Abstract
Background
HIV subtypes A and CRF01_AE (A/AE) became prevalent in Israel, first through immigration of infected people, mostly intravenous-drug users (IVDU), from Former Soviet-Union (FSU) countries and then also by local spreading. We retrospectively studied virus-transmission patterns of these subtypes in comparison to the longer-established subtype B, evaluating in particular risk-group related differences. We also examined to what extent distinct drug-resistance patterns in subtypes A/AE versus B reflected differences in patient behavior and drug-treatment history.
Methods
Reverse-transcriptase (RT) and protease sequences were retrospectively analyzed along with clinical and epidemiological data. MEGA, ClusalX, and Beast programs were used in a phylogenetic analysis to identify transmission networks.
Results
318 drug-naive individuals with A/AE or patients failing combination antiretroviral therapy (cART) were identified. 61% were IVDU. Compared to infected homosexuals, IVDU transmitted HIV infrequently and, typically, only to a single partner. 6.8% of drug-naive patients had drug resistance. Treatment-failing, regimen-stratified subtype-A/AE- and B-patients differed from each other significantly in the frequencies of the major resistance-conferring mutations T215FY, K219QE and several secondary mutations. Notably, failing boosted protease-inhibitors (PI) treatment was not significantly associated with protease or RT mutations in either subtype.
Conclusions
While sizable transmission networks occur in infected homosexuals, continued HIV transmission among IVDU in Israel is largely sporadic and the rate is relatively modest, as is that of drug-resistance transmission. Deviation of drug-naive A/AE sequences from subtype-B consensus sequence, documented here, may subtly affect drug-resistance pathways. Conspicuous differences in overall drug-resistance that are manifest before regimen stratification can be largely explained in terms of treatment history, by the different efficacy/adherence limitations of older versus newer regimens. The phenomenon of treatment failure in boosted-PI-including regimens in the apparent absence of drug-resistance to any of the drugs, and its relation to adherence, require further investigation.
Introduction
Several interrelated factors detrimentally influence the efficacy of measures to control the HIV epidemic at the individual and community levels, including risk-behaviors, sub-optimal treatment regimens, incomplete patient adherence and drug-resistance development and transmission. It is difficult to assess the relative roles of these factors: the constitution of the infected population and of the population at risk is heterogeneous and variable; drug-resistance mechanisms in non-B subtypes are incompletely understood [1], [2]; and the dependence of these mechanisms on body concentrations of specific drugs is complex [3]. Moreover, since most laboratory-based and epidemiological studies are retrospective, it is impossible to study the different factors affecting the course of the epidemic separately; an “integrative” approach is required [4], [5]. Such approach involves pooling different kinds of information together and the identification of “patterns” within the complex body of data.
Since every HIV-infected Israeli citizen has free access to cART and the collection of clinical, epidemiological and laboratory data is centralized, such comprehensive analyses have been facilitated. They enable comparisons of drug-resistance patterns in conjunction with other parameters among patients infected with different subtypes and/or belonging to different risk-groups. Recently, we were able to infer from the evolution of such patterns over time, and from the extent and character of phylogenetic clustering of HIV sequences, a striking increase in the frequency of unprotected and multi-partner sex in the gay community in Israel [5], [6]. Assessing behavioral trends usually relies on the collection of behavioral data directly from the target population, but this approach is not always feasible [4]. Studies that focus on the analysis of pooled, centrally collected laboratory and epidemiological data may replace or complement studies that require direct investigation of people while avoiding major sampling biases.
Subtypes A, A1, the recombinant virus CRF01_AE and related variants (collectively, A/AE) are widespread in the far East and Former Soviet Union (FSU), two major epicenters of the HIV pandemic today [7]–[13]. A/AE variants are common also in Israel since the late 1990s, along with subtypes B and C, first through immigration and tourism [14]–[17], but lately also because of endemic transmission. After the large outbreak of HIV-1 epidemic in the FSU in 1996–1997, mainly among intravenous-drug users (IVDU) and their partners [11]–[13], immigrants to Israel from this region [18] imported these variants, which today are carried by ∼20% of the HIV-infected population in Israel. As combined antiretroviral treatment (cART) becomes available globally, extending our current understanding of drug resistance to non-B subtypes is increasingly required. Besides, a substantial portion of those infected with the A/AE variants in Israel and elsewhere are, or were IVDU and a better understanding of behavioral trends within this group and with other groups is instrumental in the ongoing efforts to control the epidemic.
Our aim in this study was two-fold: discerning the impact of antiretroviral treatment on subtypes A/AE versus B, and inferring risk-behavior trends in IDVU versus men who have sex with men (MSM) from the different patterns of HIV transmission within these groups.
Materials and Methods
We analyzed genotypic information from 318 individuals carrying A/AE viruses along with clinical and demographic data. We compared to earlier-reported data from B-infected patients [5], in particular 254 drug-naive and 60 drug-treated diagnosed after 2001, the period in which most A/AE carriers were diagnosed and treated.
Patients and Data Collection
Demographic and clinical data, including detailed antiretroviral-treatment history, are provided on standardized forms when samples are submitted for genotyping. These data are cross-checked with the national HIV, tuberculosis (TB) and sexually transmitted disease (STD) registries and are stored in an anonymous database.
“Recent” HIV infection at diagnosis was identified retrospectively either by documented evidence that sero-conversion occurred in the preceding 12 months or when acute retroviral syndrome was documented, based on a compatible pattern of viral load, CD4 count and clinical history [19], [20].
Co-infection with Other Pathogens
Hepatitis: HIV infected patients in Israel are routinely screened biannually for co-infection with hepatitis B and C.
Tuberculosis: HIV and TB registries are cross-matched annually.
Syphilis: Infectious syphilis (primary, secondary or early-latent) was defined as previously described [5].
Genotyping
Blood samples from HIV-infected patients were sent for drug resistance evaluation as part of patients’ routine follow-up, as previously described [21]. Genotyping was performed either at the National HIV Reference Laboratory (NHRL) or at the Laboratory of Viruses and Molecular Biology, Sourasky Medical Center. Subtypes were determined using the Stanford Database Rapid Subtyping tool (www.hivdb.stanford.edu/hiv/) [22]–[24]. Resistance-conferring mutations in drug-naive patients were identified according to Bennett et al. [25].
Phylogenetic Analysis
Phylogenetic and molecular-evolution analyses of protease and reverse-transcriptase (RT) sequences were performed using MEGA, version 5.05 [26] and ClusalX (MegAlign, Lasergene version 5.01, DNASTAR Inc., Madison, WI, USA). Phylogenetic trees were drawn using FigTree v1.3.1 [27] and branch reproducibility was assessed on 1000 replicates using Seqboot. Alignments were subjected to Bayesian Monte-Carlo Markov Chain analyses using BEAST to construct phylogenies and investigate ancestral relationships. Transmission clusters were defined as distinct populations with short branch lengths and a posterior probability ≥0.95 to have a recent common ancestor [28].
Statistical Analysis
Clinical data and mutation frequencies were compared across patient groups using Chi2 test for the categorical independent variables and Student’s t-test for continuous variables. P<0.05 was considered significant. Analyses were conducted using SPSS version 19.0.
Ethics Statement
The retrospective analysis of clinical and laboratory data, which were obtained from the medical charts of HIV-1 patients attending the Sourasky and Sheba Medical Centers, was approved by the respective ethical committees. Specifically, permission was granted by the Sourasky Ethical Committee to analyze such data without the need of a signed informed consent by the patients. The samples obtained at the Sheba Medical Center that were used in this study belonged to patients who had signed an informed consent agreeing to participate in a range of studies.
Results
Demographics
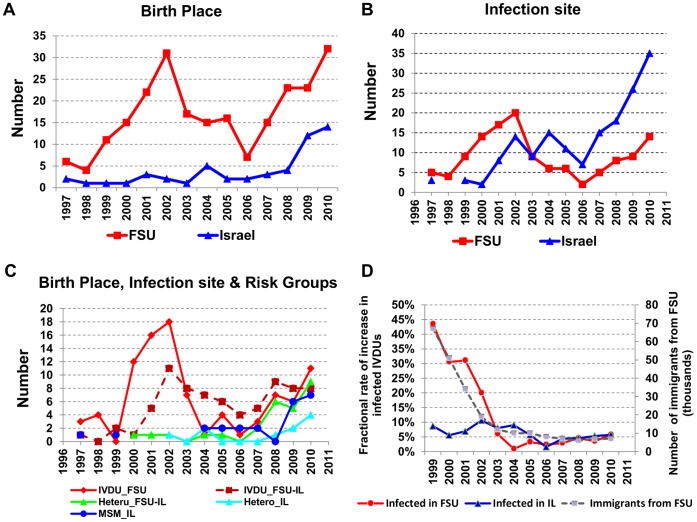
Seventy-six percent of more than 300 infected immigrants from FSU whose HIV virus was genotyped carried A/AE viruses. Most of them were IVDU (Table 1). A/AE viruses started to be detected in considerable numbers in Israel in 1996–7 (Figure 1). Before 2004, the total number of A/AE carriers diagnosed in Israel each year and the number of those among them known to have been infected with A/AE-HIV in the FSU paralleled quite closely. This correlation was weakened later, as immigration from FSU declined and the fractions of MSM and non-IVDU heterosexuals among A/AE carriers progressively increased (Figure 1A). Since 2007, the numbers of newly diagnosed individuals infected with the A/AE viruses while in Israel exceeded the numbers of those who were infected in FSU (Figure 1B). Stratifying the data by risk groups (Figure 1C, Table 1) shows a number of interesting features. First, as IVDU who were infected in FSU were diagnosed at an increasing rate between 1999 and 2002, they started to increasingly infect other FSU-born IVDU in Israel, but hardly any others. Second, the numbers of heterosexuals diagnosed with A/AE viruses each year were on the rise only since 2007, and those were mainly FSU-born females infected in Israel (Figure 1C, Table 1). Third, only in 2008 did the virus start establishing itself rapidly among Israeli-born, and those infected were almost exclusively MSM (Figure 1C). Gay men diagnosed with A/AE who were born in FSU were also mostly infected in Israel (Table 1).
Table 1. Demographic data.
| Risk Group | Hetero | IVDU | MSM | Other | Total | |
| Gender Number (%) | Male | 24 | 145 | 36 | 3 | 208 (65%) |
| Female | 50 | 54 | 0 | 6 | 110 (35%) | |
| Total | 74 (23%) | 199 (63%) | 36 (11%) | 9 (3%) | 318 | |
| Place of Birth (Number) | FSU | 46 | 185 | 7 | 6 | 244 (77%) |
| Israel | 17 | 11 | 26 | 2 | 56 (18%) | |
| Other | 11 | 3 | 3 | 1 | 18 (6%) | |
| Total | 74 | 199 | 36 | 9 | 318 | |
| Place of Infection (Number) | FSU | 20 | 103 | 2 | 4 | 129 (41%) |
| Israel | 41 | 91 | 34 | 4 | 171 (54%) | |
| Other | 13 | 5 | 0 | 1 | 18 (6%) | |
| Total | 74 | 199 | 36 | 9 | 318 | |
| Place of Birth(% in the RG) | FSU | 62% | 93% | 19% | 67% | 77% |
| Israel | 23% | 6% | 72% | 22% | 18% | |
| Other | 15% | 2% | 8% | 11% | 6% | |
| Total | 100% | 100% | 100% | 100% | 100% | |
| Place of Infection(% in the RG) | FSU | 27% | 52% | 6% | 44% | 41% |
| Israel | 55% | 46% | 94% | 44% | 54% | |
| Other | 18% | 3% | 0% | 11% | 6% | |
| Total | 100% | 100% | 100% | 100% | 100% | |
FSU – Former Soviet Union; RG – Risk Group.
Figure 1. Propagation and incidence rates of A/AE-HIV in Israel.
A. Birth sites of individuals diagnosed with A/AE by year of diagnosis. (FSU – red squares; Israel – blue triangles). B. Infection site of individuals diagnosed with A/AE by year of diagnosis. (FSU – red squares; Israel – blue triangles). C. Birth site and infection place of A/AE-patients of specified transmission groups, by year of diagnosis. Red solid line – IVDU born and infected in FSU; dashed brown line – IVDU born in FSU and infected in Israel; green solid line – heterosexuals (mostly females) born in FSU and infected in Israel; light blue solid line – heterosexsuals born and infected in Israel; blue solid line – MSM born and infected in Israel. D. Newly-diagnosed IVDU infected in Israel (blue triangles) or in FSU (red circles) per-year as a fraction of the total number of A/AE-infected IVDU in the same year. Also shown are the total numbers of immigrants from FSU to Israel per-year (dashed line). FSU – Former Soviet Union; IVDU – Intravenous drug users; Is – Israel; MSM – Men who have sex with men.
Clinical Data
For a summary of clinical parameters at diagnosis, see Tables 2 and 3.
Table 2. Clinical data.
| Clinical data | Drug-naive | Treated | p | Co-infection with other pathogens (n = 318) | |
| (n = 234) | (n = 78) | ||||
| CD4 count (median cells/µL) | 315 | 338 | NS | Hepatitis | 176 (55.3%) |
| Viral load (median copies/ml (logVL)) | 29,950 (4.48) | 15,800 (4.20) | NS | TB total | 14 (4.4%) |
| Sero-conversion | 6 | 2 | NS | TB+HCV | 12 (3.7%) |
| Age at diagnosis (years median ± S.E.M) | 31.9±0.6 | 31.1±1.4 | NS | Syphilisa total | 10 (3.1%) |
| Time from diagnosis to genotyping (Months median ± S.E.M) | 2.1±1.9 | 56.8±3.6 | >0.001 | Syphilis+HCV | 8 (2.5%) |
| Time under treatment (Months median ± S.E.M) | NA | 37.8±2.7 | NA | TB+Syphilis+HCV | 1 (0.3%) |
The Table lists various clinical parameters, pertaining to drug-naive and treatment-failing individuals (the two left columns, respectively), and specifically regarding co-infection status (on the right).
Infectious syphilis (primary, secondary or early-latent) was defined by a positive VDRL test (Venereal Disease Research Laboratory Becton-Dickenson, Shannon, Ireland) as previously described [5]. Four individuals were in a primary or secondary phase and six were in late phase of the disease. Two of the ten (20%) were MSM, who comprised 15% of all A/AE-infected individuals. At least three acquired HIV in Israel.
cART – combination antiretroviral therapy; HCV – Hepatitis C virus; MSM – Men who have sex with men; NA – Not applicable; NS – Not significant; S.E.M – Standard error of mean; TB – Tuberculosis; VL – viral load.
Table 3. Co-infection with hepatitis and demographic data on patients stratified according to their hepatitis status.
| A | Hepatitis | Number | Percent (of total) | ||||
| HCV | 148 | 46.8 | |||||
| HBV | 11 | 3.5 | |||||
| HCV+HBV | 17 | 5.4 | |||||
| Negative | 130 | 41.1 | |||||
| Not Knowna | 12 | 3.8 | |||||
| Total | 318 | 100 | |||||
| B | Hepatitis | Negative (n = 130) | Positive a (n = 176) | p | |||
| Number (% of TG) | Percent (of total) | Number (% of TG) | Percent (of total) | ||||
| Transmission Groups (percentage in the group) | Hetero | 49(74%) | 37.7 | 17(26%) | 9.7 | <0.0001 | |
| IVDU | 49(25%) | 37.7 | 141(71%) | 80.0 | <0.0001 | ||
| MSM | 30(64%) | 23.1 | 15(32%) | 8.5 | 0.001 | ||
| Other | 2(29%) | 1.5 | 3(43%) | 1.7 | 1 | ||
| Total | 130 | 100 | 176 | 100 | |||
| Birth Place (percentagein the group) | Is | 42(78%) | 32.0 | 10(19%) | 5.7 | <0.0001 | |
| FSU | 77(32%) | 59.2 | 159(65%) | 90.3 | <0.0001 | ||
| Otherb | 14(55%) | 10.8 | 7(35%) | 4.0 | 0.1 | ||
| Total | 130 | 100 | 176 | 100 | |||
176 of the 318 A/AE-HIV carriers (55.3%) were co-infected with hepatitis: 148 had HCV, 11 HBV and 17 had both. 130 were not infected with hepatitis and for 12 there was no information.
For 8 IVDU, 2 MSM and 2 Others the status of hepatitis infection was unknown.
Other birth sites were in Africa, America, Asia, Europe, or unknown.
IVDU – Intravenous drug users; MSM – men who have sex with men; TG – Transmission group.
One hundred seventy six of the 318 A/AE-HIV carriers (55.3%) were co-infected with hepatitis B and C viruses (HBV and HCV). Co-infection among IVDU and/or FSU-born individuals was significantly higher than in other transmission groups or among Israeli-born, respectively (p<0.005; Tables 2 and 3). Co-infection with HBV and/or HCV did not significantly affect viral load or CD4 count, but we identified a borderline increase (p = 0.05) in the diversity of HIV sequences (not shown; [29]–[31]), suggesting that diversification of HIV may be faster in the presence of HBV and/or HCV.
Fourteen of the 318 (4.4%) had tuberculosis, a much higher co-infection rate than in B-subtype patients (p<0.001). Thirteen (92.8% of the TB infected) immigrated from FSU and one from Kenya; eleven (78.5%) were IDVU and 12 were co-infected with HCV. Ten of the 318 (3.1%) had Syphilis (Table 2).
Evidence for “recent infection” was found at the time of diagnosis in three of 44 MSM (6.8%) and in four of 198 IVDU (2.0%; p = 0.07).
A/AE HIV Incidence Rates
Figure 1D shows steady increases in the number of infected IVDU, while the growth in infected MSM and non-IVDU heterosexuals accelerated. The latter represented mainly, and increasingly, female-spouses of infected male IVDU, as indicated by the fact that almost all heterosexuals in our study group who were infected in Israel were FSU-born women (not shown). Newly-diagnosed IVDU and MSM presumably acquired HIV mainly from infected persons within their respective transmission-groups. The relatively-constant per-capita infection rate of IVDU in Israel (number of newly-diagnosed per year divided by the number of those already-infected) was about 5% during 2007–2010 (Figure 1D). The fraction of newly-diagnosed IVDU who were infected in FSU diminished, paralleling a decline and then low-level stabilization of the immigration rate.
Phylogenetic Analysis of Viral Protease and RT Sequences
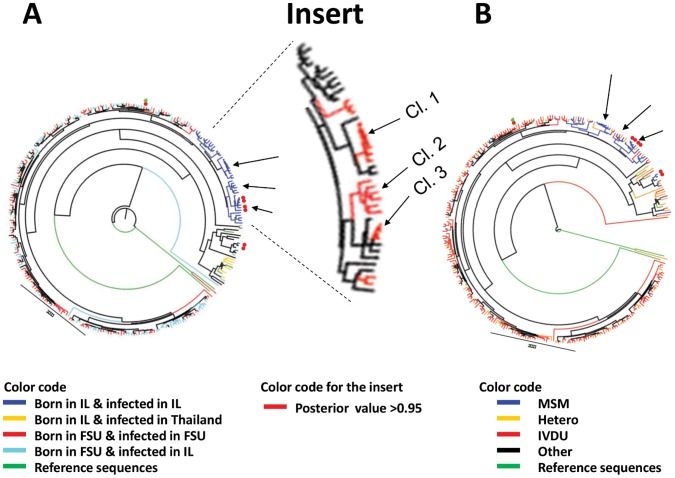
To better characterize the A/AE HIV-transmission process, a phylogenetic tree of the first available sequences of protease-RT from 281 patients (216 drug-naive and 65 drug-treated) was constructed (Figure 2). Figure 2 links transmission pathways to additional information, including birth place and infection place (Figure 2A), risk groups (Figure 2B), and resistance-conferring mutations found in drug-naive patients (Figures 2A and 2B). Clusters having more than four members with posterior probability>0.95 of having a common ancestor are marked with arrows. The tree section in which they are found was enlarged (Insert to Figure 2).
Figure 2. Phylogenetic tree of Pr-RT sequences from A/AE-HIV samples.
Neighbor-joint analysis of A/AE protease and RT sequences, combined (918 nucleotides). The first available sequence from 216 drug-naive and 65 drug-treated individuals was used. Trees were colored according to: A. Birth place and infection site: red lines – born and infected in FSU; blue lines – born and infected in Israel; turquoise lines – born in FSU and infected in Israel; yellow lines – born in Israel and infected in Thailand; green lines – reference sequences. B. Transmission groups: red lines – IVDU; blue lines – MSM; turquoise lines – Hetero; green lines – reference sequences. Insert: red lines – posterior probability>0.95 of having a common ancestor. Major drug-resistance mutations in drug naive individuals: Red circles – K103N; Green triangle – M184V; orange rhombus – protease M46I. Reference sequences used in constructing the tree: Subtype-A/AE variants: subtype A – AF193275, subtype CRF01_AE – AF197340 and AF447851.1; subtype CRF03_AB – AF193276; subtype B – K03455, subtype C – AF286233 and AY585268; subtype D – AY322189; subtype F – AJ249238. Cl. – cluster. Clusters having more than 4 members with posterior probability>0.95 of having a common ancestor are marked with arrows.
Typically, we could link with high posterior probability of having a recent common ancestor only single A/AE-infected IVDU to each other or to single heterosexuals. Large networks, of more than four infected individuals, did conspicuously occur only within A/AE-infected MSM (Figure 2, Clusters 1, 2 and 3). Interestingly, cluster 3 included four drug-naive patients infected with K103N-containing virus (Figure 2, red circles), two of whom also had syphilis, and the MSMs in cluster 1 included two sero-convertors. We found no clustering for other parameters.
cART and Drug Resistance
Antiretroviral treatment
Detailed account of treatment regimens is given in Table 4. Most patients in our study population (>85%) began cART after 2001 and more than 45% after 2007. First-regimen data were available for 165 patients (51.9% of the 318) and the treatment given while failing therapy was known for 78 (24.5%); their median treatment period was 37.8±2.7 months (range 1.2–142 months). Thirty-four failed a second or higher treatment regimen at genotyping. Only six individuals, diagnosed before 1997, had prior suboptimal therapy documented. The most common regimens included tenofovir+emtricitabine (TDF+FTC; ∼51%) or zidovudin+lamivudine (ZDV +3TC; ∼31%) as the nucleoside reverse transcriptase inhibitors (NRTI) backbone plus either boosted protease inhibitors (PI) (mainly boosted lopinovir – LPV/r) or efavirenz (EFV) as the third drug. These were also the most common regimens for treatment-failing patients, but the treatment-failure frequencies were significantly disproportionate, ∼44% for regimens including ZDV+3TC, and only ∼23% for those including TDF+FTC (p<0.01; Table 4).
Table 4. First-administrated and actual drug regimens while failing treatment.
| Drug | First cART regimens (n = 165)a | Actual regimens when genotyped (n = 78)b | |||
| Number | Percent | Number | Percent | ||
| PI | LPV/r | 53 | 32.1% | 24 | 30.8% |
| Other Boosted PI | 14 | 8.5% | 2 | 2.6% | |
| Non boosted PI | 21 | 12.7% | 8 | 10.3% | |
| 18.5±2.5 | Total | 88 | 53.3% | 34 | 43.6% |
| NRTI | TDF+FTC | 84 | 50.9% | 34 | 43.6% |
| ZDV+3TC | 51 | 30.9% | 18 | 23.1% | |
| Others | 30 | 18.2% | 26 | 33.3% | |
| 20.5±2.3 | Total | 165 | 100.0% | 78 | 100.0% |
| NNRTI | EFV | 71 | 43.0% | 33 | 42.3% |
| NVP | 6 | 3.6% | 6 | 7.7% | |
| 24±3.3 | Total | 77 | 46.7% | 39 | 50.0% |
All except 5 received NRTIs as part of the first regimen. A few received mono- or duo-therapy or combinations of PIs and NNRTIs.
All 78 treatment-failing patients received NRTI backbone. Thirty-four received also PI and 39 NNRTI as additional drug.
3TC – lamivudine; EFV – efavirenz; FTC – emtricitabine; LPV/r – lopinavir/r; NNRTI – Non-Nucleoside Reverse Trancriptase Inhibitor; NRTI – Nucleoside Reverse Trancriptase Inhibitor; NVP – nevirapine; PI – Protease inhibitor; r – ritonavir; TDF – tenofovir; ZDV – zidovudine.
Drug resistance in drug-naive patients
Pre-treatment genotyping was performed 2.1±1.9 months after diagnosis (median ± S.E.M; range 0–167 months). Resistance-conferring mutations were found in 16 individuals (6.8% of the drug-naive group), one heterosexual (of 53; 1.9%), 7 IVDU (of 121; 5.8%), and 8 MSM (of 34; 23.3%). Two carried the protease mutation M46I; three had NRTI-related mutations; 11 non-nucleoside reverse transcriptase inhibitors (NNRTI); and one was resistant to NRTI and NNRTI. The most frequent mutation was K103N, in 7 individuals. Mutation frequencies that differed significantly between drug-naive and drug-treated patients (see below) and/or between A/AE and B are shown in Tables 5 and 6.
Table 5. Resistance Mutations in the reverse transcriptase of drug-naive and drug-treated individuals.
| Amino Acids | A/AEa | B patients diagnosed from 2001 on | p A/AE vs. B | ||||||||||
| Naive n = 234 | Treated n = 78 | p Naive A/AE vs. Treated A/AE | Naïve n = 254 | Treated n = 60 | p Naive B vs. Treated B | Naive A/AE vs. Naive B | Treated A/AE vs. Treated B | ||||||
| No | % | No | % | No | % | No | % | ||||||
| NRTI resistance mutations | M41L | 2 | 1 | 4 | 5 | 0.009 | 1 | 0.4 | 3 | 5 | 0.02 | 0.6 | 1 |
| A62V | 32 | 14 | 5 | 6 | 0.003 | 1 | 0.4 | 1 | 2 | 0.3 | <0.0001 | 0.2 | |
| K65R | 0 | 0 | 4 | 5 | 0.009 | 0 | 0 | 2 | 3 | 0.04 | NA | 0.7 | |
| D67N | 1 | 0.4 | 3 | 4 | 0.03 | 0 | 0 | 4 | 7 | 0.001 | 1 | 0.5 | |
| M184V | 1 | 0.4 | 27 | 35 | <0.0001 | 0 | 0 | 17 | 28 | <0.0001 | 1 | 0.5 | |
| T215FY | 1 | 0.4 | 2 | 3 | 0.1 | 7 | 3 | 8 | 13 | 0.002 | 0.07 | 0.02 | |
| K219QE | 0 | 0 | 1 | 1 | 0.3 | 0 | 0 | 7 | 12 | <0.0001 | NA | 0.02 | |
| NRTI accessory mutations | V118I | 3 | 1 | 3 | 4 | 0.03 | 11 | 4 | 2 | 3 | 1 | 0.06 | NS |
| L228H | 0 | 0 | 1 | 1 | 0.3 | 0 | 0 | 3 | 5 | 0.007 | NA | ||
| NNRTI resistance mutations | K101E | 1 | 0.4 | 4 | 5 | 0.009 | 0 | 0 | 3 | 5 | 0.007 | 0.5 | 1 |
| K103N | 7 | 3 | 18 | 23 | <0.0001 | 10 | 4 | 11 | 18 | <0.0001 | 0.6 | 0.5 | |
| V108I | 2 | 1 | 6 | 8 | 0.001 | 0 | 0 | 3 | 5 | 0.007 | 0.2 | 0.7 | |
| Y181C | 0 | 0 | 5 | 6 | 0.003 | 0 | 0 | 3 | 5 | 0.007 | NA | 1 | |
| G190AS | 0 | 0 | 16 | 21 | <0.0001 | 0 | 0 | 5 | 8 | <0.0001 | NA | 0.06 | |
| P225H | 0 | 0 | 4 | 5 | 0.009 | 0 | 0 | 1 | 2 | 0.2 | NA | 0.3 | |
| NNRTI related polymorphisms | A98S | 3 | 1 | 1 | 1 | 0.3 | 51 | 19 | 13 | 22 | 0.6 | <0.0001 | <0.0001 |
| K101N | 1 | 0.4 | 0 | 0 | NS | 0 | 0 | 6 | 10 | <0.0001 | 0.5 | 0.006 | |
| K101RQ | 3 | 1 | 5 | 6 | 0.003 | 22 | 9 | 0 | 0 | 0.01 | <0.0001 | 0.07 | |
| K103R | 1 | 0.4 | 0 | 0 | 1 | 5 | 2 | 1 | 2 | 1 | 0.2 | 0.4 | |
| V106I | 4 | 2 | 4 | 5 | 0.009 | 7 | 3 | 1 | 2 | 1 | 0.5 | 0.4 | |
| E138A | 5 | 2 | 3 | 4 | 0.03 | 7 | 3 | 5 | 8 | 0.06 | 0.8 | 0.3 | |
| V179I | 50 | 21 | 21 | 27 | <0.0001 | 7 | 3 | 1 | 2 | 1 | <0.0001 | <0.0001 | |
| K238R | 15 | 6 | 3 | 4 | 0.03 | 0 | 0 | 1 | 2 | 0.2 | <0.0001 | 0.6 | |
| Polymorphisms characteristicfor A/AE subtype | K122E | 126 | 54 | 46 | 60 | 0.001 | 14 | 6 | 12 | 20 | 0.001 | <0.0001 | <0.0001 |
| D123SEGN | 223 | 96 | 71 | 92 | 0.6 | 74 | 2 | 18 | 30 | 0.9 | <0.0001 | <0.0001 | |
| A158S | 99 | 42 | 37 | 48 | <0.0001 | 5 | 2 | 2 | 3 | 0.6 | <0.0001 | <0.0001 | |
| K173SLA | 226 | 97 | 75 | 97 | 1 | 1 | 0.4 | 8 | 13 | <0.0001 | <0.0001 | <0.0001 | |
| Q174K | 224 | 96 | 71 | 92 | 0.4 | 4 | 2 | 2 | 3 | 0.3 | <0.0001 | <0.0001 | |
| D177E | 200 | 86 | 58 | 75 | 0.3 | 110 | 43 | 23 | 38 | 0.6 | <0.0001 | <0.0001 | |
| Q207A | 229 | 98 | 76 | 99 | 1 | 4 | 2 | 16 | 27 | <0.0001 | <0.0001 | <0.0001 | |
| R211S | 214 | 92 | 74 | 96 | 0 | 2 | 0 | 0 | 0 | 1 | <0.0001 | <0.0001 | |
250 samples from 234 drug-naive patients and 115 samples from 78 treated A/AE patients were genotyped. 31 patients were sampled both prior to treatment and after treatment failure. RT mutations found in all A/AE naive patients were compared to those found in A/AE drug-treated ones and to samples from 254 drug naive and 60 drug treated B individuals diagnosed since 2001. The first available sample from each drug-naive individual was used for analysis. For mutation-frequency analysis of drug-treated patients each mutation was counted once. Only mutations showing statistically significant differences between drug-naive and drug-treated patients and/or between A/AE and B frequencies are included.
Major NRTI related mutations included TAMs: M41L, D67N, K70R, L210W, T216Y/F and K219Q/E, as well as A62V, K65R, L74V/I, L77F, F116Y, Q151M and M184V/I. Major NNRTI mutations included A98G, L100I, K101E/P, K103N/S, V106A/M, V108I, Y181C, Y188C/H/I, G190A/S, P225H and K238T.
NNRTIs – Non-nucleosides reverse transcriptase inhibitors; NS – Not significant; NRTIs – Nucleosides reverse transcriptase inhibitors;
Subtyping was performed using the Stanford Database Rapid Subtyping Tool [23]–[24]. According to that classification 192 patients had virus containing protease of subtype A and RT most similar to CRF01_AE; for 70 both the protease and the RT were CRF01_AE; 52 were of subtype A; and four had protease classified as CRF01_AE and RT classified as A. Other subtyping tools such as Geno2Pheno (http://www.geno2pheno.org/) or the Rega Subtyping Tool (http://jose.med.kuleuven.be/subtypetool/html/) [57] vary to some extent in the classification of variants.
Table 6. Resistance Mutations in the Protease of drug-naive and drug-treated individuals.
| Amino Acids | A/AEa | B patients diagnosed from 2001 on | p A/AE vs. B | ||||||||||
| Naive n = 234 | Treated n = 78 | p Naive A/AE vs. Treated A/AE | Naïve n = 254 | Treated n = 60 | p Naive B vs. Treated B | Naive A/AE vs. Naive B | Treated A/AE vs. Treated B | ||||||
| No | % | No | % | No | % | No | % | ||||||
| Major resistance mutations | D30N | 0 | 0 | 0 | 0 | NS | 0 | 0 | 1 | 0 | NS | NS | 1 |
| N88D/S | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | NS | 1 | |||
| L90M | 0 | 0 | 0 | 0 | 6 | 2.4 | 3 | 5 | 0.03 | 0.08 | |||
| Accessory resistance mutations | K20R | 44 | 19 | 14 | 18 | NS | 8 | 3 | 6 | 10 | 0.03 | <0.0001 | 0.2 |
| I62V | 19 | 8 | 7 | 9 | 130 | 51 | 36 | 60 | 0.3 | <0.0001 | <0.0001 | ||
| L63P | 61 | 26 | 27 | 36 | 148 | 58 | 58 | 97 | <0.0001 | <0.0001 | <0.0001 | ||
| A71V | 0 | 0 | 1 | 1 | 6 | 2 | 19 | 32 | <0.0001 | 0.03 | <0.0001 | ||
| T74S | 15 | 6 | 5 | 7 | 0 | 0 | 1 | 2 | 0.2 | <0.0001 | 0.2 | ||
| V77I | 29 | 12 | 5 | 7 | 129 | 51 | 24 | 40 | 0.2 | <0.0001 | <0.0001 | ||
| I93L | 143 | 61 | 39 | 51 | 85 | 34 | 19 | 32 | 0.9 | <0.0001 | 0.04 | ||
| A/AE signature mutations | E35D | 211 | 90 | 70 | 92 | NS | 90 | 35 | 26 | 43 | 0.3 | <0.0001 | <0.0001 |
| M36I | 234 | 100 | 76 | 100 | 29 | 11 | 13 | 22 | 0.05 | <0.0001 | <0.0001 | ||
| H69K | 233 | 100 | 76 | 100 | 0 | 0 | 3 | 5 | 0.007 | <0.0001 | <0.0001 | ||
| L89M | 231 | 99 | 71 | 93 | 2 | 0.8 | 13 | 6 | <0.0001 | <0.0001 | <0.0001 | ||
| No. of patient having resistanceconferring mutations | PI-m | 2 | 0.9 | 4 | 5 | 0.02 | 9 | 3.5 | 5 | 8 | 0.003 | <0.05 | NS |
| TAMs | 1 | 0.4 | 8 | 10 | 0.001 | 9 | 3.5 | 10 | 17 | 0.001 | 0.02 | ||
| Other N | 4 | 2 | 27 | 35 | <0.0001 | 1 | 0.4 | 15 | 25 | <0.0001 | 0.2 | ||
| Any N | 4 | 2 | 30 | 38 | <0.0001 | 9 | 3.5 | 19 | 32 | <0.0001 | 0.3 | ||
| NN only | 8 | 3 | 7 | 9 | <0.0001 | 9 | 3.5 | 3 | 5 | 0.7 | 1 | ||
| Any NN | 10 | 4 | 30 | 38 | <0.0001 | 10 | 4 | 16 | 27 | <0.0001 | 1 | ||
| N+NN | 2 | 1 | 18 | 23 | <0.0001 | 1 | 0.4 | 15 | 25 | <0.0001 | 0.6 | ||
| N+NN+PI | 0 | 0 | 3 | 4 | 0.003 | 0 | 0 | 0 | 0 | NA | NA | ||
| Any | 12 | 5 | 37 | 47 | <0.0001 | 18 | 7 | 22 | 37 | <0.0001 | 0.5 | ||
250 samples from 234 drug-naive patients and 115 samples from 78 treated A/AE patients were genotyped. 31 patients were sampled both prior to treatment and after treatment failure. PI mutations found in all A/AE naive patients were compared to those found in A/AE drug-treated ones and to samples from 254 drug naive and 60 drug treated B individuals diagnosed since 2001. The first available sample from each drug-naive individual was used for analysis. For mutation-frequency analysis of drug-treated patients each mutation was counted once. Only mutations showing statistically significant differences between drug-naive and drug-treated patients and/or between A/AE and B frequencies are included.
Mutations in the Protease: The PI mutations L23I, L24I, D30N, V32I, M46I/L, I47A, G48V, I50L/V, I54V, V82A/S, I84V/A/C, N88S/T and L90M were considered major mutations. Secondary PI mutations included L10V/I/F/M, K20R, L33F, M36I, F53L, A71V/I and G73S/T/C/A.
N – NRTIs; NN – NNRTIs; NNRTIs – Non-nucleosides reverse transcriptase inhibitors; NS – Not significant; NRTIs – Nucleosides reverse transcriptase inhibitors; PI – Protease inhibitors;
Subtyping was performed using the Stanford Database Rapid Subtyping Tool [23]–[24]. According to that classification 192 patients had virus containing protease of subtype A and RT most similar to CRF01_AE; for 70 both the protease and the RT were CRF01_AE; 52 were of subtype A; and four had protease classified as CRF01_AE and RT classified as A. Other subtyping tools such as Geno2Pheno (http://www.geno2pheno.org/) or the Rega Subtyping Tool (http://jose.med.kuleuven.be/subtypetool/html/) [57] vary to some extent in the classification of variants.
Thirty-four drug-naive patients (14.5%) carried the RT mutation A62V. The frequency of this mutation (6%) decreased significantly under treatment (Table 5) and there was no difference between treated A/AE and B at this amino-acid in spite of its high prevalence in the drug-naive A/AE.
Drug resistance in drug-treated patients
Among the 78 treatment-failing patients seven (9.1%) carried major PI-related mutations (Table 6), nine (11.5%) had thymidine-analog mutations (TAMs), 29 (37.2%) other NRTI-related mutations (of those, 27 (35%) had M184V, the most prevalent mutation), and 32 (41.0%) had NNRTI-related mutations (Table 5).
Of the 78 treatment-failing patients, 55 (70.5%) were exposed to PIs and 34 (43.6%) were genotyped while failing PI treatment. Notably, only 7 (20.6%) of the 34 had any major PI mutation. Six of those had been treated previously with non-boosted PIs. All the 15 patients, who failed treatment while receiving LPV/r as first-line therapy, did not have any major mutations, either protease or RT (Tables 5, 6, 7). The picture was entirely different for NNRTIs. Thirty-five were genotyped while failing EFV or NVP. Twenty-three, including 21 for whom these drugs where part of their first-line therapy, had at least one major NNRTI mutation, with K103N and G190A/S being the most prevalent (Table 5). Only five of 26 (19.3%) failing NNRTI as first-line therapy did not have any mutations (Table 5). The use of population sequencing in this study does not allow us to exclude existence of minority populations with drug-resistance mutations, as long as such a population is below 15% of total.
Table 7. Mutations found in patients failing regiments containing LPV/r or NNRTIs.
| Subtype | Treatment(No. of patients) | Patients(samples) | Past Other PIs | Past Other regimens | PI Mutations | NRTI Mutations | NNRTI Mutations | |
| A/AE | LPV/r (22) | 2 | IDV,NFV,SQV | >2 | + | + | + | |
| 3 (4) | − | ≥1 | − | M184V (4) | K101E(1);K103N(2);G190S(1) | |||
| 1 | − | ≥1 | − | T69N | − | |||
| 1 | − | ≥1 | − | − | K103N;G179I | |||
| 15 (19) | − | 0 | − | − | − | |||
| EFV(24) or NVP(2) | 21 | − | 0 | − | + | + | ||
| 5 | − | 0 | − | − | − | |||
| B | LPV/r (24) | 2 | IDV,NFV | >2 | + | + | + | |
| 1 | − | ≥1 | − | M184V | K103N;G190S | |||
| 1 | − | ≥1 | − | D67N;T215I;K219E | − | |||
| 20 | − | 0 | − | − | − | |||
| EFV(15) or NVP(3) | 8 | − | 0 | − | + | + | ||
| 5 | − | 0 | − | − | − | |||
Mutations found in patients failing regiments containing LPV/r or NNRTIs.
The Table classifies patients failing on LPV/r or NNRTI containing regimens according to the number of mutations conferring resistance to the different drug classes. “+” indicates presence of mutations, but for some patients the actual mutations are listed. “–” indicates “no mutations” or also “no previous PI-containing regimens”.
EFV – efavirenz; IDV – indinavir; LPV/r – lopinovir/ritonavir; NFV – nelfinavir; NNRTIs – Non-nucleosides reverse transcriptase inhibitors; NRTIs – Nucleosides reverse transcriptase inhibitors; NVP – nevirapine; SQV – saquinavir; PIs – protease inhibitors.
To gain more insights into the factors shaping the above-described pattern, we compared A/AE viruses to B-viruses from all treatment-failing patients genotyped at NHRL [5] and, especially, from those diagnosed after 2001, who were given similar treatment regimens (Tables 5 and 6, right panels). The latter differed significantly in the frequencies of the major resistance RT mutations T215FY and K219QE (NRTI) and of several secondary/accessory mutations, including: the protease mutations I13V, M36I, I62V, L63P, A71V, V77I, L89M and I93L (Table 6) and the RT mutations A98S, K101N, K103R and V179I (Table 5). Significantly, however, in patients diagnosed after 2001 with B-viruses, a pattern of treatment-failure with no detectable resistance-conferring mutations, occurring much more frequently in PI-treated than in NNRTI-treated patients, was again observed (Tables 5, 6, 7) similar to what we found in the A/AE patients.
Discussion
Although A/AE are among the most rapidly-spreading HIV variants, in particular in the Far East, FSU, and recently also in the Western hemisphere, relatively little has been documented about treatment outcomes in this group relatively to other variants. Here we summarize more than 10 years of well-documented treatment of A/AE viruses in Israel, including clinical, epidemiological and laboratory surveillance data.
As shown here, A/AE variants spread in Israel in the wake of a wave of accelerated immigration from FSU in the mid-1990’s [17], [18], which paralleled an explosive spreading of these variants there, mainly among IVDU [11]–[13]. In the late 1990’s, A/AE infection began to be diagnosed in Israel in significant numbers, mainly among IVDU and their spouses who immigrated to Israel from FSU. Among A/AE carriers, 55.3% were co-infected with hepatitis viruses, probably acquired during drug-use, as they are also blood borne viruses.
A major concern has been that the use and/or sharing of contaminated needles, indicated by this clinical association and partially accounting for the rapid spread of HIV in other parts of the world, may be commonly practiced by IVDU also in Israel. Several lines of evidence reported here suggest that certain measures taken by health authorities to limit the spread of HIV via intravenous drug injection [32] did have an impact, at least as regards IVDU carrying A/AE viruses. First, the high rates of co-infection with hepatitis were found only among immigrants who were infected in the FSU but not in those infected in Israel (Table 3). Additional evidence is the relatively low incidence of drug-resistance mutations in drug-naive IVDU diagnosed in Israel with the A/AE subtypes (5.8%). Resistant A/AE viruses were found before treatment mainly among MSM (23.3%), consistent with our findings in subtype B [5]. A third line of evidence is provided by the observation that the fractional rate of local infection of IVDU by other IVDU is only 5% (doubling time = 15 years, compared to <5 years for MSM [5]). Yet, since more than two-thirds of all documented IVDU with HIV in Israel are FSU-born (data not shown), it is possible that focused screening and prevention efforts, directed at IVDU among immigrants from FSU, would have reduced the infection rate even further and perhaps delayed transmission of A/AE viruses in the community (see Figure 1).
The average fractional rate of 5% does not tell us whether infection events occur sporadically and uniformly or rather are the consequence of highly-risky behaviors of a relatively few. Behavioral factors that distinguish risk-groups from each other can be revealed through phylogenetic analyses of virus sequences. We earlier identified large clusters of B-infected MSM [5] with a posterior probability >0.95 of having a common proximal ancestor [31] (calculated using BEAST [27]). In particular, resistance-conferring mutations, including the protease mutation L90M and the RT mutations K103N and T215Y were chain-transmitted to drug-naive individuals [5]. Such observations helped us reveal a trend of risky sexual behavior in MSM [5]. In sharp contrast, most evolution-tree “clusters” of A/AE-infected patients included only two individuals, mostly IVDU and their spouses (Figure 2). Three relatively large clusters included A/AE-infected MSM (Figure 2). The emergence of these latter clusters and our earlier characterization of large clusters of B-infected MSM [5] serve as “positive controls”, indicating that the scarcity of larger-than-two clusters of IVDU is real and meaningful.
Both demographics and behavior explain the lack of A/AE clustering. A large proportion of the A/AE-IDVU patients were infected in the FSU, as indicated also by the fact that only a few were diagnosed in Israel near sero-conversion (data not shown), in sharp contrast to subtype-B MSM [5]. Even though originally multiple infections occurred in a social-group setting, potential molecular evidence for such grouping would tend to be lost upon immigration of individual member(s) from each group, given the large pool of HIV-infected IVDU who did not immigrate to Israel. Thus, with the exception of IVDU and their infected spouses, we would not expect significant clustering of patients in phylogenetic trees consisting mostly of such immigrants. In contrast, we would expect to frequently find larger clusters of IVDU infected in Israel more recently if the practice of needle sharing were prevalent here. The fact that such clusters are not found, along with the other afore-mentioned evidence, may indicate that the drug-rehabilitation services and the needle-exchange project, initiated in 2003 by the Ministry of Health [32], were probably effective.
In B-infected MSM, we had interpreted frequent evidence for “recent infection” at the time of diagnosis, including sero-conversion, as an indication of risk-awareness, and of engagement in risk-behavior despite such awareness [5]. The fact that such evidence is not found in newly-diagnosed IVDU may suggest that further efforts to enhance risk-awareness and adherence to routine HIV testing among IVDU may be worthwhile.
Comparing the overall pattern of resistance-conferring mutations in A/AE-infected patients with such mutations in subtype B revealed large differences, mainly in the frequencies of PI-associated mutations and TAMs. The frequency of certain mutations in these categories in the treatment-failing B-population reached more than 30% [5] but only a few percent in the A/AE-population. Thus, although 34 patients were genotyped, some repeatedly, while they were failing PI treatment (usually boosted PI; Table 4), we found very few mutations conferring resistance to PIs (Table 6), and those that were found had emerged in patients earlier treated with non-boosted PIs. By contrast, individuals failing EFV treatment had NNRTI-related mutations independently of subtype, in particular K103N and G190AS. The A/AE backbone has certain signal mutations absent in B, e.g. the protease mutations M36I, L89M or the RT mutation R211S, and others (Tables 5 and 6; see also Kantor et al. [33]). There are no significant differences in the frequency of those between naive and treated A/AE individuals but such polymorphisms could a-priori account for the observation of different pathways (e.g., [34], [35]). The influence of the A62V mutation and other secondary mutations on long-term cART outcomes is also still unresolved.
Most B-patient genotypic data in our database pertain to patients treated before 2001, who received non-boosted PIs, while only twelve A/AE-infected individuals started treatment before 2001. To better understand resistance-pattern differences, we next restricted the B-subtype data used for comparison to patients diagnosed between 2001 and 2011. The differences in drug-resistance mutation frequencies between the two subtypes diminished drastically (Tables 5 and 6). B-patients who were treated with boosted PIs failed treatment with no major PI-related mutations, as did the A/AE patients, in agreement with findings recently reported by other centers [36]–[41]. Moreover, B-patients failing boosted-PI regimens also usually did not have NRTI-resistance mutations, like similarly-treated A/AE patients (Table 7). In addition, mutation frequencies in A/AE-infected patients genotyped prior to 2001, who were initially given mono- or 2-drug therapy, were similar to those found in the general B-population, except that D30N was not found in any of the 9 A/AE patients failing NFV while it was found in 23% of B-patients failing this drug [42]. Finally, in both PI-failing and NNRTI-failing individuals the number of TAMs was considerably lower during 2001–2011. More patients were failing NNRTI with TAMs (12.9%) than failing boosted PIs with TAMs (4.1%), although the difference was not significant. The trend is not surprising as diminution in TAMs under modern cART is attributed to the use of TDF and FTC instead of ZDV and 3TC, resulting in emergence of mainly K65R and M184V but not of TAMs [43]–[46]. Of note, although 51% were treated with TDF+FTC and 31% with ZDV +3TC, the latter treatment was significantly more often associated with treatment failure than the first (∼44% and ∼23%, respectively; p<0.01), suggesting a higher efficacy for the TDF+FTC combination.
Recently, analyses of in-vitro pharmacokinetic and pharmacodynamic data [3], [47]–[49] showed that antiviral activity falls quickly as drug concentration is reduced for drugs with sharp dose-response curves and short half-lives, such as boosted protease inhibitors, limiting the time during which resistance can be selected for. Poor adherence to such drugs could cause treatment failure via growth of virus susceptible to the drug. However, these mono-therapy results have yet to be extended to combination therapies in which several drugs overlap and interact. Neither these studies, nor the possibility that mutations may occur outside the protease-encoding gene [50]–[52], escaping detection by common genotyping, can fully explain the puzzling clinical observation of a regular virologic failure in the absence of any mutations, including those related to NRTIs (Tables 5 and 6). Indeed, once insufficient adherence effectively removes the PI-imposed selection pressure, patients should become even more likely to develop NRTI-related resistance mutations. Moreover, we have observed that this phenomenon of failure in the absence of any detectable resistance mutations is quite common also in NNRTI-containing regimens, though it is not as regular as in the boosted PI- regimens. We have identified distinct groups of patients, those failing with relatively low viral loads in the range of a few thousand cp/ml and those in the hundred-thousand range (not shown). We are tempted to speculate that in the higher range, the most frequent cause of failure-without-mutation is that the patient’s adherence was very poor indeed, while in the low VL range, though adherence may be far from optimal, drug concentrations are sufficient to partially suppress wild-type virus replication while the development of overt drug resistance may be delayed for weeks or months, due to existing genetic barriers and poor fitness of variants [53]. Partial suppression would be related to the local non-uniformity of drug concentration in lymphoid tissues and to the physiologically structured, non-uniform distribution of activated CD4 T cells; local HIV replication at foci of CD4 T-cell activation would be more difficult to inhibit [53]. The “high” and “low” dichotomy is found also in the VL of those failing with resistance mutations, but it had already been observed that maintaining the failing drug regimen often results in lower VL as compared to pretreatment levels [54]. In any case, the results reported here and elsewhere call for efforts to promote better patient adherence and perhaps also for a reevaluation of drug dosing and scheduling.
Not surprisingly, we found almost no PI-resistance mutations in A/AE viruses from drug-naive individuals (Table 6). While K103N found its way to this population (3%), G190AS, which developed in 21% of the treated individuals, was not found prior to treatment (Table 5). By contrast, because of the high prevalence of PI mutations and TAMS in the B-infected population treated before 2001, these mutations were also transmitted to drug-naive individuals [5], [6].
Our dataset necessarily involves a degree of idiosyncrasy, as would any region-specific data, limiting generalization. An example is the discrepancy between our failure to find the mutation G190AS among drug-naive individuals, in both subtypes, and the transmission of this mutation to subtype-B infected individuals reported elsewhere [19]. We note that the unpredictable existence of particular transmission networks (“clusters”) among MSM in one region but not in the other might be sufficient to account for differences in the presence of particular mutations. Such circumstances are facilitated by the relatively small size of the populations under study.
Our comparative analysis of the patterns of drug-resistance mutations in treated patients infected with A/AE-HIV versus B does not exclude differences related to polymorphism, and understanding of such effects may be required to optimize treatment of non-B HIV infection, as indicated in other subtypes [1], [2], [33], [55]. However, the many resistance-pattern differences seen prior to regimen stratification reflected primarily the differences in treatment history, as the older treatment regimens had lower efficacy and the drug-virus interactions leading to drug resistance when a patient’s adherence was incomplete differed as well.
In summary, this study provides evidence that the practice of needle sharing among IVDU in Israel is not wide-spread as it might have been. In all classes of patients, strict adherence to treatment should be maintained to minimize both the emergence of drug-resistance and wild-type virus replication in the presence of drugs. Structural differences between subtypes A/AE and B may subtly affect drug-resistance pathways. Our study underscores the difficulties in discerning inherent viral effects from clinical, demographic, epidemiologic and behavioral factors and the need for comprehensive multidimensional analyses of consolidated data in order to weigh these factors relative to each other for the benefit of clinicians and health-policy makers.
Acknowledgments
We thank Zehuvit Wiexelboim and Ruslan Gosinov for extensive and rigorous data collection and processing, and Hagit Rudich, Fenando Mileguir, Marina Vax, and Ruth Pavel for their excellent performance of the technical laboratory work.
Several sequences documented in the present study are shared with SPREAD, a scientific surveillance program supported by the European Commission [56].
Data presented in this paper were reported in part at the XVIII International HIV Drug Resistance Workshop, June 9–13, 2009, in Fort Myers, Florida (abstract in Antiviral Therapy 2009; 14 Suppl 1: A165) and at the 15th Annual Meeting of the European Society for Clinical Virology, September 4–7, 2012, Madrid, Spain (abstract # P046).
GenBank Accession Numbers: KC184162 - KC184397.
Funding Statement
This study was supported in part by a European Commission grant (LSHP-CT -2006-518211). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1. Spira S, Wainberg MA, Loemba H, Turner D, Brenner BG (2003) Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J Antimicrob Chemother 51: 229–240. [DOI] [PubMed] [Google Scholar]
- 2. Wainberg MA, Brenner BG (2010) Role of HIV Subtype Diversity in the Development of Resistance to Antiviral Drugs. Viruses 2: 2493–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bangsberg DR, Porco TC, Kagay C, Charlebois ED, Deeks SG, et al. (2004) Modeling the HIV protease inhibitor adherence-resistance curve by use of empirically derived estimates. J Infect Dis 190: 162–165. [DOI] [PubMed] [Google Scholar]
- 4. Bengtsson L, Thorson A (2010) Global HIV surveillance among MSM: is risk behavior seriously underestimated? AIDS 24: 2301–2303. [DOI] [PubMed] [Google Scholar]
- 5. Levy I, Mor Z, Anis E, Maayn S, Leshem E, et al. (2011) Men who have sex with men, risk behavior, and HIV infection: integrative analysis of clinical, epidemiological, and laboratory databases. Clin Infect Dis 52: 1363–1370. [DOI] [PubMed] [Google Scholar]
- 6. Turner D, Amit S, Chalom S, Penn O, Pupko T, et al. (2011) Emergence of an HIV-1 cluster harbouring the major protease L90M mutation among treatment-naive patients in Tel Aviv, Israel. HIV Med 13: 202–206. [DOI] [PubMed] [Google Scholar]
- 7. Adler MW (1987) ABC of AIDS. Development of the epidemic. Br Med J (Clin Res Ed) 294: 1083–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nabatov AA, Kravchenko ON, Lyulchuk MG, Shcherbinskaya AM, Lukashov VV (2002) Simultaneous introduction of HIV type 1 subtype A and B viruses into injecting drug users in southern Ukraine at the beginning of the epidemic in the former Soviet Union. AIDS Res Hum Retroviruses 18: 891–895. [DOI] [PubMed] [Google Scholar]
- 9. Roudinskii NI, Sukhanova AL, Kazennova EV, Weber JN Pokrovsky VV, et al. (2004) Diversity of human immunodeficiency virus type 1 subtype A and CRF03_AB protease in Eastern Europe: selection of the V77I variant and its rapid spread in injecting drug user populations. J. Virol. 78: 11276–11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saad MD, Aliev Q, Botros BA, Carr JK, Gomatos PJ, et al. (2006) Genetic forms of HIV Type 1 in the former Soviet Union dominate the epidemic in Azerbaijan. AIDS Res Hum Retroviruses 22: 796–800. [DOI] [PubMed] [Google Scholar]
- 11.UNAIDS. 2010 Global Report (2011) Available: http://www.unaids.org/documents/20101123_GlobalReport/.
- 12. Hemelaar J, Gouws E, Ghys PD, Osmanov S (2011) Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25: 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mathers BM, Degenhardt L, Phillips B, Wiessing L, Hickman M, et al. (2008) Global epidemiology of injecting drug use and HIV among people who inject drugs: a systematic review. Lancet 372: 1733–1745. [DOI] [PubMed] [Google Scholar]
- 14. Slater PE, Costin C (1993) The epidemiology of adult AIDS and HIV infection in Israel. Isr J Med Sci 29: 2–6. [PubMed] [Google Scholar]
- 15. Kaplan EH, Slater PE, Soskolne V (1995) How many HIV infections are there in Israel? Reconstructing HIV incidence from AIDS case reporting. Public Health Rev 23: 215–235. [PubMed] [Google Scholar]
- 16. Chemtob D, Grossman Z (2004) Epidemiology of adult and adolescent HIV infection in Israel: a country of immigration. Int J STD AIDS 15: 691–696. [DOI] [PubMed] [Google Scholar]
- 17.Mor Z, Pinsker G (2011) HIV/AIDS in Israel, Epidemiological Periodic Report 1981–2010. Public Health Services, Department of Tuberculosis & AIDS, MOH, Israel. Available: www.health.gov.il/TBAIDS.
- 18.Central Bureau of Statistics Jerusalem, the State of Israel (2010) Statistical abstract of Israel, Jerusalem. Available: http://www1.cbs.gov.il/.
- 19. Brenner BG, Roger M, Moisi DD, Oliveira M, Hardy I, et al. (2008) Transmission networks of drug resistance acquired in primary/early stage HIV infection. AIDS 22: 2509–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schupbach J, Gebhardt MD, Tomasik Z, Niederhauser C, Yerly S, et al. (2007) Assessment of recent HIV-1 infection by a line immunoassay for HIV-1/2 confirmation. PLoS Med 4: e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grossman Z, Vardinon N, Chemtob D, Alkan ML, Bentwich Z, et al. (2001) Genotypic variation of HIV-1 reverse transcriptase and protease: comparative analysis of clade C and clade B. AIDS. 15: 1453–1460. [DOI] [PubMed] [Google Scholar]
- 22. Kantor R, Machekano R, Gonzales MJ, Dupnik K, Schapiro JM, et al. (2001) Human Immunodeficiency Virus Reverse Transcriptase and Protease Sequence Database: an expanded data model integrating natural language text and sequence analysis programs. Nucleic Acids Res 29: 296–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, et al. (2003) Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 31: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rhee SY, Kantor R, Katzenstein DA, Camacho R, Morris L, et al. (2006) HIV-1 pol mutation frequency by subtype and treatment experience: extension of the HIVseq program to seven non-B subtypes. AIDS 20: 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, et al. (2009) Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: update. PLoS One 4: e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2009) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A. FigTree v1.3.1. (2009) Available: http://tree.bio.ed.ac.uk/software/figtree/.
- 28.Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. [DOI] [PMC free article] [PubMed]
- 29. Collins KR, Quinones-Mateu ME, Wu M, Luzze H, Johnson JL, et al. (2002) Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity. J Virol 76: 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kearney M, Palmer S, Maldarelli F, Shao W, Polis MA, et al. (2008) Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS 22: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grossman Z, Avidor B, Mor Z, Turner D, Levy I, et al. (2012) Identification of HIV Transmission Networks in Israel. Antiviral Therapy 17 Suppl 1A172. [Google Scholar]
- 32. Chemtob D, Levit S, Mell H, Margolis A, Levy A, et al. (2008) [“Injecting clean or being clean?” The international and Israeli experiences of Syringe Exchange Programs among injecting drug users]. Harefuah 147: 634–638. [PubMed] [Google Scholar]
- 33. Kantor R, Katzenstein DA, Efron B, Carvalho AP, Wynhoven B, et al. (2005) Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med 2: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ariyoshi K, Matsuda M, Miura H, Tateishi S, Yamada K, et al. (2003) Patterns of point mutations associated with antiretroviral drug treatment failure in CRF01_AE (subtype E) infection differ from subtype B infection. J Acquir Immune Defic Syndr 33: 336–342. [DOI] [PubMed] [Google Scholar]
- 35. Martinez-Cajas JL, Wainberg MA, Oliveira M, Asahchop EL, Doualla-Bell F, et al. (2012) The role of polymorphisms at position 89 in the HIV-1 protease gene in the development of drug resistance to HIV-1 protease inhibitors. J Antimicrob Chemother 67: 988–994. [DOI] [PubMed] [Google Scholar]
- 36. Audelin AM, Lohse N, Obel N, Gerstoft J, Jorgensen LB (2009) The incidence rate of HIV type-1 drug resistance in patients on antiretroviral therapy: a nationwide population-based Danish cohort study 1999–2005. Antivir Ther 14: 995–1000. [DOI] [PubMed] [Google Scholar]
- 37. Lohse N, Obel N, Kronborg G, Jorgensen LB, Pedersen C, et al. (2006) Declining prevalence of HIV-infected individuals at risk of transmitting drug-resistant HIV in Denmark during 1997–2004. Antivir Ther 11: 591–600. [PubMed] [Google Scholar]
- 38. Vercauteren J, Deforche K, Theys K, Debruyne M, Duque LM, et al. (2008) The incidence of multidrug and full class resistance in HIV-1 infected patients is decreasing over time (2001–2006) in Portugal. Retrovirology 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Di Giambenedetto S, Zazzi M, Corsi P, Gonnelli A, Di Pietro M, et al. (2009) Evolution and predictors of HIV type-1 drug resistance in patients failing combination antiretroviral therapy in Italy. Antivir Ther 14: 359–369. [PubMed] [Google Scholar]
- 40. Bartmeyer B, Kuecherer C, Houareau C, Werning J, Keeren K, et al. (2010) Prevalence of transmitted drug resistance and impact of transmitted resistance on treatment success in the German HIV-1 Seroconverter Cohort. PLoS One 5: e12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lataillade M, Chiarella J, Yang R, Schnittman S, Wirtz V, et al. (2010) Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PLoS One 5: e10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grossman Z, Paxinos EE, Averbuch D, Maayan S, Parkin NT, et al. (2004) Mutation D30N is not preferentially selected by human immunodeficiency virus type 1 subtype C in the development of resistance to nelfinavir. Antimicrob Agents Chemother 48: 2159–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nikolenko GN, Delviks-Frankenberry KA, Palmer S, Maldarelli F, Fivash MJ Jr, et al. (2007) Mutations in the connection domain of HIV-1 reverse transcriptase increase 3′-azido-3′-deoxythymidine resistance. Proc Natl Acad Sci USA 104: 317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Svicher V, Alteri C, Artese A, Forbici F, Santoro MM, et al. (2010) Different evolution of genotypic resistance profiles to emtricitabine versus lamivudine in tenofovir-containing regimens. J Acquir Immune Defic Syndr 55: 336–344. [DOI] [PubMed] [Google Scholar]
- 45. Gallant JE, DeJesus E, Arribas JR, Pozniak AL, Gazzard B, et al. (2006) Tenofovir DF, emtricitabine, and efavirenz vs. zidovudine, lamivudine, and efavirenz for HIV. N Engl J Med 354: 251–260. [DOI] [PubMed] [Google Scholar]
- 46. Parikh UM, Zelina S, Sluis-Cremer N, Mellors JW (2007) Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS 21: 1405–1414. [DOI] [PubMed] [Google Scholar]
- 47. Shen L, Peterson S, Sedaghat AR, McMahon MA, Callender M, et al. (2008) Dose-response curve slope sets class-specific limits on inhibitory potential of anti-HIV drugs. Nat Med 14: 762–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sampah MES, Shen L, Jilik BL, Siliciano RF (2011) Dose-response curve slope is a missing dimension in the analysis of HIV-1 drug resistance. Proc. Natl. aca. Sci. USA 108: 7613–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosenbloom DIS, Hill AL, Rabi SA, Siliciano RF, Nowak MA (2012) Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nat. Med. 18: 1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parry CM, Kohli A, Boinett CJ, Towers GJ, McCormick AL, et al. (2009) Gag determinants of fitness and drug susceptibility in protease inhibitor–resistant human immunodeficiency virus type 1. J. Virol. 83: 9094–9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dam E, Quercia R, Glass B, Descamps D, Launay O, et al. (2009) Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 5, e1000345. [DOI] [PMC free article] [PubMed]
- 52. Gupta RK, Kohli A, McCormick AL, Towers GJ, Pillay, et al (2010) Full length HIV-1 gag determines protease inhibitor susceptibility within in vitro assays. AIDS 24: 1651–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grossman Z, Polis M, Feinberg MB, Grossman Z, Levy I, et al. (1999) Ongoing HIV dissemination during HAART. Nat Med. 5: 1099–1104. [DOI] [PubMed] [Google Scholar]
- 54. Deeks SG, Barbour JD, Grant RM, Martin JN (2002) Duration and predictors of CD4 T-cell gains in patients who continue combination therapy despite detectable plasma viremia. AIDS 25 16: 201–207. [DOI] [PubMed] [Google Scholar]
- 55. Wainberg MA, Zaharatos GJ, Brenner BG (2011) Development of antiretroviral drug resistance. N Engl J Med 365: 637–646. [DOI] [PubMed] [Google Scholar]
- 56. Wensing AM, van de Vijver DA, Angarano G, Asjo B, Balotta C, et al. (2005) Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J Infect Dis 192: 958–966. [DOI] [PubMed] [Google Scholar]
- 57. de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, et al. (2005) An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 21: 3797–3800. [DOI] [PubMed] [Google Scholar]