Abstract
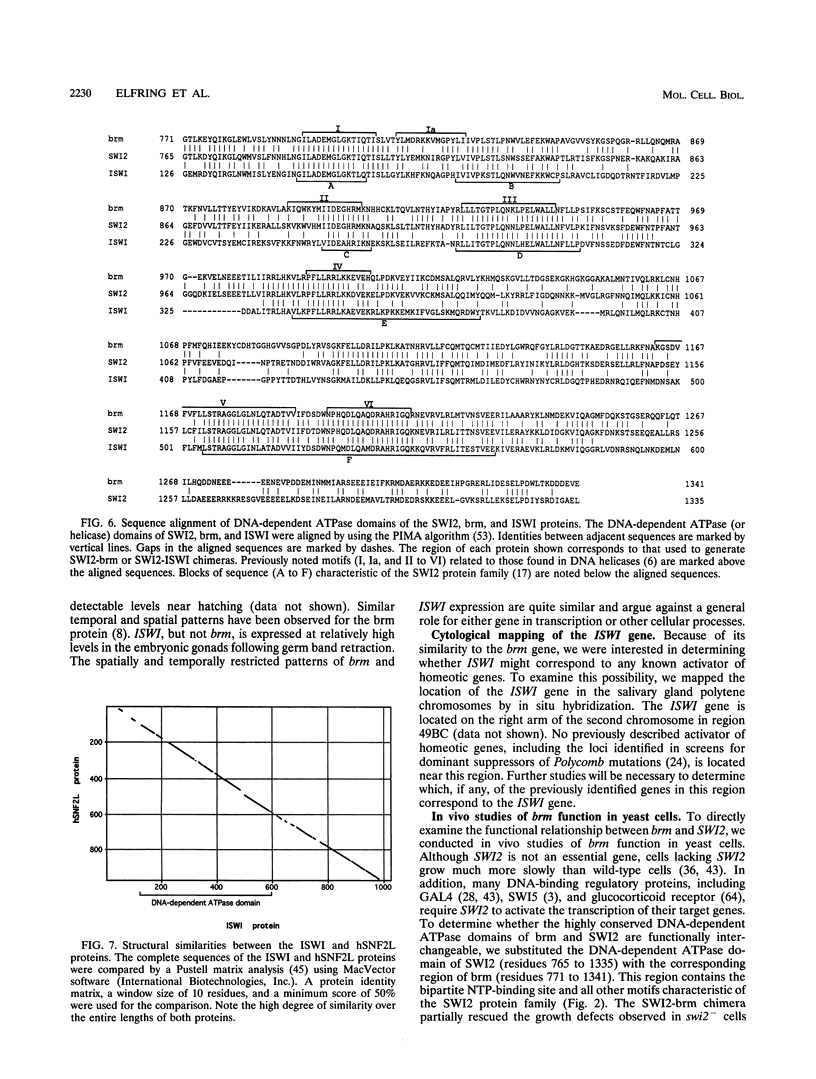
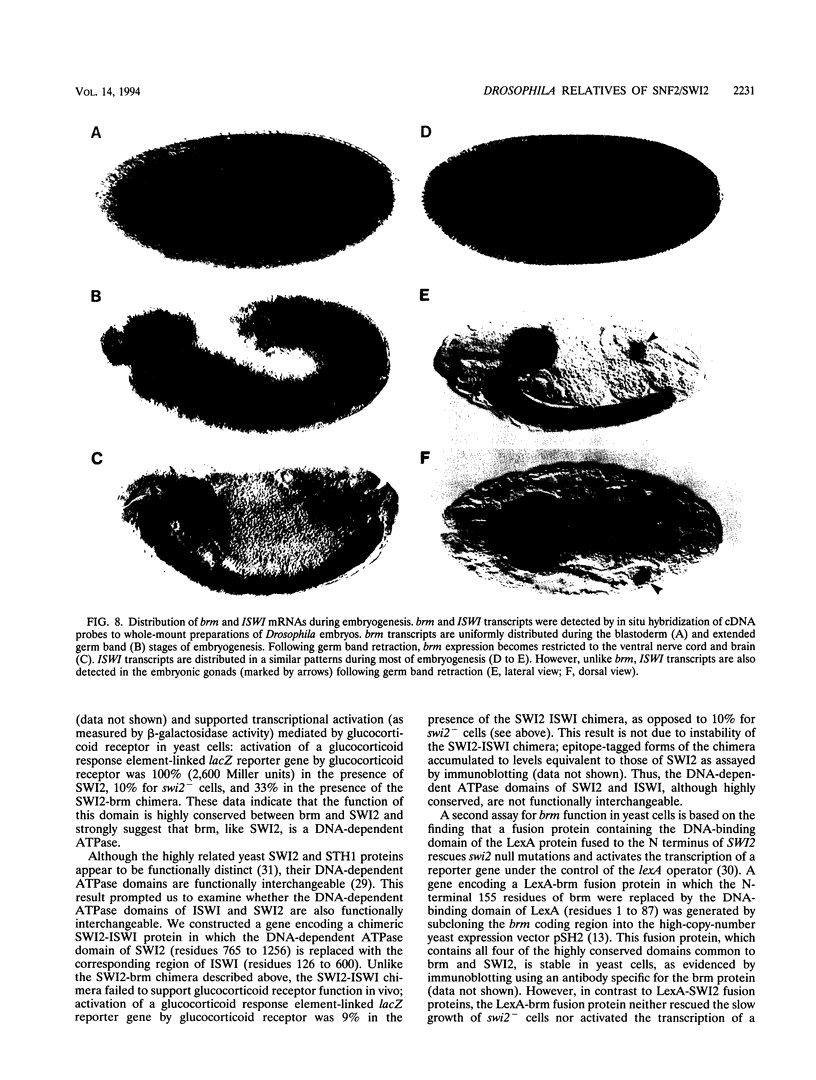
The Drosophila brahma (brm) gene encodes an activator of homeotic genes that is highly related to the yeast transcriptional activator SWI2 (SNF2), a potential helicase. To determine whether brm is a functional homolog of SWI2 or merely a member of a family of SWI2-related genes, we searched for additional Drosophila genes related to SWI2 and examined their function in yeast cells. In addition to brm, we identified one other Drosophila relative of SWI2: the closely related ISWI gene. The 1,027-residue ISWI protein contains the DNA-dependent ATPase domain characteristic of the SWI2 protein family but lacks the three other domains common to brm and SWI2. In contrast, the ISWI protein is highly related (70% identical) to the human hSNF2L protein over its entire length, suggesting that they may be functional homologs. The DNA-dependent ATPase domains of brm and SWI2, but not ISWI, are functionally interchangeable; a chimeric SWI2-brm protein partially rescued the slow growth of swi2- cells and supported transcriptional activation mediated by the glucocorticoid receptor in vivo in yeast cells. These findings indicate that brm is the closest Drosophila relative of SWI2 and suggest that brm and SWI2 play similar roles in transcriptional activation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrew D. J., Scott M. P. Downstream of the homeotic genes. New Biol. 1992 Jan;4(1):5–15. [PubMed] [Google Scholar]
- Andrews B. J., Herskowitz I. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell. 1989 Apr 7;57(1):21–29. doi: 10.1016/0092-8674(89)90168-2. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M., Freidel K., Löhning C. Characterization of trans-acting mutations affecting Ty and Ty-mediated transcription in Saccharomyces cerevisiae. Curr Genet. 1991 Dec;20(6):441–448. doi: 10.1007/BF00334769. [DOI] [PubMed] [Google Scholar]
- Davis J. L., Kunisawa R., Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992 Apr;12(4):1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan I. The bithorax complex. Annu Rev Genet. 1987;21:285–319. doi: 10.1146/annurev.ge.21.120187.001441. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Girdham C. H., Glover D. M. Chromosome tangling and breakage at anaphase result from mutations in lodestar, a Drosophila gene encoding a putative nucleoside triphosphate-binding protein. Genes Dev. 1991 Oct;5(10):1786–1799. doi: 10.1101/gad.5.10.1786. [DOI] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989 Jun 30;57(7):1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- Happel A. M., Swanson M. S., Winston F. The SNF2, SNF5 and SNF6 genes are required for Ty transcription in Saccharomyces cerevisiae. Genetics. 1991 May;128(1):69–77. doi: 10.1093/genetics/128.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K., Levine M. Gap genes define the limits of antennapedia and bithorax gene expression during early development in Drosophila. EMBO J. 1988 Jan;7(1):205–214. doi: 10.1002/j.1460-2075.1988.tb02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes S. R., Dollard C., Winston F., Beck S., Trowsdale J., Dawid I. B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992 May 25;20(10):2603–2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Transcriptional activator components and poxvirus DNA-dependent ATPases comprise a single family. Trends Biochem Sci. 1993 Aug;18(8):291–292. doi: 10.1016/0968-0004(93)90037-n. [DOI] [PubMed] [Google Scholar]
- Hirschhorn J. N., Brown S. A., Clark C. D., Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992 Dec;6(12A):2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Hisatake K., Hasegawa S., Takada R., Nakatani Y., Horikoshi M., Roeder R. G. The p250 subunit of native TATA box-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature. 1993 Mar 11;362(6416):179–181. doi: 10.1038/362179a0. [DOI] [PubMed] [Google Scholar]
- Irish V. F., Martinez-Arias A., Akam M. Spatial regulation of the Antennapedia and Ultrabithorax homeotic genes during Drosophila early development. EMBO J. 1989 May;8(5):1527–1537. doi: 10.1002/j.1460-2075.1989.tb03537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman T. C., Seeger M. A., Olsen G. Molecular and genetic organization of the antennapedia gene complex of Drosophila melanogaster. Adv Genet. 1990;27:309–362. doi: 10.1016/s0065-2660(08)60029-2. [DOI] [PubMed] [Google Scholar]
- Kennison J. A., Tamkun J. W. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J. A., Tamkun J. W. Trans-regulation of homeotic genes in Drosophila. New Biol. 1992 Feb;4(2):91–96. [PubMed] [Google Scholar]
- Kennison J. A. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet. 1993 Mar;9(3):75–79. doi: 10.1016/0168-9525(93)90227-9. [DOI] [PubMed] [Google Scholar]
- Khavari P. A., Peterson C. L., Tamkun J. W., Mendel D. B., Crabtree G. R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993 Nov 11;366(6451):170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Carlson M. Yeast SNF2/SWI2, SNF5, and SNF6 proteins function coordinately with the gene-specific transcriptional activators GAL4 and Bicoid. Genes Dev. 1992 Sep;6(9):1707–1715. doi: 10.1101/gad.6.9.1707. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Treich I., Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993 Apr;7(4):583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Laurent B. C., Treitel M. A., Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci U S A. 1991 Apr 1;88(7):2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B. C., Yang X., Carlson M. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol Cell Biol. 1992 Apr;12(4):1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer S., Franke A., Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992 Jul;6(7):1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- Muchardt C., Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993 Nov;12(11):4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Bienz M. Sharp anterior boundary of homeotic gene expression conferred by the fushi tarazu protein. EMBO J. 1992 Oct;11(10):3653–3661. doi: 10.1002/j.1460-2075.1992.tb05450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L., Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984 Dec;108(4):845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell P. O., Rosbash M. Sequence, structure, and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 1984 Jul 11;12(13):5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R., Hogness D. S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990 Dec;6(12):416–421. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- Pattatucci A. M., Kaufman T. C. The homeotic gene Sex combs reduced of Drosophila melanogaster is differentially regulated in the embryonic and imaginal stages of development. Genetics. 1991 Oct;129(2):443–461. doi: 10.1093/genetics/129.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992 Feb 7;68(3):573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A high speed, high capacity homology matrix: zooming through SV40 and polyoma. Nucleic Acids Res. 1982 Aug 11;10(15):4765–4782. doi: 10.1093/nar/10.15.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo F. M., Khavari P., Crabtree G., Tamkun J., Rossant J. brg1: a putative murine homologue of the Drosophila brahma gene, a homeotic gene regulator. Dev Biol. 1994 Jan;161(1):229–242. doi: 10.1006/dbio.1994.1023. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D., Glassner B. J., Mortimer R. K., Carlson M., Laurent B. C. Identification of RAD16, a yeast excision repair gene homologous to the recombinational repair gene RAD54 and to the SNF2 gene involved in transcriptional activation. Yeast. 1992 May;8(5):385–395. doi: 10.1002/yea.320080506. [DOI] [PubMed] [Google Scholar]
- Schmid S. R., Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992 Feb;6(3):283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., Hazelrigg T. I., Polisky B. A., Pirrotta V., Scalenghe F., Kaufman T. C. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983 Dec;35(3 Pt 2):763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- Simon J., Chiang A., Bender W. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development. 1992 Feb;114(2):493–505. doi: 10.1242/dev.114.2.493. [DOI] [PubMed] [Google Scholar]
- Smith R. F., Smith T. F. Pattern-induced multi-sequence alignment (PIMA) algorithm employing secondary structure-dependent gap penalties for use in comparative protein modelling. Protein Eng. 1992 Jan;5(1):35–41. doi: 10.1093/protein/5.1.35. [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Stern M. J., Clark I., Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987 Feb 27;48(4):567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- Tamkun J. W., Deuring R., Scott M. P., Kissinger M., Pattatucci A. M., Kaufman T. C., Kennison J. A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992 Feb 7;68(3):561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tamkun J. W., Kahn R. A., Kissinger M., Brizuela B. J., Rulka C., Scott M. P., Kennison J. A. The arflike gene encodes an essential GTP-binding protein in Drosophila. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3120–3124. doi: 10.1073/pnas.88.8.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989 Aug;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Travers A. A. The reprogramming of transcriptional competence. Cell. 1992 May 15;69(4):573–575. doi: 10.1016/0092-8674(92)90218-2. [DOI] [PubMed] [Google Scholar]
- Troelstra C., Hesen W., Bootsma D., Hoeijmakers J. H. Structure and expression of the excision repair gene ERCC6, involved in the human disorder Cockayne's syndrome group B. Nucleic Acids Res. 1993 Feb 11;21(3):419–426. doi: 10.1093/nar/21.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen C., Harding K., Levine M. Spatial regulation of Antennapedia and bithorax gene expression by the Polycomb locus in Drosophila. Cell. 1986 Mar 14;44(5):739–748. doi: 10.1016/0092-8674(86)90840-8. [DOI] [PubMed] [Google Scholar]
- White R. A., Lehmann R. A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell. 1986 Oct 24;47(2):311–321. doi: 10.1016/0092-8674(86)90453-8. [DOI] [PubMed] [Google Scholar]
- Yoshimoto H., Yamashita I. The GAM1/SNF2 gene of Saccharomyces cerevisiae encodes a highly charged nuclear protein required for transcription of the STA1 gene. Mol Gen Genet. 1991 Aug;228(1-2):270–280. doi: 10.1007/BF00282476. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S. K., Peterson C. L., Herskowitz I., Yamamoto K. R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992 Dec 4;258(5088):1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]