Abstract
Rapamycin is an antibiotic that stimulates autophagy in a wide variety of eukaryotes, including the budding yeast Saccharomyces cerevisiae. Low concentrations of rapamycin extend yeast chronological life span (CLS). We have recently shown that autophagy is required for chronological longevity in yeast, which is attributable in part to a role for autophagy in amino acid homeostasis. We report herein that low concentrations of rapamycin stimulate macroautophagy during chronological aging and extend CLS.
Keywords: autophagy, aging, chronological life span, rapamycin, Saccharomyces cerevisiae
The nutrient-sensing target of rapamycin (TOR) pathway is implicated in the aging process in many organisms.1-3 In yeast, the TOR signaling pathway has been implicated in both replicative and chronological aging as well as in life-span extension by calorie restriction.3-6 In general, reduced signaling through the TOR pathway, achieved by either genetic or pharmacological means, results in increased longevity. Two underlying mechanisms, which are not mutually exclusive, are proposed: increased stress resistance and increased respiration.4,5 The most well-known small molecule inhibitor of TOR signaling is rapamycin. Rapamycin (Sirolimus) is a macrolide antibiotic with anti-fungal and immunosuppressive properties that inhibits TOR kinase complexes, which leads to inhibition of translation and growth arrest.7 Inhibition of TOR signaling by rapamycin upregulates autophagy in many organisms, including yeast.8 However, inhibition of TOR signaling by rapamycin has an impact on many metabolic pathways other than autophagy.9 This raises the question: is autophagy required for extension of CLS by rapamycin treatment? To address this question, we have studied the effects of rapamycin on CLS in autophagy-deficient yeast and measured induction of macroautophagy by exposure to a low concentration of rapamycin that extends yeast CLS.
Autophagy is Required for Rapamycin to Extend CLS
If rapamycin extends CLS by upregulating autophagy, then rapamycin should not extend CLS in atg mutants. To test this prediction, life spans were measured in synthetic dextrose (SD) minimal medium containing concentrations of rapamycin (0.1–40 nM) that were well below the standard cytostatic dose (220 nM or 0.2 μg/ml). Four strains were compared: hoΔ, atg1Δ, atg7Δ and atg11Δ. The hoΔ strain has a “wild-type” capacity for autophagy, and is referred to as “WT” below. Atg1 is a protein serine/threonine kinase required for macroautophagy.10 Atg7 is an E1-type ubiquitin-activating enzyme that executes two steps required for autophagosome formation: conjugation of Atg12 (a ubiquitin-like modifier) to Atg5 and attachment of PE to Atg8.11 An atg11Δ mutant was used as an additional control. ATG11 is not required for macroautophagy, but is required for the Cvt pathway and selective degradation of peroxisomes by microautophagy (pexophagy).12
Rapamycin at 10, 20 and 40 nM extended CLS in the WT and atg11Δ control strains as expected, but no extension was observed in atg1Δ or atg7Δ strains (Fig. 1B). Rapamycin at 0.1 or 1 nM final concentration did not extend CLS (Fig. 1A), although 0.33 nM (300 pg/ml) and 1.1 nM (1 ng/ml) rapamycin are reported to extend CLS in YPD.5 Cultures containing 10 nM or less rapamycin reached saturation after approximately 24 h of growth on day 1 of the CLS experiment. However, 20 nM and 40 nM rapamycin treatments slowed growth such that cultures achieved saturation on day 3 and 5 of the CLS experiment, respectively (data not shown). Similarly, treatment at 100 nM and 150 nM, which also extended CLS, delayed saturation until day 5 (data not shown). Importantly, however, the effects of rapamycin on growth rate were observed in both control and autophagy-deficient strains and there was no correlation between the time required to achieve saturation and chronological longevity.
Figure 1.
Autophagy is required for extension of chronological life span (CLS) by low concentrations of rapamycin. CLS was determined for yeast strains grown in liquid synthetic dextrose (SD) minimal medium containing essential supplements (H, K, L and uracil),15 250 μg/ml G418, 0.1% ethanol (solvent), and rapamycin at final concentrations ranging from 0–10 nM (A) or 0–40 nM (B). Strains in the BY4742 background contained the deletions hoΔ (WT), atg1Δ, atg7Δ or atg11Δ.16 Viability in terms of percent of colony forming units (CFU) observed on day 1 is plotted on a log scale over the 24 day time course of this experiment. Rapamycin was added on day zero to cultures containing a low cell density (OD600 ∼ 0.01). Cultures were grown to saturation (OD600 ∼ 1) and maintained at 30°C in a drum rotator at ∼15 rpm. For methodological details, see reference 13.
Autophagy and regulation of amino acid homeostasis are important factors in determining chronological longevity in yeast.13 Macroautophagy-deficient strains exhibit reduced chronological life span (CLS) in SD minimal medium compared to control strains. Increased availability of essential and nonessential branched side chain amino acids extends the CLS of macroautophagy-deficient strains by a mechanism that likely involves downregulation of the transcription factor Gcn4, which regulates general amino acid control. Furthermore, certain growth conditions, such as media containing low glucose or galactose, extend the CLS of autophagy-deficient yeast (unpublished results). These findings indicate that autophagy-deficient yeast are not necessarily short-lived relative to controls. Thus, the inability of rapamycin to extend CLS in autophagy-deficient strains is not simply due to the fact that these strains are inherently short-lived.
Rapamycin Promotes Autophagy during Chronological Aging
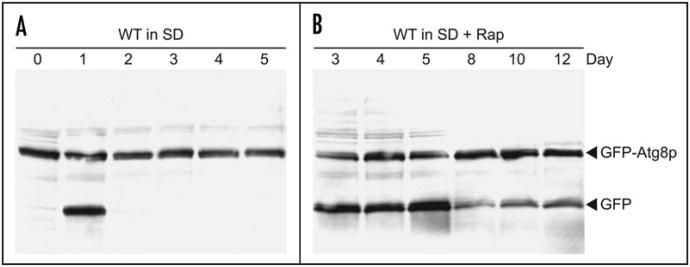
The observation that low concentrations of rapamycin extend CLS in autophagy-competent yeast raises the question: do low concentrations of rapamycin induce macroautophagy during chronological aging? To answer this question, a CLS experiment was done as described above (Fig. 1) and cells were collected and analyzed using a well-known assay for macroautophagy that is based on proteolytic conversion of a GFP-Atg8 fusion protein to GFP.14 During logarithmic growth, on day 0, macroautophagy is not induced, as expected (Fig. 2A). Macroautophagy is induced on day 1 of a CLS experiment, at which point the culture is saturated (Fig. 2A). However, during days 2–5, the level of macroautophagy was very low or not detectable (Fig. 2A). In contrast, growth in the presence of 10 nM rapamycin results in a prolongation of macroautophagic activity through day 12 of the CLS experiment (Fig. 2B). We conclude that a low concentration of rapamycin that is capable of extending CLS is also capable of upregulating macroautophagy during chronological aging.
Figure 2.
A low concentration of rapamycin induces macroautophagy. WT cells containing plasmid pCuGFPAUT7(416)17 were grown in liquid SD medium containing essential amino acids (H, K and L),15 250 μg/ml G418, and either no rapamycin (A) or 10 nM rapamycin (B). Cultures reached saturation (OD600 ∼ 1) on day 1. Cells were collected on the indicated days and equivalent amounts of whole cell extracts (based on OD600 cell density measurements) were analyzed by western blotting with the polyclonal anti-GFP antibody ab290 (Abcam, Inc.,). Macroautophagy-dependent proteolysis of the GFP-Atg8 fusion protein yields the GFP band, which is relatively stable to digestion by vacuolar proteinases. (B) shows days 3–12 to illustrate detection of the GFP band over a longer period of time than is evident in (A). Days 1 and 2 in (B) were comparable to lane 3 in (B); day 0 in (B) was equivalent to day 0 in (A) (data not shown).
Conclusions
Our findings suggest that upregulation of autophagy contributes to extension of CLS by treatment of yeast with low-dose rapamycin. It is becoming increasingly clear that modulation of autophagy contributes to chronological longevity via a number of different mechanisms, including nutrient recycling, organelle turnover and clearance of aberrant, damaged or toxic macromolecules. Small molecule interventions that upregulate autophagy offer the potential for enhancing health and life span, and an effective means for accomplishing this in vivo may be modulation of the TOR pathway.
Acknowledgments
This study was supported by NIH grant AG023719 to J.P.A.
References
- 1.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 2.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–90. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaeberlein M, Powers RW, III, Steffen KK, Westman EA, Hu D, Dang N, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–6. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 4.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–77. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers RW, III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–84. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, et al. Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Kamada Y, Sekito T, Ohsumi Y. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr Top Microbiol Immunol. 2004;279:73–84. doi: 10.1007/978-3-642-18930-2_5. [DOI] [PubMed] [Google Scholar]
- 9.De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006;25:6392–415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- 10.Stephan JS, Herman PK. The regulation of autophagy in eukaryotic cells: do all roads pass through Atg1? Autophagy. 2006;2:146–8. doi: 10.4161/auto.2.2.2485. [DOI] [PubMed] [Google Scholar]
- 11.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–6. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 12.He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–35. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell. 2009 doi: 10.1111/j.1474-9726.2009.00469.x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- 15.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 16.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–6. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]