated in a tissue culture plate for 1 h, during which fibroblasts preferentially adhere to the surface of the tissue culture plate and CM remain in suspension. If these steps are sequentially repeated, enrichment of CM to over 80% can be achieved.12 Although simple, this technique is non-specific and leads to the loss of cell viability as many pre-plating steps are repeated in sequence.
Amongst all inherent physical properties of cells, perhaps, magnetic properties are the most relevant for isolation of CM. CM contain a high amount of iron due to the presence of myoglobin. Under physiological conditions, myoglobin contains Fe2+. Upon treatment of CM with molecules such as NaNO2, the cells can be rendered transiently paramagnetic by oxidation of myoglobin to metmyoglobin. Metmyoglobin consists of a backbone of eight helices wrapping around a central pocket containing a prosthetic protoheme group, a stable compound of ferric iron, Fe3+.13 Metmyoglobin is paramagnetic from the isolated coordination complex with an unpaired d-electron.14 In principle, the realignment of the unpaired electrons in the ferric iron in the direction of an externally applied magnetic field should direct the paramagnetic CM to move along the magnetic field gradient.
Magnetic separation has been successfully implemented for the identification and isolation of red blood cells (RBCs) by taking advantage of the high level of iron in hemoglobin. Zborowski et al. used a magnetic field of 1.4 T, and mean gradient of 0.131 T/mm and showed that separation based on the magnetic properties of RBCs is possible.15 Han and Frazier, through a series of papers, reported magnetophoretic separation of RBCs,16, 17 while Huang et al. developed a multi-step microfluidic system for the isolation of nucleated RBCs from blood of pregnant women.18 The first module in their device depleted the non-nucleated RBCs. The magnetic module of their device, then, separated the white blood cells from nucleated RBCs with purity of over 99.90%. Interesting approaches, such as capture of beads by magnetotactic bacteria, to directly and actively transport the beads along the magnetic field lines were reported.19 However, this approach is not applicable to CM since they have limited surface markers and cannot be selectively attached to magnetophoretic bacteria.
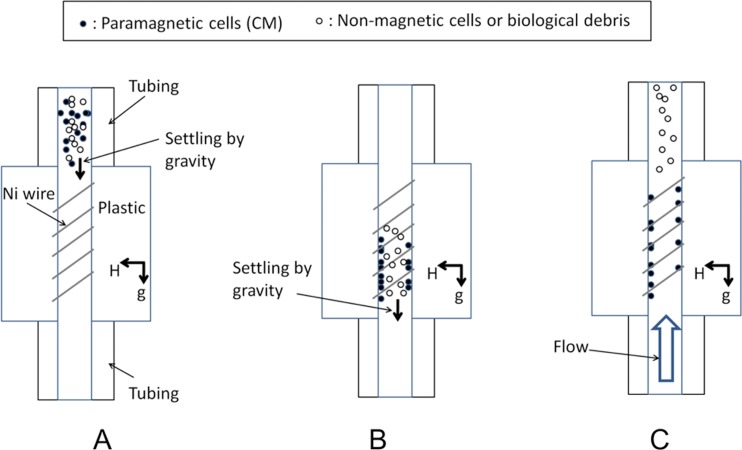
Other magnetophoretic devices, described above, also cannot be directly applied to the separation of CM from heterogeneous cell preparations since the amount of iron per volume of a CM is ∼1000 times less than the amount of iron in RBCs. Thus, it was thought in the past that CM cannot be isolated from heterogeneous cell populations using their magnetic properties and that only RBCs are suitable for this kind of separation.20 Here, it is first demonstrated that the non-magnetic CM can be rendered paramagnetic by treating the cells with a solution of NaNO2. We then report enrichment results obtained from a microfluidic device that relies on the manipulation of the settling velocity of the cells by magnetic force (Figure 1).
Figure 1.
Mechanism of action for cell separation. (a) Cell suspension is loaded into a vertical column placed in a magnetic field. (b) Paramagnetic cells are attracted to the column wall where magnetic field is concentrated due to the circumferentially positioned nickel wire. (c) Non-paramagnetic cells are rinsed out of the column by application of the flow at the bottom inlet to the column.
THEORETICAL ANALYSIS: FORCE BALANCE ON CARDIOMYOCYTES IN THE SEPARATION COLUMN
The separation mechanism is based on a high gradient magnetic separation method.21 We used magnetic force effects in conjunction with the settling velocity of the paramagnetic cells in the separation column in order to affect the paramagnetic cell trajectory and achieve capture of the target cells in the column. In our design, the column was vertical (Figure 1). The media flowed from the bottom to the top and pushed the cells upward. However, the magnet attracted CM toward the boundary layer of the flow, where the flow velocity was less than in the central parts of the column (Figure 5a). By adjusting the flow rate such that the settling velocity of CM due to gravity and the magnetic field remained larger than the upward velocity of CM within the boundary layer of the fluid flow resulted in CM being trapped within the channel, while the rest of the cells (non-myocytes) were carried out with the flow (Figure 1). The desired cell type, CM, was then released from the column in the second step by increasing the rinsing flow rate.
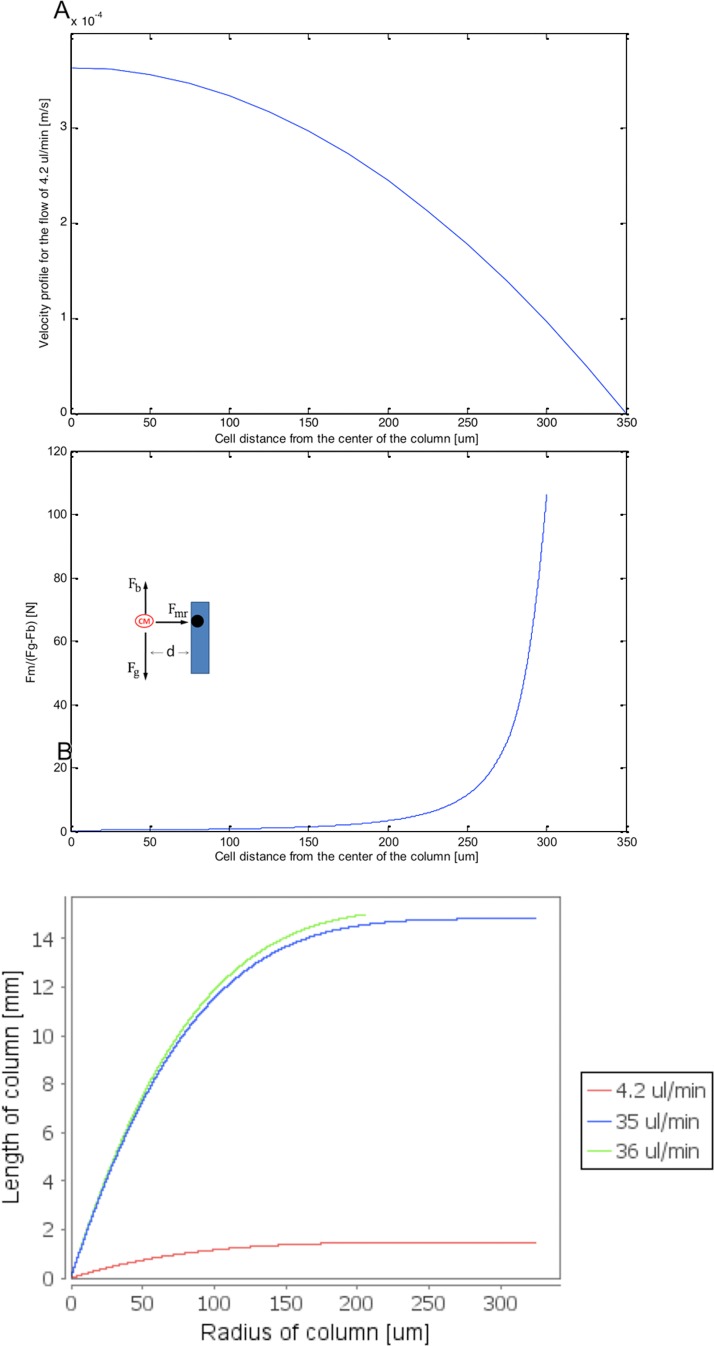
Figure 5.
Mathematical modeling results of hydrodynamic and magnetic force effects on a paramagnetic particle trajectory in the vertical separation column. (a) Theoretical velocity profile of the fluid flow at within a diameter vertical column. (b) Balance of magnetic and gravitational forces in the vertical separation column as a function of radial position in the column. (c) Trajectories of a paramagnetic particle initially placed in the center at the bottom of the column as a function of fluid flow rate (4.2, 9.5, or 10 μl/min). Column length of 15 mm is assumed.
In this study, it was necessary to generate a strong magnetic field and magnetic gradient at the vertical column wall. Calculation of an average flow rate and applied magnetic force for the paramagnetic particles inside the column was performed in order to clarify the effect of magnetic field gradient on the accumulation of the paramagnetic CM in the column.
A small paramagnetic particle that travels in a fluid stream in the presence of a wrapped magnetized wire experiences magnetic, hydrodynamic, and gravitational forces. The balance of forces which describes the particle motion is given by the following equation:
| (1) |
where is the magnetic force, the gravitational force, buoyancy, and the drag force.
The magnetic force acting on a magnetic particle is proportional to the applied magnetic field H and magnetic field gradient
| (2) |
where is the magnetic permeability of vacuum, is the particle volume, and is the particle magnetic susceptibility. In order to examine the value of magnetic force and the boundary fluid flow required for accumulation of paramagnetic particles in the column, the trajectory of a paramagnetic particle moving as a result of flow in the vertical column under the magnetic field gradient was calculated.
The general configuration of the particle motion problem and a 2D schematic of the particle control system utilized for modeling the targeting of the paramagnetic particle by the magnetic force arising from the magnet placed outside the column are represented in Figure 2a.21
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
where is the difference in magnetic susceptibility of the particles and media at room temperature, r is the distance of a particle from the nickel wire, a is the radius of the nickel wire, saturation magnetization of wire, external magnetic field strength, dynamic viscosity of fluid, and represent the velocity of the fluid and the particle. In addition, ρp is the density of the particle, ρf is the density of the fluid media, Vp is the volume of the CM, η is the dynamic viscosity of the media, and c is the effective radius of the CM.
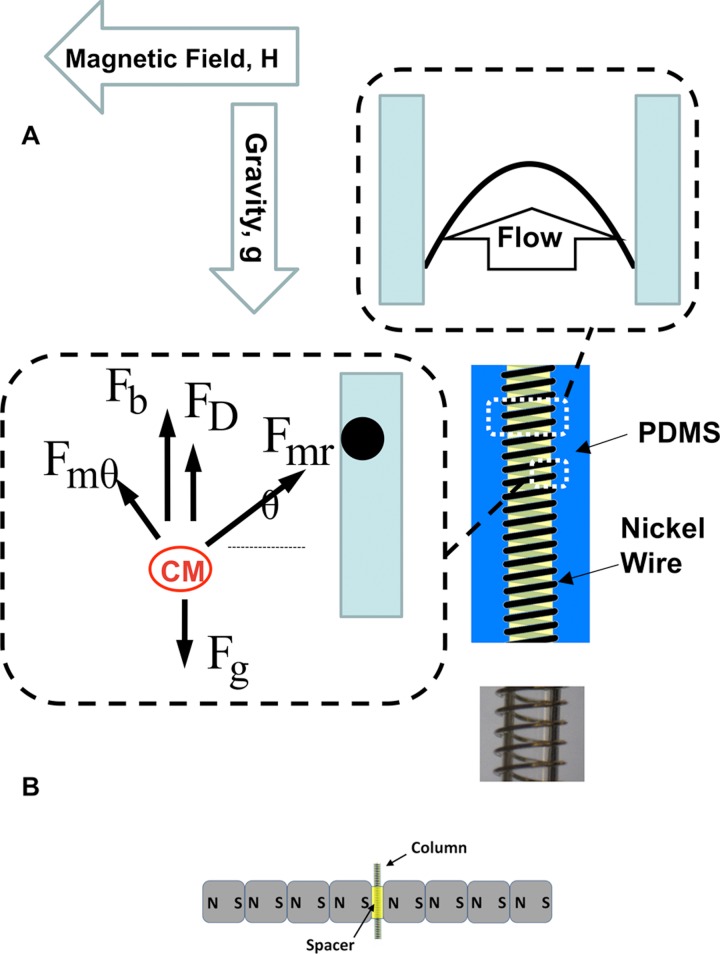
Figure 2.
Microfluidic device schematics and forces acting on the paramagnetic particle in the column. (a) Typical dimensions of the vertical column are: length of 15 mm and diameter of 700 μm. The nickel wire diameter was 69 μm. Trajectory of the paramagnetic particle in the vertical column is determined by the balance of the gravitational force (Fg), drag (FD), buoyancy (Fb), and magnetic force (Fmr, Fmθ). (b) The column is placed vertically in the field generated by four permanent nickel-iron-boron magnets on each side of the column.
The trajectory of the paramagnetic particle, positioned in the center at the bottom of the column, was calculated using Eqs. 3, 4, 5, 6, 7 assuming that the particle is at the same level as the nickel wire (. All calculations and plotting were performed using matlab. The initial velocity of the paramagnetic particle in the cylindrical column at an initial location was set as . Particle acceleration, at the position was calculated using the following equation:
| (8) |
where is the mass of the paramagnetic particle. At the second and, generally, nth position, velocity and acceleration are calculated using the following equations:
| (9) |
| (10) |
| (11) |
MATERIALS AND METHODS
Neonatal and adult mouse heart cells
All animal experimental procedures were approved by the Animal Care Committee of the University of Toronto following the Guide of Care and Use of Laboratory Animals. Ventricular CM were obtained from 8- to 12-week-old adult yellow fluorescent protein (YFP) transgenic mice (129-Tg 7AC5Nagy/J, Jackson Laboratory) of either sex using an isolation procedure described previously.22 Mice were heparinized (10 IU/g body weight) and 5 min later, anesthetized with 2.5% isoflurane as confirmed by the absence of pedal reflexes. Hearts were excised via a midline thoracic incision and transferred to ice-cold Ca2+-free Tyrode's solution ((mmol/l) 137 NaCl, 5.4 KCl, 1.0 MgCl2, 0.33 NaH2PO4, 10 d-glucose, 10 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4). The hearts were then mounted on a 20-gauge blunted stainless steel cannula and retrogradely perfused via aorta with 37 °C, oxygenated Ca2+-free Tyrode's solution for 5 min, followed by collagenase (1 mg/ml, Worthington) for 8 min. Ventricular tissues were dissected out and transferred to and stored in Krebs-bicarbonate solution ((mmol/l) 120 potassium glutamate, 20 KCl, 20 HEPES, 1.0 MgCl2, 10 d-glucose, 0.5 K-Ethylenediaminetetraacetic acid (EDTA), and 0.1% bovine serum albumin, pH 7.4) at 4 °C, with gentle trituration to dissociate the cells. The cell suspension was then subjected to separation as described below.
Neonatal mouse CM and fibroblasts were dissociated according to a standard isolation protocol.23 Briefly, neonatal mice (YFP transgenic, 129-Tg(CAG-EYFP)7AC5Nagy/J; Jackson Laboratories) were first euthanized. The hearts were removed and quartered. Quartered hearts were digested in 0.06% (w/v) solution of trypsin in Ca2+ and Mg2+ free Hank's balanced salt solution (HBSS), overnight at 4 °C. Then, collagenase II (Worthington, USA 220 units/ml) in HBSS was used to further digest the hearts at 37 °C in series of five 4-8 min digestions. After the last digestion step, the cells were centrifuged at 500 rpm for 5 min, which ensured that cardiac cells were pelleted, while red blood cells remained in the suspension. The cells were pre-plated in T75 flasks for 1 h in an incubator in cardiac culture medium. The cells that remained unattached after 1 h of pre-plating were CMs and used for quantification of myoglobin as described below. The attached cells, cardiac fibroblasts, were expanded for up to 1 week and used as a negative control. The CM culture medium and fibroblast culture medium were identical in composition, consisting of Iscove's modified Dulbecco's medium (IMDM) with L-glutamine and 25 mM HEPES, 10% certified fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin.
Myoglobin quantification by enzyme-linked immunosorbent assay
Cell lysates were prepared with NP40 lysis buffer ((mmol/l) 50 Tris-HCl, pH 7.4, 150 NaCl, 40 NaF, 0.5 Na3PO4, 1% NP40) supplemented with complete protease inhibitors without EDTA and 200 μM Na3PO4. To normalize the amount of myoglobin per number of CM, the cells in suspension were counted prior to lysis. Since myocardium is composed of a heterogeneous cell population, total CM number in the sample lysis was obtained by analyzing the percentage of the CM. Neonatal myocardium cells collected from enzymatic digestion were all spherical and vaguely distinguishable by shape or size distribution; therefore, the percentage of neonatal CMs in the cell suspension was determined by immunostaining of paraformaldehyde fixed cells for a cardiac marker Troponin I and flow cytometry as we have described previously.12 The collected adult CMs were morphologically distinguishable rod shaped cells in bright field microscopy. Thus, their percentage was quantified using hemocytometer by counting the rod shaped cells (ratio of length to width >3). Myoglobin determination by sandwich enzyme-linked immunosorbent assay (ELISA) was performed according to the manufacturer's protocol (Life Diagnostics). Briefly, the test samples were placed into microtiter wells pre-immobilized with monoclonal antibody directed against the myoglobin molecule. The polyclonal anti-myoglobin antibody conjugated to horseradish peroxidase (HRP) was added in solution. The test samples were allowed to react simultaneously with the two antibodies, resulting in the myoglobin sandwiched between the solid phase and the enzyme-linked antibodies. After 60 min of incubation at room temperature, the wells were thoroughly washed to remove unbound HRP labeled antibodies. HRP substrate, tetramethyl-benzidine (TMB), was added to allow a development of color, with the intensity directly proportional to the concentration of myoglobin. The absorbance was measured spectrophotometrically at 450 nm using a plate reader (Apollo LB911, Berthold Technologies).
Metmyoglobin evaluation using spectrophotometry
UV-vis measurements were made in cells lysed with NP40 lysis buffer for 30 min on ice, with cell debris spun down at 13 000 rpm for 10 min at 4 °C. The lysates were collected and mixed with NaNO2 solution (2.5 mM for up to 1 h on ice), while NaNO2-free solutions were used as controls. ND-1000 Nanodrop spectrophotometer (Nanodrop Technologies, Inc., Wilmington, DE, USA) was used to collect absorption spectra from 200 to 700 nm.
Microfluidic device fabrication
A schematic of the microfluidic column that was used for the separation studies is shown in Figure 2. In the developed device, nickel wire was wrapped around the circumference of a separation column (Figure 2a). The column was placed in a vertical position between the permanent magnets with the flow inlet at the bottom of the column (Figure 2a).The effects of gravity, hydrodynamic conditions, and magnetic properties of the cells were combined in the device in such a way that upon application of the magnetic field, paramagnetic CM remained in the column, while fibroblasts and other non-myocytes traveled out of the column. The columns were fabricated by embedding a core inside polydimethylsiloxane (PDMS). The core was made by wrapping a nickel wire of thickness 69 μm in diameter around a stainless steel rod. The rod was coated by an anti-sticking Teflon spray (Dupont) before the wrapping of the nickel wire. The rod length was 30 mm and the diameter varied from 300 to 1200 μm for different designs.
PDMS was prepared by a 10:1 ratio of monomer to crosslinker according to the manufacturer's instructions. The mold was then degassed in a dessicator and placed in a 65 °C convection oven for 2 h, followed by overnight curing at room temperature. After curing, the rod was pulled out. Tygon tubing was inserted to either end of the column and sealed with 5-min epoxy. To generate a uniform magnetic field, four neodymium-iron-boron permanent magnets of cubic shape (12.7 mm × 12.7 mm × 12.7 mm) were separated from four similar magnets by means of two spacers. The field strength within the space between two magnets was measured to be 1.23 ± 0.05 T. The separation column was then placed inside the space between the magnets and the spacers as shown in Figure 2b. In initial studies, it was observed that the column diameter of 700 μm resulted in the enrichment of adult CM and was selected as the column diameter thereafter. The column length within the space between the magnets was 15 mm.
Cell separation
For the enrichment tests, adult mouse heart cells were mixed with primary neonatal mouse fibroblasts at different ratios ranging from 15% to 85%, to study the effect of initial CM percentage on final enrichment. Cell concentration was determined using a hemocytometer. The concentration of the cell sample also varied from low, 0.2 × 106/ml, to high 5 × 106/ml to track the effect of sample concentration on the enrichment results. The cells were treated with NaNO2 in the concentration from 2.5 mM to 50 mM for 20 min on ice using media consisting of Ca2+ and Mg2+ free Hank's balanced salt solution with 30% FBS. Sodium nitrite was kept in solution during the entire separation process to ensure that metmyoglobin is present in the cells during separation. Initial studies demonstrated that 0.2 × 106 cells/ml and 2.5 mM of NaNO2 resulted in the highest enrichment, thus they were pursued in all subsequent studies.
To load the cells into the column, 20 μl of the sample was withdrawn by a pipette tip from the Eppendorf tube. The tip was then extracted from the pipette and placed on the top of the column. The mixture was either withdrawn into the column by a syringe pump that was connected to the bottom of the column or just by settling of the cells into the column as a result of gravity. Upon settling, the flow of 4.2 μl/min was applied from the bottom of the column to rinse the fibroblasts out, while keeping CM in the column. A pipette tip was inserted into the top of the column to collect the rinsing flow. After 200 μl, the flow was stopped, a new pipette tip was inserted and the flow rate was increased to 144 μl/min. As a result, the trapped cells inside the column were pushed out, into the pipette tip.
After collecting 200 μl of media into the pipette tip, the tip was removed and emptied into a well of a 12-well plate that was already coated with a solution of 10% Matrigel and 90% plating media for adult CM. Plating media, 2 ml, was added to the well and suspended few times. The fluid was then split into two volumes of 1 ml each in new wells. One well was used to count the enrichment of the cells and to study the cell viability right after the test, while the other well was incubated for 24 h to study the viability of the cells upon separation and culture. Bright field imaging was used to count the number of CM and fibroblasts, where rod shaped cells were considered CM.
Cell viability
Viability of adult mouse CM upon treatment with NaNO2 (2.5 mM for 20 min on ice) and upon enrichment within the microfluidic device was determined using trypan blue exclusion with untreated cells used as a control. The cells were emptied into one well of a 24-well plate and an equal volume of trypan blue was added to each well. After 1 min, 1 ml of media was added to each well and the images were taken to determine the viability with dark blue cells being considered dead and transparent cells alive.
The cell viability after 24 h of culture was determined by live/dead staining. Following the incubation, 500 μl of supernatant from each well was removed. In each well, 0.5 μl of carboxyfluorescein diacetate and 37 μl of propidium iodide were added. The plate was then incubated at 37 °C and 5% CO2 for 30 min. After 30 min, 500 μl of M199 media with 1% penicillin/streptomycin was added to each well and imaging was done to assess viability. The size of adult CM was determined from 5-Carboxyfluorescein Diacetate (CFDA) stained images using imagej, by determining the long and the short axes of each cell.
Statistical analysis
Statistical significance was determined using one-way ANOVA in conjunction with Tukey's test. Normality and equality of variance were tested. p < 0.05 were considered significant. A minimum of 3 samples were used per data point.
RESULTS AND DISCUSSION
Myoglobin quantification and paramagnetic properties of cells
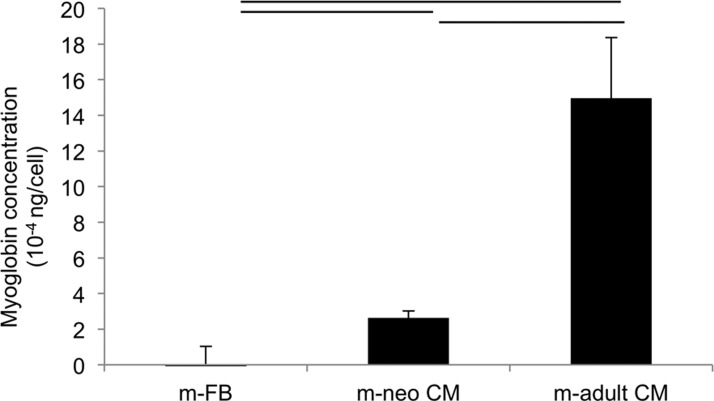
The amounts of myoglobin in the cells were confirmed by ELISA (Figure 3). While non-contractile fibroblasts consistently tested below myoglobin detection levels for the ELISA kit, this protein was detected in both neonatal mouse CM, as well as in the adult CM with significantly higher concentration in the adult CM compared to the neonatal CM. Consistent with maturation of the cells, the amounts of myoglobin per cell increased more than 5 times in adult mouse CM compared to the neonatal mouse CM.
Figure 3.
Myoglobin quantification in cells by ELISA. m-FB, neonatal mouse fibroblasts; m-neo CM, neonatal mouse cardiomyocyte; m-adult CM, adult mouse cardiomyocyte.
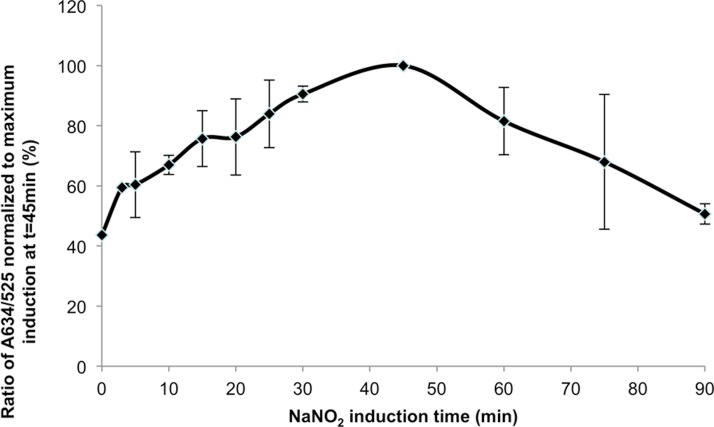
The presence of metmyoglobin was also confirmed by spectrophotometric measurements in the samples treated with NaNO2 (Figure 4). We assessed the extent of induction of metmyoglobin from the total myoglobin upon treatment with 2.5 mM NaNO2, conditions used for cell separation in this study (Figure 4). Since myoglobin derivatives differ in their absorbance spectra, the ratio of the absorbance peak for metmyoglobin at 635 nm to the isobetic point for myoglobin, oxymyoglobin, and metmyoglobin at 525 nm contains information on the amount of metmyoglobin as a fraction of the total myoglobin.24 We observed that the maximum amount of myoglobin induction upon treatment with 2.5 mM NaNO2 occurred 45 min after nitrite was introduced to the cells, motivating our decision to load the cells into the column 30 min after exposure, to ensure cardiomyocytes are rendered paramagnetic at maximal levels during separation.
Figure 4.
Metmyoglobin induction as a function of NaNO2 treatment in cardiomyocytes. Treatment of adult cardiomyocytes with 2 mM NaNO2 indicates maximal induction of metmyoglobin after 45 min.
Calculation of the trajectory of a paramagnetic CM in a cylindrical tube
The parameters in Eqs. 3, 4, 5, 6, 7 and the values for a typical experimental condition with a 700 μm diameter column are described in Table TABLE I..
TABLE I.
Experimental and mathematical model parameter values.
| Variable | Name | Value | Unit |
|---|---|---|---|
| Vacuum permeability | 4π × 10−7 | H/m | |
| Magnetic susceptibility of CM | 7.11 × 10−8 | ||
| Magnetic susceptibility of fluid | −0.9 × 10−5 | ||
| a | Nickel wire radius | 34.5 | μm |
| Saturation magnetization of nickel | 486 × 103 | A/m | |
| r | Distance of cell from the wire | 0–200 | μm |
| External magnetic field | 1.23 | T | |
| g | Acceleration of gravity | 9.98 | m/s2 |
| Θ | Angle to define the location of cell vs wire | Rad | |
| η | Dynamic viscosity of fluid | 0.001 | Ns/m2 |
| Density of CM | 1060 | kg/m3 | |
| Fluid density | 1000 | kg/m3 | |
| c | Effective radius of adult CM | 22 | μm |
The size of the adult CM was determined by image analysis. These cells were rod/elliptical shaped with the average long axis of 77 ± 14 μm and the short axis of 25 ± 3 μm (N = 10). The area of each cell was calculated using the area of the ellipse formula, based on the measured long and short axes. For each calculated area, a radius of the circle occupying the same area was calculated. This radius was termed effective cell radius. For the purpose of mathematical modeling, the effective cell radius determined as described above was 22 μm. The ratio of magnetic force to the gravitational force applied on a representative sodium nitrite treated paramagnetic CM that is at the same level as the nickel wire ( is plotted in Figure 5b by substituting values from Table TABLE I. in the equations above. The theoretical analysis shows that at the central areas of the column (farther than 100 μm from the column wall), the magnetic force is much smaller than the gravitational force due to the low magnetic susceptibility of the CM. These results indicate that the resulting velocity of the cells toward the magnet is much smaller than the cell's typical settling velocity, which is 64.4 μm/s for a cell with effective radius of 22 μm.25 Therefore, the flow rate in the column must be slow to prevent the washout of the desired cell type.
In our batch separation approach, the heterogeneous cell population was loaded into the column. Once in the column, the paramagnetic CMs were attracted towards the column wall as a result of the magnetic field gradient generated by the nickel wire and the presence of the permanent magnet. The theoretical analysis showed that the flow rate from the bottom of the column could be adjusted in such a way to keep the CM in the column and rinse out the non-paramagnetic fibroblasts. At the given column radius and the magnitude of the magnetic field, the percentage of CM trapped in the column would depend on the applied flow rate and the length of the column. In our theoretical analysis, we assumed that the paramagnetic cell was initially present at the centerline at the bottom of the column. We calculated the trajectory of such a paramagnetic cell inside the column at different flow rates. According to our analysis, the paramagnetic cell would travel upward in the column and towards the wall where the nickel wire was circumferentially positioned, reaching the wall of the column after a certain distance was traveled in the z-direction. Due to the no slip boundary condition at the column wall, we assumed that cells would be trapped inside the column once they reached the wall. Solving Eqs. 3, 4, 5, 6, 7 and plotting the cell trajectories (Figure 5c), we observed: (1) at 4.2 μl/min, the cell would fall on the column wall after 1.475 mm, and percentage of trapped cells was 100% for a column described in this paper, which was 15 mm long; (2) at 35 μl/min, the cell would fall on the column wall after 14.817 mm and percentage of trapped cells was 100%; (3) at 36 μl/min, the cell would leave the column and the percentage of trapped cells was 75%.
Microfluidic enrichment results
Cell separation experiments with paraformaldehyde fixed adult CM showed that the highest enrichment was achieved at a flow rate of 4.2 μl/min consistent with our theoretical analysis above. The average enrichment in a single pass for the fixed cell population was 93% ± 4% (N = 8); thus, 4.2 μl/min was selected for all experiments shown in Figure 6. The initial cell concentration was consistently maintained at 0.2 × 106/ml, as higher concentrations tested (up to 5 × 106/ml), resulted in lower enrichment due to overcrowding and formation of heterogeneous cell clumps, consisting of both CM and FB that hindered the separation process.
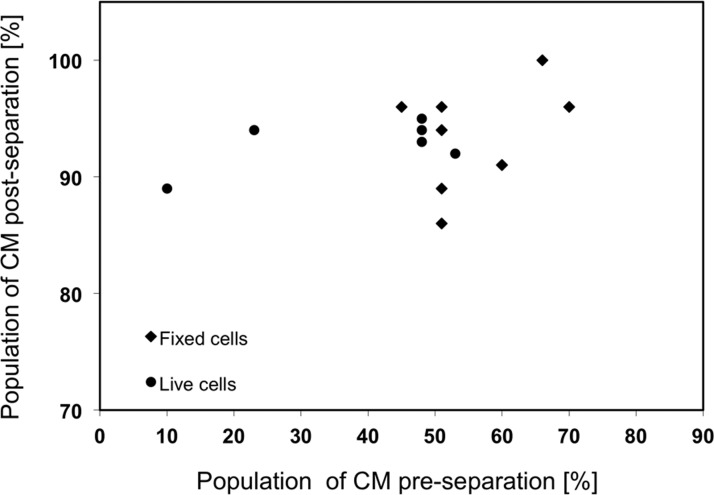
Figure 6.
Correlation between input cardiomyocyte percentage and output percentage in the cell population during the separation at 4.2 μl/min. Application of magnetic field during separation enables consistently high enrichment efficiently between 90% and 100%.
The velocity entrance length was calculated for the 700 μm diameter column at 4.2 μl/min according to the following formula:26
| (12) |
where Rc is the column radius and Re is Reynolds number. The calculated velocity entrance length of 0.42 mm indicated that the flow was laminar and fully developed over most of the column length validating our theoretical analysis at the given experimental conditions as presented above (Figure 5).
To confirm the enabling contribution of the magnetic effects and rule out the possibility that the enriched population was only due to the difference in size, density, or the difference in settling velocities alone of the adult mouse CM and neonatal fibroblasts, several tests were conducted in the absence of the magnet. For these control tests, similar to the tests with magnet, mixtures of live or fixed CM with fibroblasts were treated with NaNO2. The same media and flow rate was used for the control tests to ensure that the viscosity of the fluid was the same. In the absence of a magnet, most of the cells were rinsed out of the column, as expected.
Next, the separation tests with live cells at the flow rate of 4.2 μl/min were performed. Live adult mouse CMs were mixed with live pre-plated neonatal fibroblasts at different ratios. The enrichment efficiency of the live cells was averaged at 93% ± 2%. No significant difference was observed between the results of fixed and live cells (Figure 6).
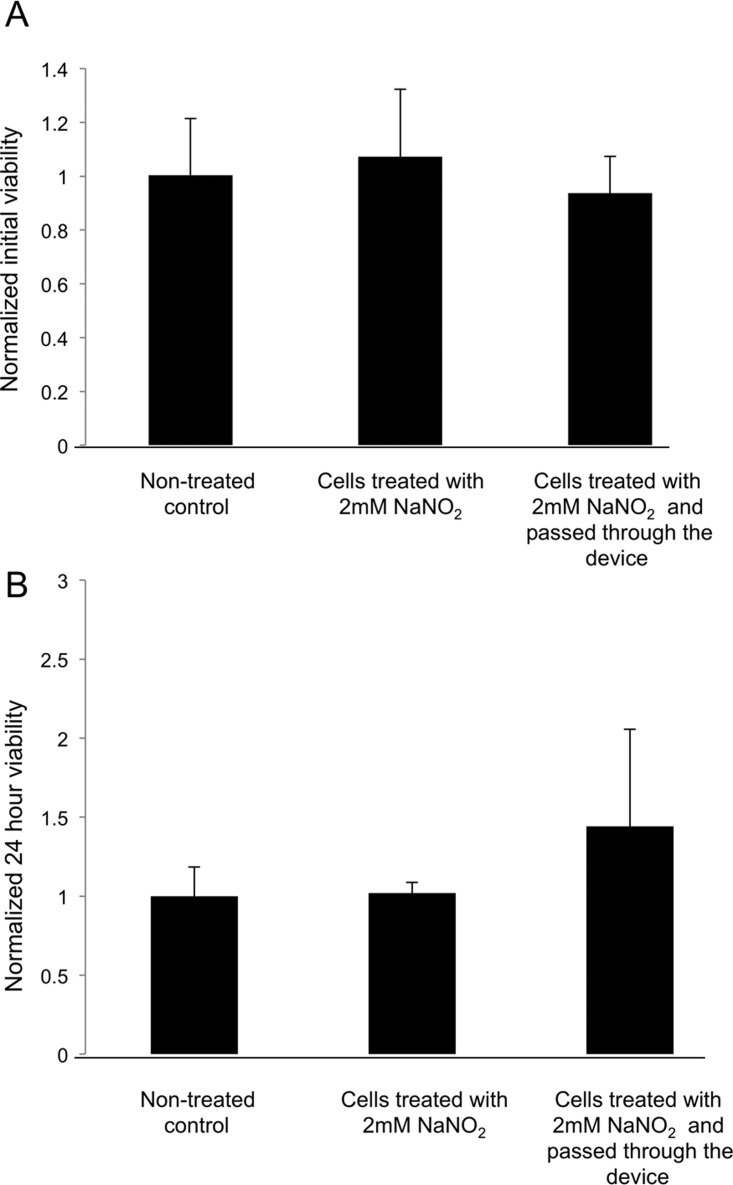
Viability of adult mouse CM after separation and subsequent culture for 24 h was not compromised by the treatment with NaNO2 or passage through the device (Figure 7). In both cases, cell viability was comparable to that of the control cell population that was not treated with NaNO2 and not separated. The treatment of cells by NaNO2 is not expected to decrease cell viability; in fact, the protective effects of NaNO2 on myocardium were reported.27, 28
Figure 7.
Viability of adult mouse cardiomyocytes (a) after separation and (b) subsequent culture for 24 h. All values are normalized to the average cell viability of the control group, at the appropriate time point. The control was kept on ice during separation and was not treated with NaNO2 (no treatment).
Each molecule of myoglobin contains a single iron atom which may bind reversibly to one molecule of O2 when the iron is in the ferrous state (Fe2+), to act as an oxygen storage and enable efficient oxygen supply under conditions of hypoxia and ischemia. Myoglobin undergoes oxidation to the inactive ferric state (Fe3+), known as metmyoglobin upon treatment with NaNO2. Studies have shown that at the concentrations required for this conversion of myoglobin to metmyoglobin, nitrite did not affect the respiration of isolated aerobic heart mitochondria, or glycolysis, and that the levels of adenosine triphosphate remained constant in both cases.29 Under normoxic conditions, in single cell suspension, such as during the microfluidic cell separation, myoglobin is of little functional significance. Post separation and upon NaNO2 removal, myoglobin can be restored freely based on the studies that reported observed reduction of metmyoglobin in isolated perfused rat and sea raven hearts following treatment with NaNO2.30 This reduction involves enzymes normally present in the cells such as endogenous metmyoglobin reductase.31
Here, a microfluidic approach for label-free magnetic separation of CM was presented. The basis for the separation was the higher amount of iron in cardiomyocytes found in myoglobin when compared to the iron level of other cardiovascular cells. After collagenase digestion, the heart isolate contained CM, fibroblasts, smooth muscle cells, and endothelial cells. The ratio of these cells in the native neonatal rat heart after collagenase digestion was 47% cardiomyocytes, 48% fibroblasts, 3% smooth muscle cells, and 2% endothelial cells as we32, 33 and others34 reported. Myoglobin is found in both CM and smooth muscle cells. However, the myoglobin content of smooth muscle cells (∼0.2 mg/g wet weight) is significantly smaller than that of CM (2.6-5.4 mg/g wet weight), due to the high demand for oxygen in contracting CM.35
A fraction of the non-CMs, which were found adjacent to the vertical walls of the column at the beginning of separation, as a result of random distribution of cells in the column, would remain inside the column during the rinsing stage. These non-CM represent the impurity which is reflected in the experimental results.
The yield of the separation process in experiments presented in this paper is limited by the capacity of the column. Loading the cells with too concentrated cell mixture would result in the rinsing of CM out of the column, and consequently losing the target cells. It was observed that maximum yield of over 80% was achieved when the total cell number that was loaded into the column was less than 10 000. In each experiment described here, between 3000 and 5000 adult CM were collected. Using only one column and given that each experiment takes 30 min, the through-put of the experiments in this study was between 6000 and 10 000 collected CM/h. Future studies will focus on improving the yield of the separation process by using multiple columns in parallel.
Alternatively, the yield of the separation process could be improved by switching to a continuous separation process, with several interesting examples described in the recent literature. In one case, the authors used a U shaped device with permanent magnet located at one arm of the separation column. As the cells passed through the U turn, and entered the other arm of the U shaped column, their trajectory was affected as a function of particle size, bulk flow rate in the column, and the magnetic properties of the particles and the surrounding fluid, thus enabling cell separation without the use of the sheath fluid.36 In another approach, the cell separation through-put was enhanced in the microfluidic device by utilizing two electromagnets with antiparallel current flow, placed around the separation channel. The two magnetic fields acted in such a way to push the magnetically tagged cells into the center of the channel, where the flow rate of the sheath fluid was the fastest.37 Additionally, the use of electromagnets vs. permanent magnets in this configuration enabled simple and fast tuning of the magnetic field strength on the same chip.37 The trajectory of magnetic particles in a microfluidic channel could also be affected by an array of stripes, whose orientation, width, and spacing, together with the positioning and strength of the external magnets, would determine the particle trajectory, potentially enabling continuous separation.38 The contaminating fibroblasts could also potentially be removed from the final cell preparation by techniques akin to resistive pulse sensing in tunable or non-tunable pores.39
In this paper, we achieved enrichment of the adult mouse CM from a heterogeneous cell population. This is not a target cell type for regenerative medicine applications due to the non-human species and the fact that adult CMs are terminally differentiated. We used this cell type here in order to demonstrate the proof of concept that live CM can be enriched using magnetophoresis in a label-free manner. Adult CM have the highest myoglobin content compared to other developmental states (Figure 3), thus simplifying the separation problem. In future studies, the column will be further refined (e.g., smaller diameter) to enable enrichment of neonatal rodent CM as well as CM derived from human pluripotent stem cells. The CM derived from human embryonic stem cells or induced pluripotent stem cells represent a true target cell type for applications in regenerative medicine, tissue engineering, and drug testing.
CONCLUSIONS
We developed here a new method to enrich live CM in a label-free manner starting from a heterogeneous cell population containing CM and fibroblasts. CM were rendered transiently paramagnetic by application of NaNO2. Manipulation of CM settling velocity in a vertical column by application of permanent magnetic field and field concentration to the column walls via a nickel wire enabled enrichment of live CM up to 93%. The separation process did not compromise cell viability immediately after separation as well as after 24 h in culture.
ACKNOWLEDGMENTS
This work was supported by an NSERC I2I, NSERC Discovery Grant and NSERC Discovery Accelerator Supplement to M.R. The authors would like to thank Moniba Mirkhani, Samy Makary, and Dr. Peter Backx for help with isolation of adult cardiomyocytes.
References
- Soonpaa M. H. and Field L. J., Circ. Res. 83, 15–26 (1998). 10.1161/01.RES.83.1.15 [DOI] [PubMed] [Google Scholar]
- Dimmeler S., Zeiher A. M., and Schneider , M. D. J. Clin. Invest. 115, 572–583 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M. A. and Murry C. E., Nat. Biotechnol. 23, 845–856 (2005). 10.1038/nbt1117 [DOI] [PubMed] [Google Scholar]
- Yang L., Soonpaa M. H., Adler E. D., Roepke T. K., Kattman S. J., Kennedy M., Henckaerts E., Bonham K., Abbott G. W., Linden R. M., Field L. J., and Keller G. M., Nature 453, 524–528 (2008). 10.1038/nature06894 [DOI] [PubMed] [Google Scholar]
- Zhang J., Wilson G. F., Soerens A. G., Koonce C. H., Yu J., Palecek S. P., Thomson J. A., and Kamp T. J., Circ. Res. 104, e30–e41 (2009). 10.1161/CIRCRESAHA.108.192237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois N. C., Craft A. M., Sharma P., Elliott D. A., Stanley E. G., Elefanty A. G., Gramolini A., and Keller G., Nat. Biotechnol. 29, 1011 (2011). 10.1038/nbt.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori F., Chen H., Yamashita H., Tohyama S., Satoh Y. S., Yuasa S., Li W., Yamakawa H., Tanaka T., Onitsuka T., Shimoji K., Ohno Y., Egashira T., Kaneda R., Murata M., Hidaka K., Morisaki T., Sasaki E., Suzuki T., Sano M., Makino S., Oikawa S., and Fukuda K., Nat. Methods 7, 61–66 (2009). 10.1038/nmeth.1403 [DOI] [PubMed] [Google Scholar]
- Perin E. C., Dib N., Silva G. V., DeMaria A. N., Marroquin O. C., Huang P. P., Traverse J. H., Krum H., Skerrett D., Jagger S. C., Taylor N., Bartels K., Campbell A., Zheng Y., Willerson J. T., Itescu S., and Henry T. D., presentation at American Heart Association Conference, Orlando, FL, 2011.
- Pethig R., Biomicrofluidics 4, 022811 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S. K., Sethu P., Vunjak-Novakovic G., Toner M., and Radisic M., Biomed. Microdevices 8, 231–237 (2006). 10.1007/s10544-006-8169-5 [DOI] [PubMed] [Google Scholar]
- Zhang B., Green J. V., Murthy S. K., and Radisic M., PLoS One 7, e37619 (2012). 10.1371/journal.pone.0037619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R. K., Chiu L. L., and Radisic M., J. Biomed. Mater. Res. A 89, 616–631 (2009) 10.1002/jbm.a.32014. [DOI] [PubMed] [Google Scholar]
- Kendrew J. C., Bodo G., Dintzis H. M., Parrish R. G., Wyckoff H., and Phillips D. C., Nature 181, 662–666 (1958). 10.1038/181662a0 [DOI] [PubMed] [Google Scholar]
- George P., Beetlestone J., and Griffith J. S., Rev. Mod. Phys. 36, 441 (1964). 10.1103/RevModPhys.36.441 [DOI] [Google Scholar]
- Zborowski M., Ostera G. R., Moore L. R., Milliron S., Chalmers J. J., and Schechter A. N., Biophys. J. 84, 2638–2645 (2003). 10.1016/S0006-3495(03)75069-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K. H. and Frazier A. B., IEE Proc.: Nanobiotechnol. 153, 67–73 (2006). 10.1049/ip-nbt:20050019 [DOI] [PubMed] [Google Scholar]
- Han K. H. and Frazier A. B., Lab Chip 6, 265–273 (2006). 10.1039/b514539b [DOI] [PubMed] [Google Scholar]
- Huang R., Barber T. A., Schmidt M. A., Tompkins R. G., Toner M., Bianchi D. W., Kapur R., and Flejter W. L., Prenat. Diagn. 28, 892–899 (2008). 10.1002/pd.2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Chen C., Wei S., Chen C., Wu L. F., and Song T., Biomicrofluidics 6, 024107-12 (2012). 10.1063/1.3702444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville D., Paul F., and Roath S., IEEE Trans. Magn. 18, 1680–1685 (1982). 10.1109/TMAG.1982.1062171 [DOI] [Google Scholar]
- Svoboda J., Magnetic Techniques for the Treatment of Materials (Kluwer Academic, Dordrecht, Netherlands, 2004). [Google Scholar]
- Costantini D. L., Arruda E. P., Agarwal P., Kim K. H., Zhu Y., Zhu W., Lebel M., Cheng C. W., Park C. Y., Pierce S. A., Guerchicoff A., Pollevick G. D., Chan T. Y., Kabir M. G., Cheng S. H., Husain M., Antzelevitch C., Srivastava D., Gross G. J., Hui C. C., Backx P. H., and Bruneau B. G., Cell 123, 347–358 (2005). 10.1016/j.cell.2005.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisic M., Park H., Shing H., Consi T., Schoen F. J., Langer R., Freed L. E., and Vunjak-Novakovic G., Proc. Natl. Acad. Sci. U.S.A. 101, 18129–18134 (2004). 10.1073/pnas.0407817101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkman K. A., Marble D. R., Burns D. H., and Feigl E. O., J. Appl. Physiol. 82, 86–92 (1997). [DOI] [PubMed] [Google Scholar]
- Chalmers J. J., Haam S., Zhao Y., McCloskey K., Moore L., Zborowski M., and Williams P. S., Biotechnol. Bioeng. 64, 509–518 (1999). [DOI] [PubMed] [Google Scholar]
- Deen W. M., Analysis of Transport Phenomena (Oxford University Press, New York, 1998). [Google Scholar]
- Duranski M. R., Greer J. J., Dejam A., Jaganmohan S., Hogg N., Langston W., Patel R. P., Yet S. F., Wang X., Kevil C. G., Gladwin M. T., and Lefer D. J., J. Clin. Invest. 115, 1232–1240 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefer D. J., Arch. Pharmcal. Res 32, 1127–1138 (2009). 10.1007/s12272-009-1804-y [DOI] [PubMed] [Google Scholar]
- Doeller J. E. and Wittenberg B. A., Am. J. Physiol. 261, H53–H62 (1991). [DOI] [PubMed] [Google Scholar]
- Bailey J. R. and Driedzic W. R., J. Exp. Biol. 135, 301–315 (1988). [DOI] [PubMed] [Google Scholar]
- Hagler L., R. I.Coppes, Jr., and Herman R. H., J. Biol. Chem. 254, 6505–6514 (1979). [PubMed] [Google Scholar]
- Radisic M., Park H., Martens T. P., Salazar-Lazaro J. E., Geng W., Wang Y., Langer R., Freed L. E., and Vunjak-Novakovic G., J. Biomed. Mater. Res. A 86(3), 713–724 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. A., Iyer R. K., and Radisic M., Biotechnol. Prog. 24, 907–920 (2008). 10.1002/btpr.11 [DOI] [PubMed] [Google Scholar]
- Naito H., Melnychenko I., Didie M., Schneiderbanger K., Schubert P., Rosenkranz S., Eschenhagen T., and Zimmermann W. H., Circulation 114, I72–I78 (2006). [DOI] [PubMed] [Google Scholar]
- Qiu Y., Sutton L., and Riggs A. F., J. Biol. Chem. 273, 23426–23432 (1998). 10.1074/jbc.273.36.23426 [DOI] [PubMed] [Google Scholar]
- Liang L. and Xuan X., Biomicrofluidics 6, 044106 (2012). 10.1063/1.4765335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe B. D., Lewis L. H., and Murthy S. K., Biomicrofluidics 5, 013413 (2011). 10.1063/1.3553239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donolato M., Dalslet B. T., and Hansen M. F., Biomicrofluidics 6, 024110-6 (2012). 10.1063/1.4704520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmott G. R., Platt M., and Lee G. U., Biomicrofluidics 6, 014103-15 (2012). 10.1063/1.3673596 [DOI] [PMC free article] [PubMed] [Google Scholar]