Abstract
Swarming motility and hemolysis are virulence-associated determinants for a wide array of pathogenic bacteria. The broad host-range opportunistic pathogen Serratia marcescens produces serratamolide, a small cyclic amino-lipid, that promotes swarming motility and hemolysis. Serratamolide is negatively regulated by the transcription factors HexS and CRP. Positive regulators of serratamolide production are unknown. Similar to serratamolide, the antibiotic pigment, prodigiosin, is regulated by temperature, growth phase, HexS, and CRP. Because of this co-regulation, we tested the hypothesis that a homolog of the PigP transcription factor of the atypical Serratia species ATCC 39006, which positively regulates prodigiosin biosynthesis, is also a positive regulator of serratamolide production in S. marcescens. Mutation of pigP in clinical, environmental, and laboratory strains of S. marcescens conferred pleiotropic phenotypes including the loss of swarming motility, hemolysis, and severely reduced prodigiosin and serratamolide synthesis. Transcriptional analysis and electrophoretic mobility shift assays place PigP in a regulatory pathway with upstream regulators CRP and HexS. The data from this study identifies a positive regulator of serratamolide production, describes novel roles for the PigP transcription factor, shows for the first time that PigP directly regulates the pigment biosynthetic operon, and identifies upstream regulators of pigP. This study suggests that PigP is important for the ability of S. marcescens to compete in the environment.
Introduction
Serratia marcescens is an important opportunistic pathogen of humans [1]. Scores of articles describe hospital outbreaks caused by S. marcescens, and clinical studies indicate that it is a major cause of hospital-acquired pneumonia, blood stream infections, surgical site infections, and urinary tract infections [2]–[4]. For example, S. marcescens was shown to be the fourth most common cause of early onset pneumonia of intensive care unit (ICU) patients when no antibiotic was administered, and the number one cause of early onset pneumonia of ICU patients to whom systemic antibiotics had been administered [5]. Additionally, S. marcescens frequently causes community acquired infections [6] including vision threatening microbial keratitis [7], [8]. Beyond humanity, this gram-negative bacterium is capable of infecting a very wide range of hosts including coral, insects, mammals, nematodes and plants [1]. These varied hosts represent different environmental niches in which S. marcescens may compete against other microorganisms.
Swarming motility is a surface-associated group behavior that confers antibiotic resistance [9], [10]. This mode of motility has been noted as a virulence determinant for other gram-negative bacteria such as Pseudomonas aeruginosa [11] and Proteus mirabilis [12]–[15]. S. marcescens flagella and surfactants, known as serrawettins, contribute to swarming motility [16]–[18]. Serratamolide (also known as serrawettin W1), one of these biosurfactants, is a small amino-lipid necessary for swarming in many S. marcescens strains, and is co-regulated with the red pigment prodigiosin with respect to temperature and growth phase [19]–[22]. In several organisms, including S. marcescens, swarming motility and hemolysis are co-regulated [23]–[29].
Hemolysins are major virulence factors for a wide variety of bacterial pathogens [30]–[32]. A recent study showed that serratamolide, originally characterized for its broad-spectrum antimicrobial activity [33], [34], can be a cytotoxic hemolysin [22]. Biosynthesis of serratamolide is catalyzed by the non-ribosomal peptide synthetase SwrW [20]. Known serratamolide regulators are the LysR family regulator, HexS [21], and the cAMP-receptor protein, CRP [22]. These are both inhibitors of swrW transcription, with HexS shown to be a direct inhibitor that binds to the swrW promoter [21]. Interestingly, both HexS and CRP also regulate prodigiosin production [21], [35].
Given the importance of serratamolide in swarming motility and hemolysis, we hypothesized the existence of a positive transcriptional regulator(s). Since serratamolide and prodigiosin can be co-regulated, we predicted that positive regulators of prodigiosin would also positively regulate serratamolide production. Relatively little is known about the transcriptional regulators of prodigiosin in S. marcescens, with the exception of CRP [35], HexS [21], RssAB [36], and SpnR [37] - all negative regulators. Genetic studies have provided a much more thorough understanding of prodigiosin regulation in Serratia species ATCC 39006, an atypical Serratia that was isolated in a salt marsh in New Jersey, USA [38]–[44]. Like S. marcescens, it produces many secondary metabolites and secreted enzymes that may be of medical and industrial importance [45].
Among the positive regulators of prodigiosin production in Serratia sp. ATCC 39006 is PigP, a predicted transcription factor of the XRE family [46]. The PigP protein was reported as a positive regulator of a carbapenem antibiotic and prodigiosin [46]. A pigP transposon mutant displayed additional defects in production of cellulase activity, modest reductions in pectinase, and altered swimming motility zones [46]. The use of a transposon with a lacZ reporter allowed Fineran and colleagues to show that loss of PigP led to reduced expression of other putative prodigiosin regulators [46]. These include pigS and pigX, the expression of which were strongly reduced in the pigP mutant, as well as pigQ, pigR, and rap whose expression was more modestly reduced. Expression of pigP was shown to be independent of quorum sensing, and mutation of pigP led to a surprisingly small reduction in pigA transcription (∼25–50% reduction in transcript) that manifested in a ∼95% reduction in pigment levels [46]. In the second report regarding PigP, it was shown that mutation of pigP did not alter expression of prodigiosin regulator pigZ [47]. In the third report that addressed pigP, Gristwood and colleagues showed that the absence of PigS, an Ars/SmtB family transcriptional regulator, lead to increased expression of the gene for a pigment inhibitor, blhA, and wild-type levels of blhA expression could be restored by mutation of the pigP gene. However, mutation of pigP alone did not significantly alter blhA expression [48].
As the study of PigP is relatively new, there are many gaps in knowledge that need to be filled. For example, there are no known upstream regulators of pigP expression [45], and it is not known whether PigP can directly regulate transcription of the prodigiosin biosynthetic operon. In this study, we tested the hypothesis that an uncharacterized PigP homolog from S. marcescens regulates swarming and hemolysis through serratamolide production. Additionally, it is not clear whether genes identified as secondary metabolite regulators in Serratia sp. ATCC 39006 are important in S. marcescens. The exception is the hexS gene of S. marcescens [21], which was named pigU in Serratia sp. ATCC39006 [46]. Therefore, we tested whether observed phenotypes conferred by pigP mutation in Serratia sp. ATCC39006 were conserved in S. marcescens in addition to our central hypothesis.
Data from the current study indicate a novel role for PigP from environmental, laboratory and clinical strains of S. marcescens as a positive regulator of swarming motility and hemolysis through indirect regulation of serratamolide production. S. marcescens PigP was found to be a positive regulator of pigment production through direct regulation of the prodigiosin biosynthetic operon, pigA-N, as well as being a direct positive regulator of pigP. Importantly, upstream regulators for pigP were determined; HexS and CRP were observed to be direct and indirect regulators, respectively, of pigP transcription.
Like serratamolide, prodigiosin may have a role in competition for environmental niches as it has anti-microbial activity [49]–[51]. Prodigiosin has also been correlated with hydrophobicity-mediated bacterial adhesion that may be important for colonization and distribution of bacteria [52], [53] and has roles in pH and energy homeostasis [54], [55]. The evidence here presents PigP as a key regulator of both an antimicrobial biosurfactant that promotes antibiotic resistance and an antimicrobial pigment, supporting a model where PigP is a key regulator controlling the interplay between S. marcescens and other organisms.
Materials and Methods
Bacterial Strains, Media, and Growth
Microbial strains used in this study are listed in Table 1. All bacteria were grown with LB (0.5% yeast extract, 1% tryptone, 0.5% NaCl). LB broth was supplemented with adenosine 3′, 5′-cyclic monophosphate (cAMP) where noted; cAMP powder was dissolved in LB and filtered. Antibiotics used to select for plasmids were gentamicin (10 µg/ml) and kanamycin (100 µg/ml). Tetracycline was used at 10 µg/ml to select against Escherichia coli in conjugations.
Table 1. Strains and plasmids used in this study.
| Strain | Description | Reference or source |
| InvSc1 | Saccharomyces cerevisiae strain uracil auxotroph | Invitrogen |
| S17-1 λpir | Escherichia coli strain used for conjugation and cloning | [71] |
| EC100D | E. coli strain used for cloning and protein purification | Epicentre |
| ER2566 | E. coli strain used for protein purification | New England Biolabs |
| MZ100 | Staphylococcus aureus laboratory strain | [58] |
| K950 | S. aureus clinical keratitis isolate (MRSA) | [59] |
| K2315 | Proteus mirabilis keratitis isolate | Regis Kowalski |
| CMS376 | Serratia marcescens wild type, PIC strain number 3611 | Presque Isle Cultures |
| CMS524 | cyaA-2– transposon mutation in CMS376 | [72] |
| CMS635 | swrW – transposon mutation in CMS376 | [22] |
| CMS786 | crp-23– transposon mutation in CMS376 | [72] |
| CMS836 | pigP-1 (pigP::pMQ118) in CMS376 | This study |
| CMS1033 | SMA3565::pMQ118 in CMS376 | This study |
| CMS1613 | hexS::pMQ118 in swrW mutant (CMS635) | This study |
| CMS1687 | Δcrp, CMS376 with crp-Δ4 allele | [35] |
| CMS1713 | ΔpigP, CMS376 with pigP-Δ allele | [73] |
| CMS1742 | Δcrp ΔpigP in CMS376 | This study |
| CMS1744 | hexS::pMQ118 in ΔpigP (CMS1713) | This study |
| CMS1779 | hexS::pStvZ3 in WT (CMS376) | This study |
| CMS1781 | hexS::pStvZ3 in ΔpigP (CMS1713) | This study |
| CMS1785 | PpigP-pStvZ3 in WT (CMS376) | This study |
| CMS2210 | ΔhexS in WT (CMS376) | [63] |
| Nima | S. marcescens ATCC 29632 laboratory strain | This study |
| CMS2980 | pigP-1 mutation in Nima | This study |
| CHASM | Environmental S. marcescens isolate | [35] |
| CMS2981 | pigP-1 in CHASM | This study |
| K904 | S. marcescens clinical keratitis isolate | [35] |
| CMS2982 | pigP-1 in K904 | This study |
| CMS2234 | swrW-transposon mutation in swrW | This study |
| K997 | S. marcescens clinical isolate, non-pigmented | This study |
| CMS2983 | pigP-1 in K997 | This study |
| CMS3408 | PpigP-pStvZ3 in ΔhexS (CMS2210) | This study |
| UC1SER | S. marcescens isolate from human neonate gut | [64] |
Mutagenesis, Plasmid Construction, and ß-galactosidase Analysis
Chromosomal DNA and plasmid DNA were isolated with commercial kits (Achieve pure DNA cell/tissue, 5 Prime; GenElute Plasmid, Sigma). PCR was performed using a high-fidelity polymerase (Phusion, New England Biolabs). All cloning was performed using yeast in vivo cloning [56]. Plasmid details are listed in Table 2.
Table 2. Plasmids used in this study.
| Plasmid | Description | Reference or source |
| pMal-C2 | Maltose binding protein fusion construct | New England Biolabs |
| pStvZ3 | oriR6K lacZ nptII promoter probe | [35] |
| pMQ118 | suicide vector nptII, rpsL, oriT, URA3, CEN6/ARSH4 | [73] |
| pMQ124 | oriColE1, oripRO1600, aacC1, PBAD-lacZa, oriT, URA3, CEN6/ARSH4 | [73] |
| pMQ125 | orip15a, oripRO1600, aacC1, PBAD-lacZa, oriT, URA3, CEN6/ARSH4 | [73] |
| pMQ131 | oripBBR1, aphA-3, Plac-lacZa, oriT, URA3, CEN6/ARSH4 | [73] |
| pMQ132 | oripBBR1, aacC1, Plac-lacZa, oriT, URA3, CEN6/ARSH4 | [73] |
| pMQ179 | pMQ118 with internal pigP fragment | This study |
| pMQ196 | pMQ118 with internal hexS fragment | This study |
| pMQ200 | oriR6K, nptII, PBAD-lacZa, oriT, URA3, CEN6/ARSH4 | [73] |
| pMQ212 | pMQ125 with the pigP open reading frame | This study |
| pMQ242 | pMQ124+ His8-CRP | [35] |
| pMQ248 | pMQ131+ PflhD-lacZ (flhD promoter) | This study |
| pMQ253 | pStvZ3+ PpigP (pigP promoter) | This study |
| pMQ268 | pStvZ3+ pigA internal fragment | [35] |
| pMQ272 | pStvZ3+ hexS internal fragment | [22] |
| pMQ302 | pMQ124+ His9-pigP | This study |
| pMQ367 | pMQ125+ swrW | [22] |
| pMQ368 | pMQ200+ swrW | This study |
| pMQ376 | oripBBR1-based plasmid with PswrW-tdtomato reporter | [22] |
| pMQ402 | pMAL-C2+ hexS (MBP-HexS fusion construct) | [63] |
Insertional mutation of the predicted pigP homolog (SMA3564), SMA3565, and hexS: Internal regions of pigP, SMA3565 and hexS open reading frames (ORF) were amplified and cloned in the suicide-vector pMQ118 [57]. Primer pairs 1238–1239, 1479–1480, and 1337–1338 respectively, were used to amplify and clone the internal region of pigP, SMA3565, and hexS. Primers are listed in Table S1. The pigP and SMA3565 constructs were introduced into S. marcescens as previously described [57]. Briefly, pMQ118 with internal fragments recombine with the respective chromosomal gene yielding a disruption of the gene. In pigP-1, the pMQ118 insertion is at base pair 466 out of 615 base pairs ORF. For SMA3565, pMQ118 inserts after base pair 525 out of 897 base pairs for the entire ORF. The hexS insertion construct integrates at base pair 400 out of 945. Mutations were verified using PCR. All insertional mutations generated in this manner were grown in kanamycin (100 µg/ml) to maintain the mutation. Controls were performed using CMS376 (wild-type strain) with a kanamycin resistance marker bearing plasmid to ensure that antibiotics alone did not affect the studied phenotypes (data not shown).
The pigP-lacZ transcriptional reporter was generated using the pStvZ3 plasmid as previously described [35], using primers 1444 and 1445. Briefly, a 491 base pair promoter region immediately upstream of the pigP ORF was amplified and cloned upstream of lacZ in pStvZ3 to generate a transcriptional fusion, resulting in plasmid pMQ253. Integration of pMQ253 creates a transcriptional lacZ fusion with the native promoter of pigP, and places the wild-type pigP gene under transcriptional control of the 491 base pair region upstream of pigP.
The pMQ248 plasmid has the flhD promoter driving expression of lacZ, and was used here as a source of flhD promoter DNA for controls in electrophoretic mobility shift assays noted below. The plasmid was generated using yeast homologous recombination in which an oxyR promoter (to be published elsewhere) was replaced with the flhD promoter in a pMQ131 background. Primers for amplification of the flhD promoter are listed in Table S1 as 1851 and 1852.
The full-length pigP gene was amplified and cloned into pMQ132 under control of the E. coli Plac promoter using primers 1645 and 1646. The resulting plasmid, pMQ221, was used for complementation analysis. An inducible pigP expression plasmid, pMQ212, was made by amplifying the pigP ORF from CMS376 and placing it under control of the E. coli PBAD promoter in vector pMQ125 using primers 1483–1484.
An N-terminal His9-tagged version of pigP was generated under control of the E. coli PBAD and recombined into pMQ124 using primer sets: 2093 and 2094, generating plasmid pMQ302.
Full-length swrW was amplified using primers detailed previously [22], and cloned into pMQ200 under control of the E. coli PBAD promoter, generating plasmid pMQ368.
Detection of pigP in S. marcescens Isolates
Bacteria from frozen stocks of ocular clinical isolates obtained from the Charles T. Campbell Laboratory of Ophthalmic Microbiology or other strains listed in Table 1 were streaked to single colonies on LB or TSA blood agar plates. DNA was extracted from a single colony using Quick Extract (Epicentre) according to the manufacturers specifications. PCR was performed using standard Taq polymerase (New England Biolabs), and standard conditions using the following primer sets to detect the pigP gene (1230–1231 and 1238–1239). S. marcescens (CMS376) and Staphylococcus aureus (MZ100 and K950) [58], [59] or Proteus mirabilis (K2315) chromosomal DNA were used as positive and negative controls respectively. As an additional control for false positive PCR amplicons, the amplified DNA from five randomly chosen isolates was sequenced and all were pigP amplicons. Analysis was performed twice with each primer set and any reproducibly generated amplicon of the expected size for any strain was considered a positive result. A quality control PCR reaction was also performed on each DNA preparation to eliminate false negative results using previously described primers, 736– 737, that amplify the oxyR gene [57].
Prodigiosin Production Assays
Single colonies were inoculated in 5 ml of LB medium ± antibiotics and incubated for 18–20 hours (h) on a rotary shaker (TC-7, New Brunswick) at speed setting “8”, (62 rpm). Prodigiosin was extracted from bacterial cells using acidified ethanol, and levels were determined by measuring absorbance at 534 nm, based upon the method of Slater, et. al. [60]. Absorbances of extracted prodigiosin and turbidity (OD600 nm) of the original culture were read with a spectrophotometer (Molecular Devices, Spectramax Plus) using 1 cm2 cuvettes, and the ratio was determined.
Transcriptional Analysis
ß-galactosidase (ß-gal) assays: after growth of bacterial cultures in LB medium at 30°C to a desired optical density, culture aliquots were pelleted, washed with Z-buffer, and analyzed for ß-gal activity [61]. Lysates were prepared by sonication in Z-buffer and were clarified by centrifugation at 16,100×g for 10 minutes (m). Protein concentration was determined by Bradford analysis, and the same amount of protein from each sample in a given experiment was added to microtiter plate wells and the volume was adjusted to 100 µl with Z-buffer. ONPG (25 µl at 0.2 mg/ml) was added as a substrate, and A410 readings were taken with a plate reader after incubation for 10–30 m (Biotek, Synergy 2). Activity was expressed as A410/(mg protein×minutes).
Tdtomato fluorescence as a reporter for swrW promoter activity was measured from 0.15 ml aliquots of bacterial cultures grown in LB broth with kanamycin using a Synergy 2 plate reader as previously described [22]. The excitation filter for fluorescence was 545/40 nm, the emission filter used measured fluorescence at 590/20 nm. Background fluorescence was equivalent in both strains and the fluorescence was normalized to culture optical density, measured at 600 nm. The experiment was repeated on two different days with similar results.
RNA and cDNA preparation and reverse transcriptase–PCR (RT-PCR) was performed as previously described [22]. The methodology for semi-quantitative RT-PCR and analysis detailed by Marone and colleagues was followed [62]. Primers sequences 2638 and 2639 to detect 16S rDNA were taken from Lin, et al., [28]. Primers 1230 and 1231 were used to detect pigP. Primers 2911 and 2912 were used to detect pigA. Primers 2917–2918 were used to detect swrW. A no RT control was performed for every RNA sample and was used to ensure the absence of contaminating chromosomal DNA in cDNA samples (data not shown). Experiments were performed at least three times with two or more independent RNA preparations.
Operon analysis was performed by generating cDNA from RNA from wild-type (WT) cultures harvested at OD600 = 2.0 and converted to cDNA with Superscript III reverse transcriptase (Invitrogen) or without RT as a control for chromosomal DNA contamination. Primers to amplify an internal region of pigP were 1238 and 1239. Primers to amplify between pigP and SMA3565 were 2701 and 2702. Primers to amplify between SMA3565–3566 were 2705 and 2706.
Protein purification and electrophoretic mobility shift assays (EMSA) were performed as previously described using the same reagents [35]. Recombinant His8-CRP, expressed from pMQ242, was previously purified [35]. Recombinant His9-PigP was purified by nickel-affinity chromatography. Briefly, cultures of EC100D containing empty vector pMQ124 or the His9-pigP expression vector, pMQ302 were grown overnight in LB medium with gentamicin. Bacteria were diluted to OD600 = 0.1 in LB medium with gentamicin and grown with aeration at 30°C until cultures reached OD600 = 0.5, at which point L-arabinose (10 mM) was added and cultures were grown for 3 h. Cells were pelleted, washed, and suspended in lysis buffer: sodium phosphate buffer (50 mM), NaCl (300 mM), imidazole (10 mM), triton X-100 (0.1%), pH 8. Clarified lysates were loaded onto columns with HisPur cobalt resin (Pierce), washed twice with wash buffer (same as lysis buffer, but with imidazole at 20 mM). Protein was eluted with elution buffer (as wash buffer without triton X-100, and with imidazole at 100–500 mM). Protein concentration was determined by Bradford analysis. Protein purity was assessed by PAGE analysis where there were no additional bands in the His9-PigP eluted fractions, and no purified band in the negative control purification (data not shown).
MBP-HexS and MBP were both purified using maltose-agarose according to the manufacturers specifications (pMAL Protein Fusion and Purification System, New England Biolabs). ER2566 bearing pMAL-C2 for purification of MBP, or pMQ402 [63] for purification of MBP-HexS, grown overnight, subcultured with aeration at 30°C until cultures reached OD600 = 0.5, induced with IPTG (0.3 mM) for 3 h, pelleted and frozen. Pellets were washed and suspended in column buffer [Tris-HCl (20 mM), NaCl (200 mM), EDTA (1 mM)], lysed by sonication, clarified by centrifugation, and lysates were loaded onto columns with amylose resin. The resin was washed with 12 volumes of column buffer, and eluted with column buffer containing maltose (10 mM). Protein purity was assessed by PAGE analysis and was greater than 80% pure when analyzed by ImageJ (data not shown).
Labeled DNA amplicons were made with 5′-biotinylated oligonucleotide primers (Integrated DNA Technologies), gel purified and verified by sequencing. A commercial EMSA kit was employed as specified by the manufacturer (Lightshift Chemiluminescent EMSA kit, Pierce) using biotinylated target DNA (1–3 ng), purified His8-tagged CRP (≥50 ng), poly-dIdC (500 ng), cAMP (500 µM), and non-labeled competitor DNA (500 ng) in a 20 µl reaction. A 10 µl aliquot of the reaction was separated on a 5% PAGE, TBE gel (Bio-Rad) with running buffer containing 500 µM cAMP. His9-PigP and MBP-HexS EMSAs were performed as above, except using poly-dIdC at 1 µg, MgCl2 (5 mM), NP-40 (0.05%), 4 ng of biotinylated target DNA, and 500 ng of unlabeled target DNA, 25 µg of MBP-HexS and 33 µg of MBP or 5–20 µg of His9-PigP as indicated. EMSA experiments were performed at least three times on different days with similar results. Primers for the pigA promoter were 1665 and 1713 (Table S1) and WT chromosomal DNA was used as a template to amplify a 635 bp amplicon that includes 426 bp upstream of the pigA start codon as previously described [35]. Primers for the pigP promoter were 1346 and 1883 and were used with pMQ253 as a template to amplify a 650 bp amplicon including 492 bp upstream of the pigP start codon. Primers for the swrW promoter were 2737 and 2738 using WT chromosomal DNA as a template to amplify a 379 bp amplicon including 352 bp of DNA upstream of the swrW start codon. Primers for the flhD promoter were 1671 and 1672 and pMQ248 was used as a template to amplify a 288 bp amplicon of DNA upstream of the flhDC operon as previously described [35]. Primers for the oxyR promoter were 1673 and 1675 and used WT chromosomal DNA as a template to amplify a 248 bp amplicon upstream of the oxyR gene. Primers to amplify a 345 bp amplicon upstream of the hexS gene from WT chromosomal DNA were 2781 and 2782. Primers to amplify a 223 bp amplicon including 199 bp upstream of the pswP gene were 2926 and 2928.
Chromatin affinity purification (ChAP) assays were performed as follows. For each strain (WT+pMQ124, WT+pMQ242, and WT+pMQ302), individual single colonies were placed into three, 5 ml cultures consisting of LB medium. These were grown overnight at 30°C with aeration, subcultured in LB medium, grown to OD600 = ∼0.5 at 30°C, supplemented with L-arabinose (13.3 mM), and grown to OD600 = ∼2.0. The three cultures for each group were combined and incubated with formaldehyde (1% final concentration) for 10 m at room temperature. Cross-linking was stopped by the addition of glycine (125 mM) and cells were washed with phosphate-buffered saline (PBS). Cells were resuspended in 1 ml lysis/equilibrium buffer (50 mM sodium phosphate, pH 8, 300 mM NaCl, 10 mM imidazole, and 0.1% triton X-100, 6 µg/ml RNase A), and sonicated for a time and intensity, determined in pilot assays, that sheared most DNA to 500–1000 bp. The lysate was centrifuged at 16,100×g for 10 m at 4°C. DNA was purified from a 100 µl aliquot of the lysate using a Qiagen PCR purification kit and the DNA concentration was determined with a spectrophotometer (Nanodrop N-1000) to normalize input DNA during the affinity purification. A sample was separated on an agarose gel to ensure uniform shearing among samples. A protease inhibitor cocktail (Halt, Pierce Thermo Scientific) was added to the remaining lysate to the manufacturers specifications. An aliquot was kept to represent the total input DNA. Affinity purification was performed on the remaining lysates, normalized by DNA concentration to 100 ng, using nickel-coated paramagnetic beads (PureProteome, Millipore). After two rounds of washing the lysate-incubated beads with lysis/equilibrium buffer, the protein-DNA complexes were separated from the beads using imidazole (1 M) and the eluate was incubated at 65°C for 14–16 h and passed through a Qiagen PCR purification column. The DNA was eluted in the kit-provided elution buffer (50 µl) and PCR was performed on samples using Taq polymerase (New England Biolabs) for 26–32 rounds to ensure amplification in the exponential range [62]. Digital images of non-saturated DNA bands from agarose gels were taken. Primers for amplifying pigA were 1665 and 1713, primers for amplifying pigP were 1444 and 1445, primers for amplifying oxyR were 1432 and 1433.
Biofilm Formation, Swimming, Swarming and Surfactant Zone Analysis
Static biofilm assays were performed as previously described [57] using polyvinyl chloride as a substrate, and incubation for 20 h at 30°C in LB medium. Biofilms were stained with crystal violet (0.1%); dye was solubilized with glacial acetic acid (33%), and measured spectrophotometrically (A590) with a plate reader. Swimming motility was measured with LB agar with 0.3% agar concentration. Surface swarming motility was assessed with LB agar (0.6% agar). Surfactant zones were measured using swarming agar plates. Assays were performed at 30°C. Where noted, serratamolide in DMSO (10 µl of 50 µg/ml) or DMSO (10 µl) were added to the center of swarming agar plates, allowed to dry, and test bacteria were spotted on the center of the plate.
To analyze serratamolide production in the WT and mutant strains of Serratia marcescens, 50 ml of overnight cultures were centrifuged. The supernatant was collected and extracted three times with 50 ml of ethyl acetate. The ethyl acetate layer was combined and dried over sodium sulfate and evaporated in vacuo. The resulting crude residue was re-dissolved in methanol and analyzed via Shimadzu LCMS-2020 using DIONEX Acclaim 120® C18 column (3 µm particle size, 120 Å pore size, 2.1 × 150 mm dimensions). The mobile phase gradient used for this analysis was as follows: 40% AcCN/60% H2O (0 min), 40% AcCN/60% H2O (1 min), 90% AcCN/10% H2O (15 min), 90% AcCN/10% H2O (35 min), 40% AcCN/60% H2O (40 min), 40% AcCN/60% H2O (45 min). The column oven temperature was set at 40°C and the flow rate was 0.2 ml/min. Serratamolide was monitored at m/z = 515 (for [M+H]+) using an ESI-MS detector at positive mode. Authentic serratamolide was isolated as previously reported [22] and used as a positive control in this analysis.
Statistical Analysis
Two-tailed Student’s t-tests and one-way ANOVA with Tukey’s pair-wise post-test analysis with significance set at p<0.05 was performed using Graphpad Prism 5 software.
Results
The S. marcescens pigP Gene is Required for Full Levels of Prodigiosin Production in Different Strain Backgrounds
The putative pigP gene from strain CMS376 used in this study was sequenced from plasmids (pMQ212 and pMQ221) containing the pigP ORF (GenBank accession number FJ041060). The predicted protein was found to be 69.1% identical to PigP of Serratia sp. ATCC 39006 [46] and 100% amino acid identity to the ORF predicted to code for PigP (SMA3564 ORF) from the sequenced S. marcescens strain, Db11.
To test the hypothesis that an S. marcescens PigP homolog controls prodigiosin production and promotes swarming and hemolysis, mutations in the pigP gene were generated. Because of the inherent artifacts associated with deletion mutations, i.e. deletion of cis-acting regulatory elements for adjacent genes, and with insertion mutations, where polar effects are likely, both types of mutation were used to assess PigP function. Mutant strains of a pigmented laboratory wild-type (WT) strain, CMS376, were generated. One had an insertion mutation in pigP (pigP-1 allele, strain CMS836) and another had an in-frame chromosomal deletion of the pigP (pigP-Δ allele, strain CMS1713, notated here as ΔpigP) in which the first 593 base pairs out of the 615 base pair ORF were deleted (Table 1). A severe defect in pigmentation was measured in both mutant strains (Table 3, 4, Figure 1A, Figure S1A). Both pigP mutant strains also exhibited a striking reduction in swarming motility and hemolysis (described below).
Table 3. Prodigiosin production in various genetic backgrounds.
| Straina | Prodigiosinb |
| WT (CMS376) | 0.17±0.04 |
| pigP (CMS1713) | 0.02±0.01 |
| SMA3565::pMQ118 (CMS1033) | 0.20±0.10 |
| crp (CMS1687) | 1.61±0.26 |
| crp pigP (CMS1742) | 0.11±0.01 |
| hexS (CMS2210) | 1.29±0.07 |
| hexS pigP (CMS1744) | 1.11±0.16 |
Strain numbers are defined in Table 1; all are isogenic to WT (CMS376).
A534/OD600, measured at 16–18 h, mean of n≥6 independent biological replicates per data point ± one standard deviation.
Table 4. Effect of pigP insertional mutation on prodigiosin production by different S. marcescens isolates.
| Straina | Prodigiosinb |
| WT (CMS376) | 0.11±0.02 |
| WT pigP-1 (CMS826) | 0.02±0.01 |
| CHASM | 0.10±0.01 |
| CHASM pigP-1 (CMS2981) | <0.01 |
| Nima | 0.76±0.11 |
| Nima pigP-1 (CMS2980) | 0.26±0.07 |
| K904 | 0.63±0.05 |
| K904 pigP-1 (CMS2982) | 0.02±0.02 |
Strain numbers are defined in Table 1.
A534/OD600, measured at 16–18 h, mean of n≥6 independent biological replicates per data point ± one standard deviation.
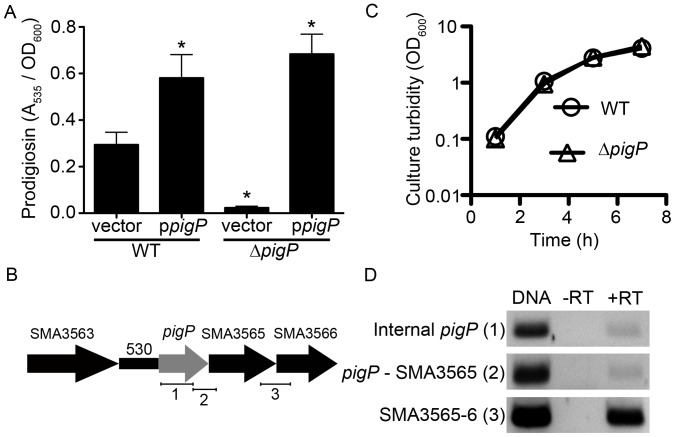
Figure 1. S. marcescens PigP positively regulates prodigiosin production.
A. Complementation analysis of a pigP mutant strain. The vector is pMQ132, and ppigP refers to pMQ221 (Table 1). Error bars = one standard deviation. * = p<0.05 compared to WT by ANOVA with Tukey’s post-test. B. Genetic organization of the chromosome proximal to the pigP gene including the predicted pigP promoter. Enumerated bars under the genes indicate the regions amplified for operon analysis in panel D. C. Growth curve analysis shows similar growth rates for the WT and isogenic pigP mutant strains. The average of four biological replicates is shown. D. Analysis of the pigP operon supports that pigP is in a polycistronic message with SMA3565 and SMA3566. RNA isolated from stationary phase cells was treated with reverse transcriptase (+RT) or without reverse transcriptase (−RT) as a negative control. Positive control DNA and experimental samples were assessed with PCR for the presence of amplicons internal to pigP as a positive control and that span the genes indicated in the figure. Regions amplified by primers are indicated in by numbered brackets in panel B. Primers for analysis of pigP-SMA3565 co-expression extend 144 base pairs into the SMA3565 open frame; those for SMA3565-SMA3566 extend 407 base pairs into the SMA3566 open reading frame.
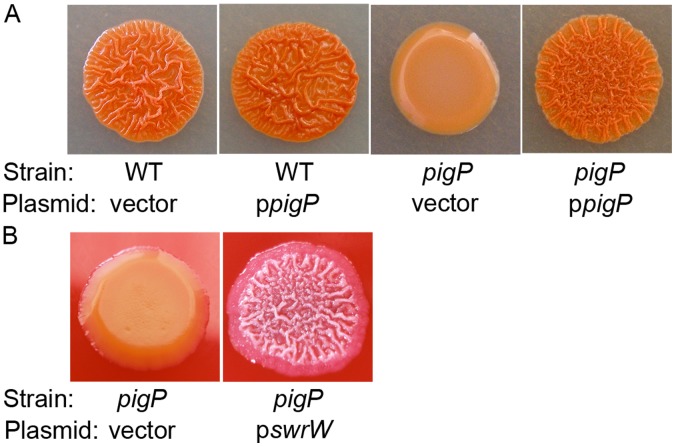
The pigP gene is the first gene in a predicted operon with three ORFs (Figure 1B); therefore, the prodigiosin defect could originate from a polar effect on the subsequent genes or from the absence of pigP. The pigP ORF was cloned under control of the E. coli Plac promoter on a medium copy plasmid (pMQ221) and introduced into the WT and ΔpigP strains. A significant increase in prodigiosin production was observed in both the WT and ΔpigP strains expressing pigP in trans (pMQ221), compared to the vector alone supporting that PigP positively regulates prodigiosin production (p<0.05, ANOVA with Tukey’s Post-test) (Figure 1A). The ΔpigP mutant with the vector alone made significantly less prodigiosin (p<0.05, ANOVA with Tukey’s post test) than the WT with the empty vector (8.4±3.7% of WT levels). Importantly, the ΔpigP mutant phenotype could be complemented by the ORF pigP on a plasmid (pMQ221) (Figure 1A). The pigment phenotype conferred by insertional mutation of pigP was also complemented by the intact pigP gene on a plasmid (Figure S1A–B, and data not shown). Furthermore, mutation of the subsequent uncharacterized ORF (SMA3565, Figure 1B) did not result in a reduction in prodigiosin (Table 3). Together, these data support that the pigP mutant pigment phenotype is due to lack of PigP rather than a polar effect, and that PigP has a positive role in biosynthesis of the secondary metabolite, prodigiosin, similar to what was observed with Serratia sp. ATCC 39006 [46].
To determine whether the reduced pigment production was a result of reduced growth, we analyzed growth and recorded identical growth curves for the WT and ΔpigP strains under the same conditions used to analyze secondary metabolites (Figure 1C), as was observed with Serratia sp. ATCC 39006 [46]. This suggests that there is an active role for PigP in promoting prodigiosin production, rather than an indirect effect associated with a reduced growth rate.
Analysis of the pigP Operon
As noted above, the pigP gene is in a predicted operon with SMA3565-SMA3566 based on the alignment and proximity of open reading frames and Softberry FGENESB operon prediction software (http://linux1.softberry.com) (Figure 1B), however, this has not been previously studied. To determine whether these genes are in a polycistronic message, we prepared DNase-treated RNA, converted it to cDNA with reverse transcriptase, and tested for co-transcribed messages using PCR (Figure 1D). Primers internal to pigP (Figure 1B - 1) were able to direct amplification from chromosomal DNA and reverse transcriptase treated RNA (+RT), but not from no-reverse transcriptase control RNA samples (−RT) indicating that there was undetectable chromosomal DNA contamination (Figure 1D). We reproducibly observed a faint amplicon when using primers that bridge pigP and SMA3565 (Figure 1B - 2) and an amplicon bridging both SMA3565 and SMA3566 (Figure 1B - 3) suggesting that these two ORFs are co-transcribed (Figure 1D). The stronger band between SMA3565 to SMA3566 could indicate the presence of another promoter independent of pigP expression.
Figure 2. PigP transcriptional regulation of the pigment biosynthetic operon.
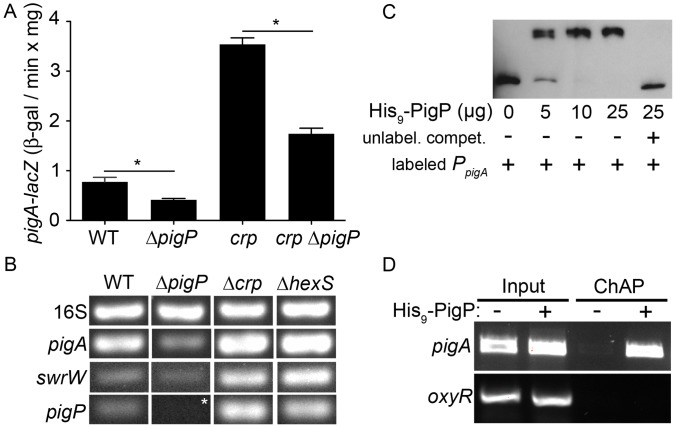
A. Expression of the pigA promoter measured using a chromosomal lacZ transcriptional fusion at early stationary phase. The average of 4 biological replicates is shown. Error bars indicate one standard deviation. One asterisk indicates a significant difference from (p<0.05, ANOVA Tukey’s post-test). B. RT-PCR of cDNA from stationary phase cells (OD600 = 3.5) with the 16S target as a control to show equal input cDNA. Genotypes are listed from left to right, and target cDNAs are listed from top to bottom, with 16S rDNA serving as an internal loading control. Representative images are shown. Asterisk indicates that there is no signal here because the pigP gene is deleted in this strain; this experiment serves as a negative control. C. EMSA assay with biotinylated pigA promoter DNA (4 ng) with or without recombinant PigP protein (His9-PigP) and with (+) or without (−) unlabeled competitor pigA promoter DNA (500 ng). D. Chromatin affinity purification (ChAP) enrichment of pigA promoter DNA, but not oxyR promoter DNA in cells expressing a functional His9-PigP (+), but not the vector alone negative control (−). “Input” indicates sheared DNA before affinity purification and shows similar levels of starting DNA.
Figure 3. Direct regulation of pigP expression by PigP. A.
Expression of a chromosomal pigP-lacZ transcriptional reporter shows reduced expression in the ΔpigP mutant strain (n = 6 biological replicates per time point). B. EMSA assay with biotinylated pigP promoter DNA (2 ng) as a target that had been incubated with or without recombinant PigP protein (His9-PigP) and with (+) or without (−) unlabeled competitor pigP promoter DNA (500 ng).
S. marcescens PigP Mediates Prodigiosin Production and the pigP gene is Found in Environmental, Clinical and Laboratory Isolates
To determine whether its role in pigment regulation was strain specific, the pigP gene was mutated in other pigmented strains by integration of pMQ118 (pMQ179) into the pigP ORF of laboratory strain Nima, environmental isolate CHASM, and clinical keratitis isolate K904. We observed a clear decrease in prodigiosin production in all three strains (Table 4) indicating that the role of PigP in prodigiosin regulation is not specific to strain CMS376. Complementation analysis using pigP on a plasmid restored pigment to these mutant strains supporting that mutation of pigP and not unknown mutations elsewhere in the chromosome were responsible for the observed phenotype (Figure S1B and data not shown).
To determine whether pigP was present in clinical strains, we tested a library of 51 pigmentless and 4 pigmented human keratitis isolates and contact lens case contaminants from the Charles T. Campbell laboratory of Ophthalmic Microbiology and one isolate, UC1SER, from a human neonate colon [64] for the presence of the pigP gene using PCR. All 56 strains tested exhibited PCR amplicons consistent with the pigP gene, a subset are shown in Figure S2, indicating that the gene is conserved among isolates from a variety of niches.
PigP Directly Regulates Transcription of the Pigment Biosynthetic Operon
To further characterize the role of PigP in prodigiosin biosynthesis, we measured pigA transcription, the first gene of the prodigiosinbiosynthetic operon. Transcription of a chromosomal pigA-lacZ reporter was significantly reduced in the ΔpigP mutant (CMS1713) compared to WT (CMS376) (Figure 2A). This reduction in pigA RNA was confirmed using semi-quantitative RT-PCR analysis (Figure 2B). These results show a positive role for PigP in regulation of S. marcescens pigment biosynthesis. It is not known whether PigP directly regulates the prodigiosin biosynthesis operon for any organism. To determine whether the positive role in pigA transcription is direct or indirect, recombinant poly-histidine tagged PigP (His9-PigP) was used in an EMSA assay with pigA promoter DNA.
EMSA experiments showed that His9-PigP could reproducibly bind to biotin-labeled pigA promoter DNA, and the interaction could be inhibited by an excess of unlabeled pigA promoter DNA (Figure 2C). As a negative control, His9-PigP was unable to bind to the oxyR promoter DNA in an EMSA reaction performed under the same conditions (data not shown). To test whether PigP interacts with the pigA promoter in vivo, a chromatin affinity purification assay (ChAP) was performed, and pigA promoter DNA was reproducibly enriched in affinity purified S. marcescens cellular fractions from cells expressing His9-PigP (Figure 2D, ChAP “+ His9-PigP”), but not from fractions with an empty vector control (Figure 2D, ChAP “- His9-PigP”). As a negative control, oxyR promoter and fimC-internal DNA amplicons were not enriched in His9-PigP affinity purified fractions (Figure 2D, and data not shown). Together, these results suggest that PigP directly regulates expression of the pigA-N biosynthetic operon.
PigP Positively Regulates pigP
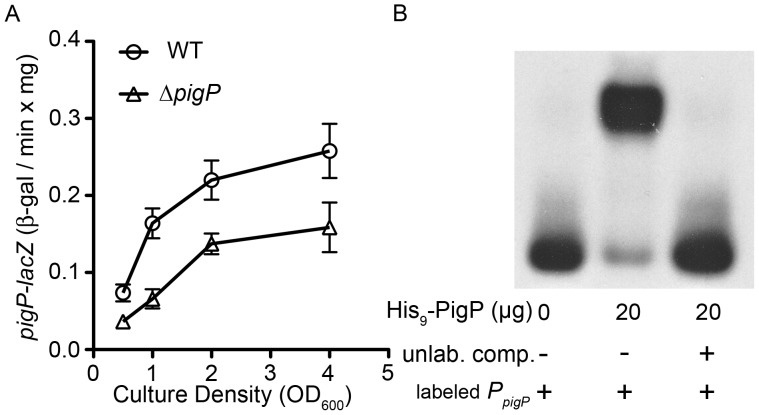
It is not known whether PigP regulates expression of the pigP promoter. A lacZ-transcriptional fusion to the pigP promoter was devised to test pigP expression in the WT (CMS376) and ΔpigP (CMS1713) background. We observed a rapid increase in pigP expression between mid- and late- exponential phase in the WT strain and that overall ß-galactosidase levels were up to a maximum difference of 3-fold lower in the ΔpigP strain (Figure 3A). Reproducible EMSA assays indicate that His9-PigP associates with the pigP promoter but not negative controls (oxyR promoter) (Figure 3B and data not shown). Together, these data suggest a direct and positive role for PigP in regulation of the S. marcescens pigP promoter, suggesting that PigP may directly or indirectly detect the secondary metabolites that it regulates.
PigP is Necessary for the Hyper-pigmented Phenotype of crp Mutants
We previously showed that cAMP-CRP negatively regulates prodigiosin production, but cAMP-CRP did not directly bind to the pigA promoter [35]. We performed an epistasis experiment to test the hypothesis that CRP regulates pigment production through PigP. The double crp pigP mutant (CMS1742) exhibited prodigiosin production levels similar to the WT (CMS376), but more than the pigP mutant (CMS1713) (Table 3).
Using a pigA-lacZ reporter construct, we measured the impact of pigP mutation on transcription of the prodigiosin biosynthetic operon in a crp mutant strain (CMS1687). Similar to previously published results [35], mutation of crp confers an increase in pigA transcription (Figure 2A–B). The elevated pigA-lacZ expression in the crp mutant was partially suppressed in the crp pigP double mutant (CMS1742) (Figure 2A). These results suggest a regulatory relationship between CRP and PigP, where pigA transcriptional control by cAMP-CRP occurs largely, but not completely through PigP. An alternative model is that PigP and CRP independently regulate pigA transcription.
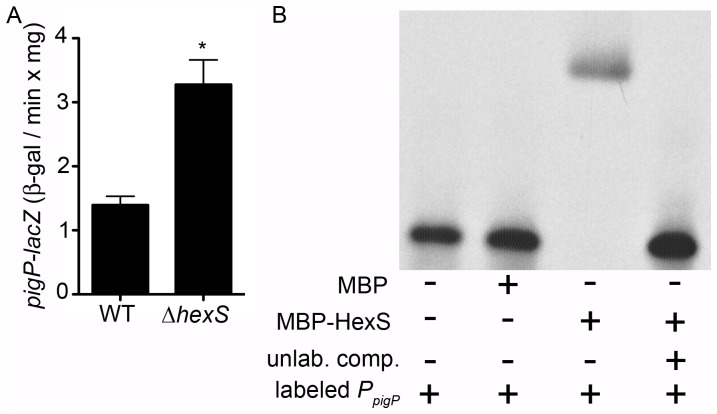
CRP Mediates pigP Expression
Upstream regulators of pigP are unknown [45], [46]. The observation that the crp mutant’s hyper-red phenotype could be partially suppressed by mutation of pigP suggested that the two genes are in a regulatory pathway; therefore, we tested whether pigP was transcriptionally regulated by CRP (Figure 2B, 4A–B). A significant (p<0.05) 3.6-fold increase in pigP-promoter-driven ß-galactosidase activity was observed in crp mutants relative to WT levels at OD600 = 0.8 (Figure 4A), and RT-PCR analysis supports that mutation of crp increases pigP transcript (Figure 2B). A cyaA mutant is expected to behave like a crp mutant because CyaA catalyzes synthesis of cAMP that mediates CRP function [35]. Similar to the crp mutant, expression of pigP-lacZ was higher in a cyaA mutant (CMS524) (Figure 4B). Furthermore, exogenous cAMP was able to reduce pigP expression in a dose-dependent manner in the cyaA mutant (Figure 4B). These results support a model that cAMP-CRP negatively regulates pigP transcription either directly or indirectly, and that crp and cyaA mutants exhibit elevated prodigiosin production partially because of increased pigP expression.
Figure 4. Impact of cAMP-CRP on pigP transcription.
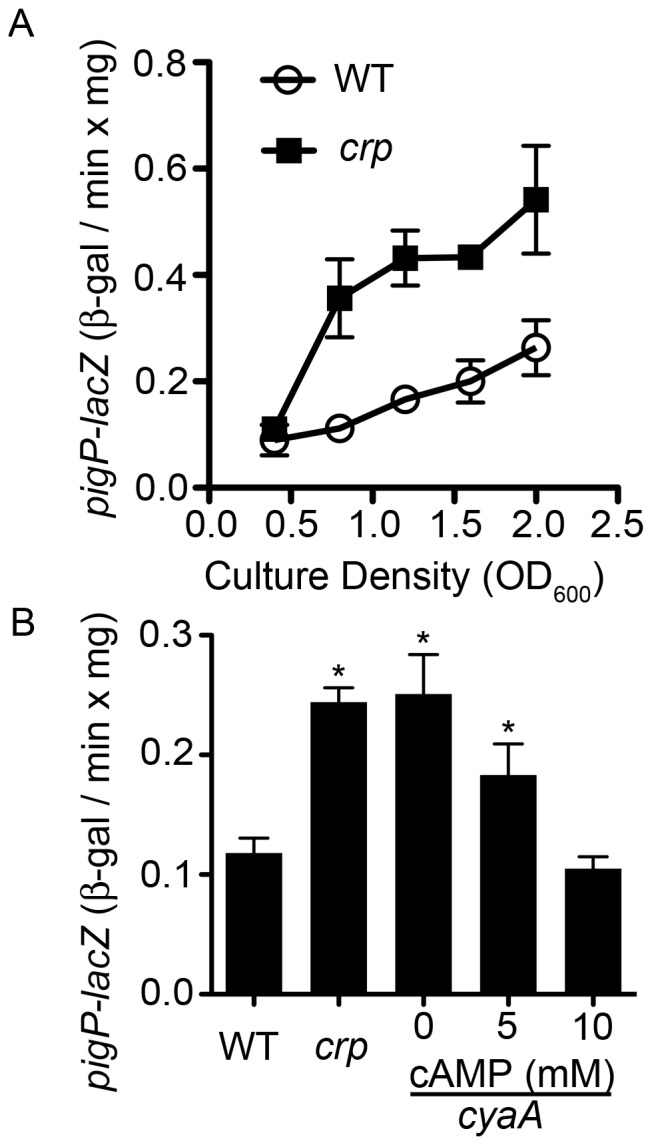
A. ß-galactosidase activity as expressed from the chromosomal pigP promoter as a function of culture density. This representative experiment shows the average of 3 biological replicates per genotype. B. ß-galactosidase activity from a chromosomal pigP reporter in early stationary phase. WT and crp strains were grown without exogenous cAMP. The isogenic cyaA mutant was grown with 0, 5, or 10 mM cAMP dissolved in the growth medium. This experiment shows the average of 6 biological replicates per cAMP concentration, performed on two different days. Asterisk = significantly different than WT (p<0.05). In this figure “crp” refers to the crp-23 transposon mutant. Error bars = one standard deviation.
Although there were no predicted CRP binding sites directly upstream of the pigP ORF in our strain background, gel shift assays (EMSA) were performed to determine whether CRP directly or indirectly interacts with these target genes. We did not find evidence that purified CRP bound to a 491 base pair region upstream of pigP under the conditions that we used, although a positive control promoter, flhDC, was readily bound under the same array of tested conditions (data not shown). Together these data suggest that CRP indirectly regulates pigP transcription.
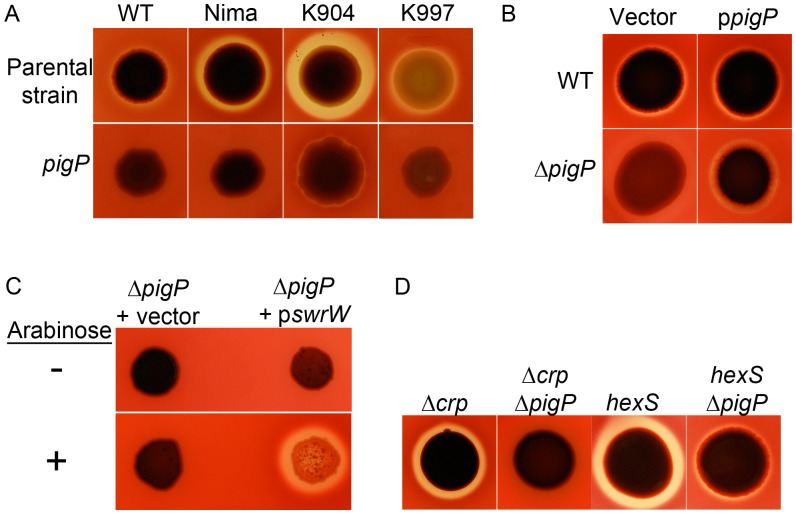
PigP is Required for Surface Swarming Motility and Serratamolide Production
Swarming motility over the surface is a virulence associated group behavior [65]–[67]. We observed that the ΔpigP strain (CMS1713) was unable to swarm (0%, n = 15, performed on 3 different days), whereas WT was capable of swarming (100%, n = 15) (Figure 5A). The pigP mutant swarming deficiency could be complemented by WT pigP on a plasmid (Figure 5B).
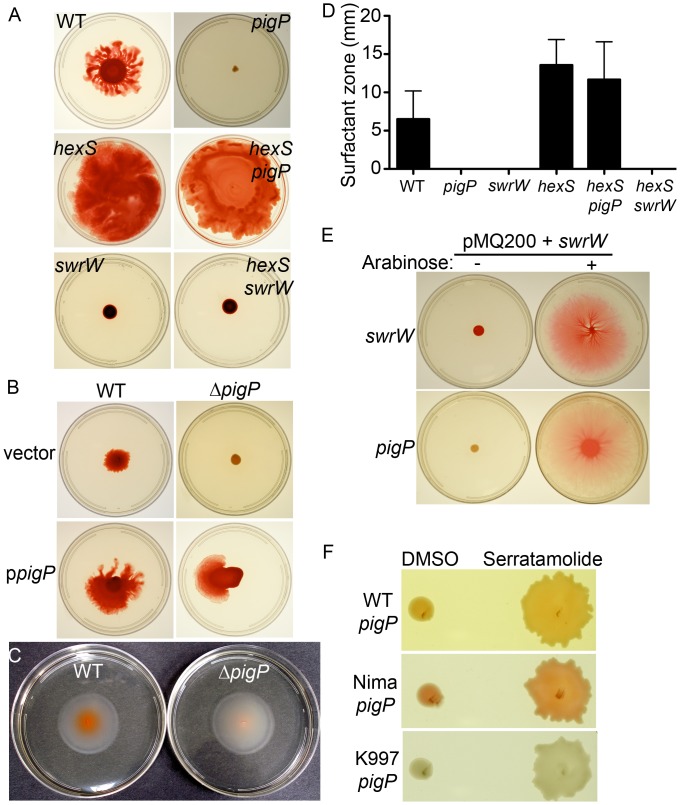
Figure 5. PigP is necessary for swarming motility and serratamolide production, but not swimming motility.
A. Swarming motility plates show that the pigP mutant is defective in surface motility. Mutation of the serratamolide inhibitor hexS restores swarming to the pigP mutant (hexS pigP), and hexS requires serratamolide to swarm (hexS swrW). B. The pigP swarming defect can be complemented by the wild-type pigP gene provided in trans. Vector refers to pMQ132, and ppigP is pMQ221. C. Swimming motility plates show similar zones (in all cases “zones” indicates the measurement from the edge of the colony to the outer edge of the observed phenotype in mm) between the WT and pigP mutant. D. Surfactant zones are absent in strains without pigP and the serratamolide biosynthetic gene swrW. Mutation of hexS restores surfactant zones to the pigP mutant. N ≥6 biological replicates per strain. E. Arabinose-induced expression of swrW is sufficient to restore swarming to a swrW and pigP mutant. pMQ200+ swrW refers to pMQ368. F. Purified serratamolide is sufficient to restore swarming to different strain backgrounds with pigP mutations, whereas the serratamolide vehicle, DMSO, is not.
Factors required for swarming motility include flagella and surface wetting agents, known as serrawettins. Swimming motility assays indicate that the pigP mutant (CMS1713) makes functional flagella (Figure 5C). We observed no significant difference (p = 0.14, Student’s t-test) in swim zones between the WT (18.7±2.0 mm, n = 11) and ΔpigP (19.9±1.65 mm, n = 11) strains.
The surfactant serratamolide (serrawettin W1) generates a visible and transparent halo on the top of swarming agar around the WT strain used in this study [22]. The surfactant zone around the WT culture extended approximately 5 mm beyond the edge of the colony after 24 h, whereas the ΔpigP mutant produced no surfactant zone (Figure 5D), and this defect could be complemented by pigP on a plasmid (surfactant zones in mm at 18 h: WT+empty vector = 3.7; WT+ppigP = 15.2, ΔpigP+empty vector = 0, ΔpigP+ppigP = 13.3). Note that the surfactant zone made by the WT strain was >4-fold larger when pigP was expressed on a multicopy plasmid than when the strain had the empty vector (p = 0.02, Student’s t-test), suggesting that PigP positively regulates surfactant production. Similar to pigP, mutation of the gene coding for SwrW, a non-ribosomal peptide synthetase necessary for production of serratamolide, confers swarming and surfactant zone deficiencies (Figure 5A,D), as has previously been shown [22].
As these data suggest that the ΔpigP mutant (CMS1713) does not swarm because it is defective in serratamolide production, we tested whether induced production of serratamolide in a pigP mutant could restore swarming motility. Expression of swrW from the arabinose-inducible PBAD promoter (pMQ368) rescued the swarming defect of the ΔpigP strain in an arabinose-dependent manner (Figure 5E). Arabinose does not rescue the pigP swarming effect without swrW on a plasmid (data not shown). Consistent with a lack of serratamolide being the sole reason why pigP mutants cannot swarm, the addition of purified serratamolide to ΔpigP mutants (50 µg/ml in DMSO) but not by the vehicle control (DMSO) restored swarming motility (Figure 5F).
The swarming defect was also observed when pigP was mutated by insertional mutagenesis in the WT strain (CMS376), the laboratory strain Nima, and ocular clinical isolates including a non-pigmented isolate K997 (Figure 5F). These mutants were also restored to swarming by the addition of exogenous serratamolide (Figure 5F).
These data suggest that PigP is an important factor in swrW regulation. Semi-quantitative RT-PCR support that there is reproducibly ∼50% more swrW transcript in the WT than the isogenic ΔpigP mutant in stationary phase cells (OD600 = 3.5) (Figure 2B). A previously described tdtomato-reporter based swrW promoter-probe [22], pMQ376 (Table 2), confirmed that there was less swrW expression in the ΔpigP mutant (83,476±5117 RFU), compared to the WT (133,221±10670 RFU, p<0.05). EMSA analysis suggests that His9-PigP does not directly bind to the swrW promoter under a variety of conditions that supported the binding of His9-PigP to the pigP promoter (Figure S3A), supporting indirect regulation of swrW by PigP.
The relatively modest reduction in swrW transcript was somewhat surprising given the absence of serratamolide zones around pigP mutants on agar plates (Figure 5D), and suggests that growth or media may alter serratamolide production. To test this we measured serratamolide from liquid cultures using liquid chromatograph-mass spectrometry under similar conditions used to assay swrW transcript noted above. We observed a reproducible ∼50% reduction in serratamolide in the ΔpigP strain compared to the WT, and a complete absence of serratamolide in the swrW mutant negative control supernatant (Figure S4).
PigP Regulates Hemolysis
A recent report indicates that serratamolide can be hemolytic [22]. We tested the prediction that pigP mutants would be defective in hemolysis. The ΔpigP strain (CMS1713) made no zone of clearing on blood agar plates, whereas the WT strain did, and this defect could be complemented by WT pigP on a plasmid (Figure 6A–B). A reduction or elimination of secreted hemolytic activity was also observed when pigP was disrupted in the WT and Nima laboratory strains, and clinical isolates K904 and K997 (Figure 6A). Arabinose-inducible expression of swrW from the ΔpigP mutant was able to restore hemolysis to the pigP mutant (Figure 6C), supporting that a reduction of swrW expression was responsible for the hemolysis defect of pigP mutants.
Figure 6. PigP is necessary for hemolysis in laboratory and clinical isolates. A.
. Hemolytic strains grown on TSA plates with sheep blood show a zone of clearing around colonies indicative of hemolysis. Isogenic pigP mutant strains show highly reduced zones of hemolysis. B. The hemolysis defect of pigP mutants can be complemented by wild-type pigP on a plasmid (pMQ221); vector refers to pMQ132. C. Arabinose inducible expression of swrW is sufficient to restore hemolysis to the pigP mutant. The swrW gene was expressed from plasmid pMQ367 (pswrW), and vector refers to pMQ125. D. Mutation of pigP reduces the hyper-hemolytic phenotypes of crp and hexS mutants.
The hexS (CMS2210) mutant produces more serratamolide than the WT (Figure 5D) as has been previously shown for a hexS mutant in multiple strain backgrounds [20], [22] and for a crp mutant in the CMS376 strain background [20], [68] (data not shown), resulting in large zones of clearing on blood agar plates (Fig. 6D). Double mutant analysis shows that the crp mutant hyper-hemolysis phenotype is eliminated in the crp pigP double mutant (CMS1742), whereas the pigP hexS double mutant (CMS1744) has intermediate hemolysis levels (Figure 6D). The WT and pigP hemolysis zones were equivalent for the experiment shown in Figure 6D as Figure 6B (data not shown). These data support a model in which PigP and CRP share a common regulatory pathway, where PigP acts downstream of CRP, and HexS and PigP act independently or have a more complex relationship for serratamolide regulation. Together these data support that PigP mediates hemolysis through control of serratamolide biosynthesis.
Mutation of hexS Suppresses the Swarming Deficiency of pigP Mutants
It was previously shown that hexS mutants produce highly elevated amounts of serratamolide [20], [22]. A ΔpigP hexS double mutant was constructed to provide insight into whether HexS and PigP regulate swrW through a common pathway. The hexS mutant exhibited surfactant zones >2-fold larger than the WT strain (Figure 5D), consistent with its role as a negative regulator of swrW expression (Figure 2B). The ΔpigP hexS double mutant generated large surfactant zones and a swarming phenotype like the hexS mutant (Figure 5A,D). These data indicate that for serratamolide production and swarming, the hexS mutation is epistatic to the pigP mutation and suggest that HexS acts downstream or independently of PigP. As a control, we generated a hexS swrW double mutant that did not produce surfactant zones and was swarming deficient, confirming that serratamolide is required for the hexS mutant surfactant zone and swarming phenotypes for the strain used in this study (Figure 5A, D).
Similar to what we observed with swarming, a hexS mutation was epistatic to a pigP mutation with respect to prodigiosin production (Table 3). Whereas the pigP mutant strain (CMS1713) exhibited significantly reduced prodigiosin compared to WT (CMS376), hexS (CMS2210) and pigP hexS (CMS1744) strains both generated elevated levels of prodigiosin compared to the WT (Table 3).
Given the common control of serratamolide and prodigiosin production by both PigP and HexS, we predicted that these two transcription factors are in a regulatory pathway that controls swrW expression. It was reported that HexS directly binds to the swrW and pigA promoters, but not to the promoter of pswP, whose gene product is involved in secondary metabolite production [21]. Our data above suggest that PigP does not bind to the swrW promoter; therefore, we hypothesized that PigP directly regulates hexS. Using a chromosomal hexS-lacZ reporter, we measured a modest increase (56±16%) in hexS expression measured from over-night stationary phase ΔpigP cultures compared to the WT (OD600 = ∼4). However, His9-PigP was not observed to bind to the hexS promoter using in vitro gel shift assays (Figure S3A). Conversely, RT-PCR suggests that HexS is a negative regulator of pigP transcription, with elevated pigP transcript in the ΔhexS mutant (Figure 2B). Similar results were measured with a chromosomal lacZ transcriptional reporter for pigP expression, where >2-fold more activity was measured in the ΔhexS mutant compared to the WT (Figure 7A). EMSA analysis indicates that MBP-HexS can bind to the pigP promoter in vitro (Figure 7B), similar to positive control promoters, pigA and swrW, however MBP-HexS did not bind to negative control promoter pswP or the hexS promoter (Figure S3B). As a control to ensure that the observed shifts were due to the HexS portion of the fusion, MBP alone was included as a control, and failed to elicit the shift of any promoter tested. These data support the model that HexS is a direct negative regulator of PigP.
Figure 7. HexS regulates pigP expression.
A. Mutation of hexS leads to an increase in output from a chromosomal pigP reporter (strains CMS1785 and CMS3408 respectively). Cells were harvested at OD600 = 4.0 and ß-gal activity is reported. Data is the mean from 7 biological replicates per genotype. Error bars = one standard-deviation and the asterisk indicates a statistical difference from the WT (p<0.05, Student’s t-test). B. EMSA analysis indicates that a recombinant maltose binding protein-HexS fusion (MBP-HexS, 25 µg) binds to the labeled pigP promoter (2 ng) in vitro, whereas the recombinant maltose binding protein (MBP, 33 µg) control does not bind to the pigP promoter. Unlabeled pigP promoter region competitor DNA (500 ng) was able to inhibit the MBP-HexS induced shift suggesting a specific interaction.
The Rugose Colony Phenotype of an Environmental Isolate Requires PigP
Another pigP mutant phenotype was noted with an environmental isolate of S. marcescens, CHASM. We report here that CHASM exhibits a dramatic rugose colony phenotype (Figure 8A). When pigP was mutated in CHASM, the colonies changed from rugose and red to smooth and pink (Figure 8A). The rugose phenotype of the CHASM pigP mutant was complemented by the wild-type pigP gene on a plasmid (Figure 8A). A plasmid with the swrW gene (pMQ367) was able to restore the rugose phenotype to the CHASM pigP mutant (CMS2982), whereas the vector alone (pMQ125) did not (Figure 8B), suggesting that the rugose phenotype is mediated by serratamolide. At this time the mechanism for serratamolide in this colony morphology phenotype is unknown; however, in other organisms, rugose colony phenotype has been linked to biofilm formation. Static biofilm assays indicate that mutation of pigP does not confer a significant biofilm defect under the conditions used (data not shown).
Figure 8. PigP mediates rugose colony morphology. A.
The CHASM rugose phenotype is absent in the pigP mutant (CMS2981) and can be complemented by wild-type pigP on a plasmid (ppigP = pMQ221). The vector alone is pMQ132. B. The rugose colony morphology defect of the CHASM pigP mutant (CMS2981) can be restored through expression of swrW from a plasmid (pMQ367), but not from the vector alone (pMQ125).
Discussion
The goal of this study was to determine whether the homolog of a secondary metabolite master regulator protein, PigP, from an atypical environmentally isolated species of Serratia positively regulates S. marcescens swarming motility and hemolysis and whether it is conserved in prodigiosin regulation. In short, we found that like Serratia sp. ATCC 39006, S. marcescens uses PigP as a key positive regulator of prodigiosin biosynthesis. Beyond what has been shown in Serratia sp. ATCC 39006, we report for the first time that PigP was involved in promoting surfactant production, hemolysis, swarming motility, and a novel rugose colony morphology. Unlike Serratia sp. ATCC 39006, mutation of pigP in S. marcescens did not have a major effect on production of secreted enzymes (Stella and Shanks, unpublished observations), nor upon swimming motility under the tested conditions. One possible mechanism for the observed difference in secreted enzyme production between species is that Serratia sp. ATCC 39006 may have a PigP-regulated secretion system not found in S. marcescens. This study illustrates that there can be measurable differences in the role of a transcriptional regulator between Serratia species.
The swarming defect of pigP mutants correlated with a loss of surfactant production and, like the prodigiosin defect, was evident in clinical, environmental and laboratory isolates. Other data suggested that the mechanism for the pigP mutant swarming defect is reduced serratamolide production. This model is based on the observations that pigP mutants exhibited reduced swrW transcript compared to the WT, the swarming defect could be bypassed by inducible expression the swrW gene, and that exogenous purified serratamolide could restore swarming to the pigP mutant. As serratamolide can act as a hemolysin [22], we tested whether PigP regulated hemolysis. The pigP mutant was defective in hemolysis, and inducible expression of swrW restored hemolytic zones to the pigP mutant. These results suggest that PigP is an important factor in mediating serratamolide production, and therefore swarming motility and hemolysis, by clinical and laboratory strains of S. marcescens.
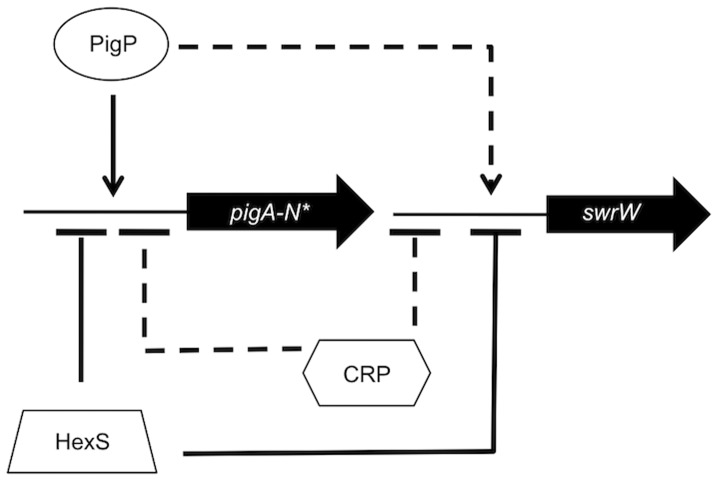
While it has been previously shown that mutation of the pigP gene leads to altered transcription of a number of genes in Serratia sp. ATCC 39006, it has not been shown that the PigP protein directly binds to the promoters that it regulates. Here we provide evidence that the S. marcescens PigP homolog directly regulates expression of the prodigiosin biosynthetic operon, pigA-N, and the pigP gene. Our data indicate that mutation of pigP leads to a reduction of swrW expression, but recombinant PigP was not shown to bind to the swrW promoter in vitro. It is possible that the PigP promoter can bind to swrW in vivo by itself or in conjunction with another protein, and that the conditions of our EMSA reaction were not appropriate, although a battery of reaction conditions were employed. These data support the model that PigP controls secondary metabolite production directly (pigA) and indirectly (swrW) through unknown regulator(s) (Figure 9). Consistent with this model, there is evidence that the Serratia sp. ATCC 39006 PigP protein regulates a number of other transcriptional regulators involved in secondary metabolite control [45].
Figure 9. Model for regulation of secondary metabolite biosynthesis genes by the transcription factors described in this study.
The secondary metabolite genes (pigA-N for prodigiosin and swrW for serratamolide) and the pigP regulator gene (not shown) are negatively (bar) and directly (solid line) regulated by HexS. They are negatively and indirectly (dashed line) regulated by CRP. The pigA-N operon and pigP are positively (arrow) and directly regulated by PigP, whereas swrW is indirectly regulated by PigP. The asterisk indicates that the same pattern of regulation for the pigA-N operon is observed for the pigP gene.
It may be noted that some of the changes in gene expression elicited by mutation of pigP are not particularly dramatic, e.g. ∼50% reduction in pigA transcript. However, this is similar to what has been reported for other regulators that effect prodigiosin and other secondary metabolite production, where modest changes in transcript correlate with large phenotypic changes [36], [46].
It was previously reported that there are no known regulators of pigP [45], [46]. Here we provide data that cAMP-CRP is an indirect upstream regulator of pigP (Figure 9). The pigP gene was found necessary for the hyper-pigment and hyper-hemolysis phenotypes of a crp mutant, and increased expression of the pigP gene in crp and cyaA mutants, suggesting that PigP acts downstream of cAMP-CRP to regulate secondary metabolite genes. The absence of cAMP-CRP binding to the pigP promoter suggests that there is an intermediate regulators(s). Multiple proteins involved in carbon utilization also contribute to pigment regulation including the transcription factor PigT [38] and components of bacterial electron transport chains [41], [69] underscoring that prodigiosin may be an important factor in control of energy homeostasis [55].
In addition to cAMP-CRP, we present data supporting that HexS is a direct regulator of pigP expression (Figure 9). A hexS pigP double mutant exhibited a hexS mutant-like phenotype with respect to swarming motility and prodigiosin levels but not hemolysis, where an intermediate phenotype was observed. The discordant phenotypes from the same strain in different assays may be due to different thresholds of serratamolide necessary to elicit each phenotype, or differential production of serratamolide under the different experimental conditions used in these assays. Nevertheless, these genetic data imply either a direct relationship with PigP acting upstream of HexS, for which we have little evidence (mutation of pigP had little effect on hexS expression and PigP did not bind to the hexS promoter), or through an independent relationship in which HexS and PigP can both independently regulate target genes. This second model is supported by evidence that both PigP and HexS regulate and bind to swrW and pigA promoters. Our transcriptional and EMSA data support the model that HexS can further influence the pathway by directly regulating pigP expression. Together supporting the model that HexS can influence secondary metabolite production both directly through control of pigA-N and swrW and indirectly as a direct upstream regulator of pigP.
Interestingly, we observed that PigP was able to directly regulate expression of its own promoter in a positive manner suggesting that it may directly or indirectly respond to the secondary metabolites that it regulates. Future studies will be focused on determining the binding site of HexS and PigP and elucidating the mechanism by which these opposing regulators mediate transcription of the prodigiosin biosynthetic operon and determining the environmental stimuli that these regulators respond to.
PigP regulates carbapenem antibiotic in Serratia sp. ATCC 39006 and prodigiosin biosynthesis in both S. marcescens and ATCC 39006 [45], [46]. Prodigiosin has antibiotic properties [49] and is excreted from cells in extracellular vesicles [70]. Similarly, serratamolide has antimicrobial properties [33], [34] and is thought to be necessary for the generation of extracellular vesicles [70]. One possible function of PigP is to regulate production of antibiotics to compete for limited nutrients against other organisms, consistent with our observation that PigP is necessary to inhibit the growth of a gram-positive organism adjacent to S. marcescens colonies (Shanks, unpublished observations). cAMP-CRP is best known for regulation of metabolic pathways in response to environmental carbon sources, but it also regulates production of adhesins, flagella and other factors involved in interacting with the environment. Therefore, PigP’s control of competitive factors ties in well with the function of cAMP-CRP in adaptation of bacteria to the environment based upon nutrient cues. However, given the number of transcription factors that contribute to secondary metabolism further research is necessary to discover and characterize the other inputs for regulating secondary metabolites in S. marcescens.
Supporting Information
Complementation of prodigiosin phenotype conferred by insertional mutation of pigP . A. Photograph of WT (CMS376) or the pigP mutant (CMS836) with the vector (pMQ125) or ppigP (pMQ212) grown on LB agar supplemented with arabinose (4 mM). B. Prodigiosin production by the environmental isolate, CHASM, and isogenic pigP-insertion mutant (CMS2981) being either the empty vector (pMQ132) or ppigP (pMQ221) grown in LB medium. The average of six independent biological replicates is shown.
(PDF)
Amplification of pigP from S. marcescens isolates. PCR was used to amplify a 138 base pair amplicon from a variety of S. marcescens strains. Db11 and CMS376 served as positive controls, whereas Proteus and Staphylococcus chromosomal DNA served as negative controls. Amplicons were separated on TBE-PAGE gels and stained with ethidium bromide.
(PDF)
EMSA analysis of His9-PigP and MBP-HexS with promoters of interest. A) His9-PigP exhibited a gel shift of the pigP promoter but not the swrW or hexS promoters. B) MPB-HexS retarded migration of pigA and swrW promoters (positive controls), but not the pswP promoter (negative control) or the hexS promoter. MBP alone did not induce a gel shift of any promoter. Excess unlabeled promoter DNA could compete successfully for MPB-HexS binding to pigA and swrW promoters. Biotin-labeled promoters (Label. Promoter) were used at 2 ng per reaction and unlabeled promoters (Unlab promoter) were used at 500 ng per reaction.
(PDF)
Analysis of serratamolide from S. marcescens culture supernatants. LC-MS was used to measure serratamolide levels in culture supernatants from the WT (CMS376) the ΔpigP strain (CMS1713) and the negative control swrW mutant (CMS635). Purified serratamolide was used as a positive control. The serratamolide peaks are boxed in red the scale of the trace is shown on the left hand side. A representative experiment is shown.
(PDF)
Primers used in this study.
(DOCX)
Acknowledgments
The authors gratefully thank Grace Altimus and James Fender for technical support and critical reading of the manuscript.
Funding Statement
This study was funded by National Institutes of Health (NIH) grant AI085570 and a Research to Prevent Blindness Career Development Award to RS and support from NIH grant EY08098, the Eye and Ear Foundation of Pittsburgh, and unrestricted funds from Research to Prevent Blindness. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mahlen SD (2011) Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24: 755–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Diekema DJ, Pfaller MA, Jones RN, Doern GV, Winokur PL, et al. (1999) Survey of bloodstream infections due to gram-negative bacilli: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, and Latin America for the SENTRY Antimicrobial Surveillance Program, 1997. Clin Infect Dis 29: 595–607. [DOI] [PubMed] [Google Scholar]
- 3. Richards MJ, Edwards JR, Culver DH, Gaynes RP (2000) Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect Control Hosp Epidemiol 21: 510–515. [DOI] [PubMed] [Google Scholar]
- 4. Jones RN (2010) Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis 51 Suppl 1S81–87. [DOI] [PubMed] [Google Scholar]
- 5. Verhamme KM, De Coster W, De Roo L, De Beenhouwer H, Nollet G, et al. (2007) Pathogens in early-onset and late-onset intensive care unit-acquired pneumonia. Infect Control Hosp Epidemiol 28: 389–397. [DOI] [PubMed] [Google Scholar]
- 6. Laupland KB, Parkins MD, Gregson DB, Church DL, Ross T, et al. (2008) Population-based laboratory surveillance for Serratia species isolated in a large Canadian health region. Eur J Clin Microbiol Infect Dis 27: 89–95. [DOI] [PubMed] [Google Scholar]
- 7. Hume EB, Conerly LL, Moreau JM, Cannon BM, Engel LS, et al. (1999) Serratia marcescens keratitis: strain-specific corneal pathogenesis in rabbits. Curr Eye Res 19: 525–532. [DOI] [PubMed] [Google Scholar]
- 8. Hume EB, Willcox MD (2004) Emergence of Serratia marcescens as an ocular surface pathogen. Arch Soc Esp Oftalmol 79: 475–477. [PubMed] [Google Scholar]
- 9. Butler MT, Wang Q, Harshey RM (2010) Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci U S A 107: 3776–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kearns DB (2010) A field guide to bacterial swarming motility. Nat Rev Microbiol 8: 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Overhage J, Bains M, Brazas MD, Hancock RE (2008) Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190: 2671–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Allison C, Lai HC, Hughes C (1992) Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis . Mol Microbiol 6: 1583–1591. [DOI] [PubMed] [Google Scholar]
- 13. Fraser GM, Claret L, Furness R, Gupta S, Hughes C (2002) Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiology 148: 2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walker KE, Moghaddame-Jafari S, Lockatell CV, Johnson D, Belas R (1999) ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol Microbiol 32: 825–836. [DOI] [PubMed] [Google Scholar]
- 15. Wang WB, Chen IC, Jiang SS, Chen HR, Hsu CY, et al. (2008) Role of RppA in the regulation of polymyxin b susceptibility, swarming, and virulence factor expression in Proteus mirabilis . Infect Immun 76: 2051–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsuyama T, Sogawa M, Nakagawa Y (1989) Fractal spreading growth of Serratia marcescens which produces surface active exolipids. FEMS Microbiol Lett 52: 243–246. [DOI] [PubMed] [Google Scholar]
- 17. O’Rear J, Alberti L, Harshey RM (1992) Mutations that impair swarming motility in Serratia marcescens 274 include but are not limited to those affecting chemotaxis or flagellar function. J Bacteriol 174: 6125–6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matsuyama T, Bhasin A, Harshey RM (1995) Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J Bact 177: 987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sunaga S, Li H, Sato Y, Nakagawa Y, Matsuyama T (2004) Identification and characterization of the pswP gene required for the parallel production of prodigiosin and serrawettin W1 in Serratia marcescens . Microbiol Immunol 48: 723–728. [DOI] [PubMed] [Google Scholar]
- 20. Li H, Tanikawa T, Sato Y, Nakagawa Y, Matsuyama T (2005) Serratia marcescens gene required for surfactant serrawettin W1 production encodes putative aminolipid synthetase belonging to nonribosomal peptide synthetase family. Microbiol Immunol 49: 303–310. [DOI] [PubMed] [Google Scholar]
- 21. Tanikawa T, Nakagawa Y, Matsuyama T (2006) Transcriptional downregulator HexS controlling prodigiosin and serrawettin W1 biosynthesis in Serratia marcescens . Microbiol Immunol 50: 587–596. [DOI] [PubMed] [Google Scholar]
- 22. Shanks RM, Stella NA, Lahr RM, Wang S, Veverka TI, et al. (2012) Serratamolide is a hemolytic factor produced by Serratia marcescens . PLoS One 7: e36398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allison C, Coleman N, Jones PL, Hughes C (1992) Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect Immun 60: 4740–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carpinella MC, De Bellis L, Joray MB, Sosa V, Zunino PM, et al. (2011) Inhibition of development, swarming differentiation and virulence factors in Proteus mirabilis by an extract of Lithrea molleoides and its active principle (Z,Z)-5-(trideca-4′,7′-dienyl)-resorcinol. Phytomedicine 18: 994–997. [DOI] [PubMed] [Google Scholar]
- 25. Dacheux D, Goure J, Chabert J, Usson Y, Attree I (2001) Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol Microbiol 40: 76–85. [DOI] [PubMed] [Google Scholar]
- 26. Givaudan A, Lanois A (2000) flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J Bacteriol 182: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hsueh YH, Somers EB, Lereclus D, Ghelardi E, Wong AC (2007) Biosurfactant production and surface translocation are regulated by PlcR in Bacillus cereus ATCC 14579 under low-nutrient conditions. Appl Environ Microbiol 73: 7225–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin CS, Horng JT, Yang CH, Tsai YH, Su LH, et al. (2010) RssAB-FlhDC-ShlBA as a major pathogenesis pathway in Serratia marcescens . Infect Immun 78: 4870–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ulrich RL, Hines HB, Parthasarathy N, Jeddeloh JA (2004) Mutational analysis and biochemical characterization of the Burkholderia thailandensis DW503 quorum-sensing network. J Bacteriol 186: 4350–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhakdi S, Tranum-Jensen J (1991) Alpha-toxin of Staphylococcus aureus . Microbiol Rev 55: 733–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Braun V, Focareta T (1991) Pore-forming bacterial protein hemolysins (cytolysins). Crit Rev Microbiol 18: 115–158. [DOI] [PubMed] [Google Scholar]
- 32. Schnupf P, Portnoy DA (2007) Listeriolysin O: a phagosome-specific lysin. Microbes Infect 9: 1176–1187. [DOI] [PubMed] [Google Scholar]
- 33. Dwivedi D, Jansen R, Molinari G, Nimtz M, Johri BN, et al. (2008) Antimycobacterial serratamolides and diacyl peptoglucosamine derivatives from Serratia sp. J Nat Prod 71: 637–641. [DOI] [PubMed] [Google Scholar]
- 34. Wasserman HH, Keggi JJ, Mckeon JE (1961) Serratamolide, a metabolic product of Serratia . J Am chem Soc 83: 4107–4108. [Google Scholar]
- 35. Kalivoda EJ, Stella NA, Aston MA, Fender JE, Thompson PP, et al. (2010) Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens . Res Microbiol 161: 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horng YT, Chang KC, Liu YN, Lai HC, Soo PC (2010) The RssB/RssA two-component system regulates biosynthesis of the tripyrrole antibiotic, prodigiosin, in Serratia marcescens . Int J Med Microbiol 300: 304–312. [DOI] [PubMed] [Google Scholar]
- 37. Horng YT, Deng SC, Daykin M, Soo PC, Wei JR, et al. (2002) The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens . Mol Microbiol 45: 1655–1671. [DOI] [PubMed] [Google Scholar]
- 38. Fineran PC, Everson L, Slater H, Salmond GP (2005) A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology 151: 3833–3845. [DOI] [PubMed] [Google Scholar]
- 39. Fineran PC, Williamson NR, Lilley KS, Salmond GP (2007) Virulence and prodigiosin antibiotic biosynthesis in Serratia are regulated pleiotropically by the GGDEF/EAL domain protein, PigX. J Bacteriol 189: 7653–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gristwood T, Fineran PC, Everson L, Williamson NR, Salmond GP (2009) The PhoBR two-component system regulates antibiotic biosynthesis in Serratia in response to phosphate. BMC Microbiol 9: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McNeil MB, Clulow JS, Wilf NM, Salmond GP, Fineran PC (2012) SdhE is a conserved protein required for flavinylation of succinate dehydrogenase in bacteria. J Biol Chem 287: 18418–18428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramsay JP, Salmond GP (2012) Quorum sensing-controlled buoyancy through gas vesicles: Intracellular bacterial microcompartments for environmental adaptation. Commun Integr Biol 5: 96–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wilf NM, Salmond GP (2012) The stationary phase sigma factor, RpoS, regulates the production of a carbapenem antibiotic, a bioactive prodigiosin and virulence in the enterobacterial pathogen Serratia sp. ATCC 39006. Microbiology 158: 648–658. [DOI] [PubMed] [Google Scholar]
- 44. Wilf NM, Williamson NR, Ramsay JP, Poulter S, Bandyra KJ, et al. (2011) The RNA chaperone, Hfq, controls two luxR-type regulators and plays a key role in pathogenesis and production of antibiotics in Serratia sp. ATCC 39006. Environ Microbiol 13: 2649–2666. [DOI] [PubMed] [Google Scholar]
- 45. Williamson NR, Fineran PC, Leeper FJ, Salmond GP (2006) The biosynthesis and regulation of bacterial prodiginines. Nat Rev Microbiol 4: 887–899. [DOI] [PubMed] [Google Scholar]
- 46. Fineran PC, Slater H, Everson L, Hughes K, Salmond GP (2005) Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol Microbiol 56: 1495–1517. [DOI] [PubMed] [Google Scholar]
- 47. Gristwood T, Fineran PC, Everson L, Salmond GP (2008) PigZ, a TetR/AcrR family repressor, modulates secondary metabolism via the expression of a putative four-component resistance-nodulation-cell-division efflux pump, ZrpADBC, in Serratia sp. ATCC 39006. Mol Microbiol 69: 418–435. [DOI] [PubMed] [Google Scholar]
- 48. Gristwood T, McNeil MB, Clulow JS, Salmond GP, Fineran PC (2011) PigS and PigP regulate prodigiosin biosynthesis in Serratia via differential control of divergent operons, which include predicted transporters of sulfur-containing molecules. J Bacteriol 193: 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gerber NN (1975) Prodigiosin-like pigments. Crit Rev Microbiol 3: 469–485. [DOI] [PubMed] [Google Scholar]
- 50.Vining LC (1990) Functions of secondary metabolites. Annu Rev Microbiol 44. [DOI] [PubMed] [Google Scholar]
- 51. Williamson NR, Fineran PC, Ogawa W, Woodley LR, Salmond GP (2008) Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a post-transcriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia . Environ Microbiol 10: 1202–1217. [DOI] [PubMed] [Google Scholar]
- 52. Rosenberg M, Blumberger Y, Judes H, Bar-Ness R, Rubinstein E, et al. (1986) Cell surface hydrophobicity of pigmented and nonpigmented clinical Serratia marcescens strains. Infect Immun 51: 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rosenberg M, Kjelleberg S (1986) Hydrophobic interactions: role in bacterial adhesion. Adv Microbial Ecol 9: 353–393. [Google Scholar]
- 54. Rius N, Sole M, Fancia A, Loren JG (1994) Buffering capacity of pigmented and nonpigmented strains of Serratia marcescens . Appl Environ Microbiol 60: 2152–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haddix PL, Jones S, Patel P, Burnham S, Knights K, et al. (2008) Kinetic analysis of growth rate, ATP, and pigmentation suggests an energy-spilling function for the pigment prodigiosin of Serratia marcescens . J Bacteriol 190: 7453–7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O’Toole GA (2006) Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72: 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shanks RM, Stella NA, Kalivoda EJ, Doe MR, O’Dee DM, et al. (2007) A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J Bacteriol 189: 7262–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shanks RM, Donegan NP, Graber ML, Buckingham SE, Zegans ME, et al. (2005) Heparin stimulates Staphylococcus aureus biofilm formation. Infect Immun 73: 4596–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kowalski RP, Romanowski EG, Mah FS, Shanks RM, Gordon YJ (2010) Topical levofloxacin 1.5% overcomes in vitro resistance in rabbit keratitis models. Acta Ophthalmol 88: e120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Slater H, Crow M, Everson L, Salmond GP (2003) Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol Microbiol 47: 303–320. [DOI] [PubMed] [Google Scholar]
- 61.Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 62. Marone M, Mozzetti S, De Ritis D, Pierelli L, Scambia G (2001) Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online 3: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stella NA, Fender JE, Lahr RM, Kalivoda EJ, Shanks RM (2012) The LysR transcription factor, HexS, is required for glucose inhibition of prodigiosin production by Serratia marcescens Advances in Microbiology in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morowitz MJ, Denef VJ, Costello EK, Thomas BC, Poroyko V, et al. (2011) Strain-resolved community genomic analysis of gut microbial colonization in a premature infant. Proc Natl Acad Sci U S A 108: 1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liaw SJ, Lai HC, Ho SW, Luh KT, Wang WB (2003) Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis . J Med Microbiol 52: 19–28. [DOI] [PubMed] [Google Scholar]
- 66. Reimmann C, Ginet N, Michel L, Keel C, Michaux P, et al. (2002) Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology 148: 923–932. [DOI] [PubMed] [Google Scholar]
- 67. Lai HC, Soo PC, Wei JR, Yi WC, Liaw SJ, et al. (2005) The RssAB two-component signal transduction system in Serratia marcescens regulates swarming motility and cell envelope architecture in response to exogenous saturated fatty acids. J Bacteriol 187: 3407–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shanks RMQ, Stella NA, Fender JE (2012) Mutation of crp mediates Serratia marcescens serralysin and global secreted protein production. Res Microbiol in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fender JE, Bender CM, Stella NA, Lahr RM, Kalivoda EJ, et al. (2012) Serratia marcescens quinoprotein glucose dehydrogenase activity mediates medium acidification and inhibition of prodigiosin production by glucose. Appl Environ Microbiol 78: 6225–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Matsuyama T, Murakami T, Fujita M, Fujita S, Yano I (1986) Extracellular vesicle formation and biosurfactant production by Serratia marcescens . Journal of General Microbiology 132: 865–875. [Google Scholar]
- 71. Miller VL, Mekalanos JJ (1988) A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR . J Bacteriol 170: 2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kalivoda EJ, Stella NA, O’Dee DM, Nau GJ, Shanks RM (2008) The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl Environ Microbiol 74: 3461–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shanks RM, Kadouri DE, MacEachran DP, O’Toole G A (2009) New yeast recombineering tools for bacteria. Plasmid 62: 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complementation of prodigiosin phenotype conferred by insertional mutation of pigP . A. Photograph of WT (CMS376) or the pigP mutant (CMS836) with the vector (pMQ125) or ppigP (pMQ212) grown on LB agar supplemented with arabinose (4 mM). B. Prodigiosin production by the environmental isolate, CHASM, and isogenic pigP-insertion mutant (CMS2981) being either the empty vector (pMQ132) or ppigP (pMQ221) grown in LB medium. The average of six independent biological replicates is shown.
(PDF)
Amplification of pigP from S. marcescens isolates. PCR was used to amplify a 138 base pair amplicon from a variety of S. marcescens strains. Db11 and CMS376 served as positive controls, whereas Proteus and Staphylococcus chromosomal DNA served as negative controls. Amplicons were separated on TBE-PAGE gels and stained with ethidium bromide.
(PDF)
EMSA analysis of His9-PigP and MBP-HexS with promoters of interest. A) His9-PigP exhibited a gel shift of the pigP promoter but not the swrW or hexS promoters. B) MPB-HexS retarded migration of pigA and swrW promoters (positive controls), but not the pswP promoter (negative control) or the hexS promoter. MBP alone did not induce a gel shift of any promoter. Excess unlabeled promoter DNA could compete successfully for MPB-HexS binding to pigA and swrW promoters. Biotin-labeled promoters (Label. Promoter) were used at 2 ng per reaction and unlabeled promoters (Unlab promoter) were used at 500 ng per reaction.
(PDF)
Analysis of serratamolide from S. marcescens culture supernatants. LC-MS was used to measure serratamolide levels in culture supernatants from the WT (CMS376) the ΔpigP strain (CMS1713) and the negative control swrW mutant (CMS635). Purified serratamolide was used as a positive control. The serratamolide peaks are boxed in red the scale of the trace is shown on the left hand side. A representative experiment is shown.
(PDF)
Primers used in this study.
(DOCX)