Abstract
The heart holds the monumental yet monotonous task of maintaining circulation. Although cardiac function is critical to other organs and to life itself, mammals are not equipped with significant natural capacity to replace heart muscle that has been lost by injury. This deficiency plays a role in leaving millions worldwide each year vulnerable to heart failure. By contrast, certain other vertebrate species like zebrafish are strikingly good at heart regeneration. A cellular and molecular understanding of endogenous regenerative mechanisms, combined with advances in methodology to transplant cells, together project a future in which cardiac muscle regeneration can be therapeutically stimulated in injured human hearts. This review will focus on what has been discovered recently about cardiac regenerative capacity and how natural mechanisms of heart regeneration in model systems are stimulated and maintained.
Keywords: heart regeneration, regenerative capacity, myocardial infarction, heart failure, dedifferentiation, transdifferentiation, stem cells, progenitor cells, cardiomyocytes, epicardium, endocardium
INTRODUCTION
The heart maintains rhythmic contractions to support blood circulation, and is thus essential for vertebrate life. The heart has from two to four chambers, and can display marked differences in structure among vertebrate classes, and between species within a class. The key component of most hearts is a muscular wall comprised of contractile cardiac myocytes supplemented with connective tissue, and typically vascularized and innervated. The inside of this wall is enveloped by a thin endothelial cell layer, the endocardium; and the outside by a mesothelial cell layer called the epicardium. The rhythmic contraction of cardiac muscle is controlled through organized electric stimuli generated and maintained by the conduction system. Among those cellular constituents of the heart, cardiomyocytes are not necessarily the dominant population in number but provide the key function.
Acute myocardial infarction (MI), typically caused by coronary artery occlusion and ischemia, is a leading cause of death worldwide. For those fortunate enough to survive myocardial infarction, necrotic myocardium that might represent one billion lost cardiomyocytes provokes an inflammatory response and recruitment of local fibroblasts. In the next few weeks, a large, collagen-rich scar tissue forms in its place. The scar provides a rapid solution; however, it is not contractile. Indeed, cardiac scarring, though to be irreversible, weakens the heart while also increasing susceptibility to compensatory pathology, aneurysm, additional MI events, and organ failure. Many scientists and clinicians believe that MI patients represent the greatest potential beneficiaries from regenerative medicine, which aims to restore functional tissue through stem or progenitor cell manipulation. Indeed, the development of therapies that can facilitate survival or regenerative replacement of destroyed myocardium would be of enormous social and economic impact.
Cardiac regeneration is a multidisciplinary research area, enlisting specialists in physiology, stem cell and developmental biology, and biomaterial and tissue engineering, with the ultimate goal of achieving regenerative medicine that can prevent or reverse heart failure. Given the poor cardiac regenerative capacity of adult mammals, considerable efforts have been made in attempts to somehow stimulate endogenous or transplanted cellular sources to create new muscle within the environment of an injured heart.
New and exciting advances in in vitro cardiomyocyte production and cell transplantation are not the subject of this review; instead, we refer interested readers to recent reviews (Laflamme & Murry 2011, Leri et al 2011, Musunuru et al 2010, Rubart & Field 2006, Yi et al 2010). A complementary approach is to identify successful examples of natural cardiac regeneration, dissect how this success is achieved, and then attempt to apply this information to humans via provision of the appropriate regenerative stimuli. The general regenerative capacity of organs, including the heart, is remarkably elevated in certain lower vertebrates like urodele amphibians and teleost fish (Poss 2010). A recent study indicated that the mammalian heart also maintains the capacity of regeneration during a short period after birth (Porrello et al 2011b). Already, we have learned information from these studies that brings a new perspective on human heart disease and potential regenerative therapies. In this review, we will overview the intrinsic capacity of cardiac cells for regeneration. We will discuss endogenous mechanisms underlying cardiac regeneration observed in experimental model animals, as well as recent findings of how regenerative responses are initiated and maintained.
CARDIAC REGENERATIVE CAPACITY IN VERTEBRATES
Mammalian hearts
In experiments performed many decades ago, adult mammals were probed for the capacity to regenerate cardiac muscle after several models of injury, including MI, burning, freezing, mechanical injury, and chemical injury (Rumyantsev 1977). Most in the field agree that this body of work, as well as subsequent experiments to date involving modern capabilities to detect bona fide regeneration, generated little evidence to conclude that there is significant myocardial regeneration after cardiac injury. Most also agree that the key limitation to cardiac muscle regeneration is likely to be the poor ability of adult mammalian cardiomyocytes to enter the cell cycle and undergo division (see below). Cardiomyocytes in the fetal mammalian heart are mononucleated and proliferative; but shortly after birth the vast majority of cardiomyocyte DNA replication occurs without cytokinesis or karyokinesis. Thus, most cardiomyocytes are binucleated with diploid nuclei in the adult mouse heart, and mononucleated with polyploid nuclei in the adult human heart (Laflamme & Murry 2011). After this postnatal switch, it is rare for cardiomyocytes to enter the cell cycle (Figure 1).
Figure 1.
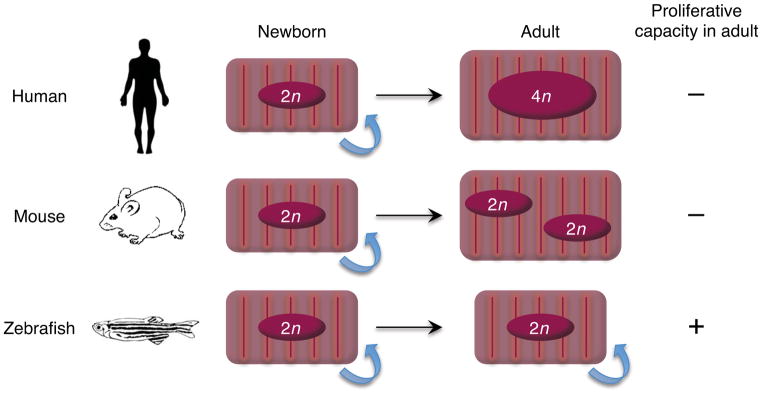
Nuclear dynamics and proliferative capacity of cardiomyocytes during growth. Cardiomyocytes in fetal humans and mice typically have a single nucleus with a diploid genome (2n) and increase mass through cell division. Human cardiomyocytes can proliferate for the first few months after birth, but are believed to lose this capacity early in life. These cells typically undergo rounds of DNA replication without karyokinesis or cytokinesis, resulting largely in mononucleated cardiomyocytes with tetraploid (4n) or higher DNA content. Murine cardiomyocytes can divide robustly until the first few days after birth, after which the majority withdraw from the cell cycle. These cells undergo additional DNA replication with karyokinesis but not cytokineis, resulting in binucleated cardiomyocytes that are diploid (2n) in each nucleus (Laflamme & Murry 2011). By contrast, most cardiomyocytes in zebrafish hearts are mononucleated with a diploid genome (2n) throughout life, with significant proliferative capacity (Wills et al 2007).
Early observations suggest that injury may influence the propensity for adult mammalian cardiomyocyte proliferation. In injured rodent ventricles, histological examination of 3H-thymidine incorporation identified detectable DNA replication in nuclei of myofibers bordering necrotic tissue (Rumyantsev 1977). Similar analysis was carried out with better resolution using transgenic mice in which cardiomyocytes were labeled by a nuclear-localized lacZ reporter protein (Soonpaa & Field 1997), although no distinction between karyokinesis and cytokinesis was provided. These labeled cardiomyocytes were detected near the border zone of myocardial damage at exceptionally low levels (~0.0083%).
The capacity for cardiomyocyte renewal in the human heart has not been assessed until recently, given the experimental limitations of research with human tissues. Frisen and colleagues overcame this hurdle by taking advantage of the high levels of the radiocarbon 14C released from nuclear bomb tests during the Cold War (Bergmann et al 2009). In the atmosphere, 14C reacts with oxygen to form 14CO2, which is then captured in plants and eventually incorporated in humans through the food chain. The genomic 14C concentration was quantified in cardiomyocyte nuclei purified by flow cytometry, facilitating retrospective dating of cardiomyocytes from recently deceased people of various ages. Mathematical modeling of the radiocarbon data suggested that human cardiomyocytes renew throughout life with a capacity that gradually decreases from ~1% annual turnover at the age of 25 to 0.45% at the age of 75. Thus, inferred from these data is that nearly 50% of cardiomyocytes are replenished during a normal life span (Bergmann et al 2009). In a different study with a similar goal, Anversa and colleagues made use of human tissue samples collected from cancer patients who had received infusion of iododeoxyuridine, a thymidine analog used as radiosensitizer for therapy, and estimated that 22% of cardiomyocytes in the human heart are renewed annually (Kajstura et al 2010). With their calculations, 13% of endothelial cells and 20% of fibroblasts undergo turnover each year in the heart, suggesting that cardiomyocytes have the highest renewal capacity among cardiac cell types examined in the study.
Overall, these results suggest that the mammalian heart possesses a measurable capacity for renewal. It is not yet clear whether cardiomyocytes are renewed through differentiation from a stem/progenitor population or through cell division by existing cardiomyocytes. However, the results provide rationale for the development of potential manipulations that might enhance myocardial regeneration after injury.
Amphibian hearts
The possibility of natural cardiac regeneration in amphibians has been examined in frogs, newts, and axolotls, with much of the original literature reporting work of Soviet scientists in the 1960’s (Rumyantsev 1977). Salamanders are the champions of regeneration among vertebrates, able to renew removed or injured body parts like lens, retina, spinal cord, jaws, portions of intestine, brain tissue, and major appendages (Brockes & Kumar 2005, Eguchi 1988, Ghosh et al 1994, Keefe 1973, Minelli et al 1987, O’Steen & Walker 1962, Piatt 1955). This body of work indicated that amphibians survive massive mechanical injury to the ventricle, including removal of as much as one-quarter of the chamber. This resilience is a feat in itself, and is likely to reflect a lesser reliance than mammalian species on vigorous circulation.
Descriptions of the regenerative capacity of the amphibian heart have varied in the literature. Resection injury penetrates the ventricular lumen, releasing a large amount of blood. In the newt heart, the eventual formation of connective scar tissue appears to be a dominant response after resection of the ventricular apex, and there is only minor replacement of cardiac muscle (Oberpriller & Oberpriller 1974). However, when the resected myocardium was minced and grafted back to the wound area, the tissue graft could assemble into a contiguous, contractile mass (Bader & Oberpriller 1978). More recently, resection injuries at the base of the newt heart were reported to regenerate with much less scarring (Witman et al 2011). These results indicated that the outcome of regeneration could be influenced by the level and type of tissue damage, a warning of sorts that heart regeneration should be assessed in multiple injury contexts. While there is some variability in reports and reviews on the extent of regeneration, there is definitive evidence for proliferative activity in newt and axolotl cardiomyocytes. These include the presence of mitotic figures in cardiomyocyte nuclei as visualized by standard histology and transmission electron microscopy (Oberpriller J 1971, Rumyantsev 1977), and multiple indicators of DNA synthesis (Flink 2002, Oberpriller & Oberpriller 1974, Rumyantsev 1973, Witman et al 2011).
Teleost hearts
The zebrafish (Danio rerio) is highly amenable to genetic approaches, and has become a popular model system for understanding vertebrate embryonic development. This popularity has recently encouraged the exploration of its regenerative capacity. Like salamanders, adult zebrafish effectively regenerate multiple structures that mammals fail to regenerate, including retinae, brain tissues, spinal cord, and major appendages (fins) (Becker et al 1997, Kroehne et al 2011, Poss et al 2003, Vihtelic & Hyde 2000). Thus, it made sense a decade ago to be curious about their natural capacity for heart regeneration. Today it is fair to argue that zebrafish display the most robust and best-characterized cardiac regenerative response known to date, and thus we give this system more attention here.
An initial study examined the effects of removing ~20% of the ventricle by surgical resection (Poss et al 2002). As with the mechanically injured amphibian heart, the injury seals by a quick clotting mechanism and the organ sustains sufficient contractile force to continue to drive circulation. Over the next month, a series of events occurs in response to ventricular resection. First, the clot that seals the apex matures within several days into a complex, fibrin-rich milieu containing serum factors and degenerated erythrocytes. In the infarcted mammalian ventricle, fibrin deposition attracts fibroblasts and inflammatory cells, and is a precursor to scarring. The fibrin clot is typically not replaced by scar tissue during cardiac repair in zebrafish; in fact, little or no collagen is retained by 1–2 months after resection injury. Instead, the clot is supplanted by cardiac muscle, restoring a contiguous wall of vascularized cardiac muscle. While the pyramidal shape of the uninjured ventricle may not be perfectly replicated, the process restores approximately the amount of muscle that was lost. Elevated indices of cardiomyocyte proliferation were detectable at the end of the first week after injury, and observable for weeks beyond this (Poss et al 2002).
As discussed for amphibian model systems, it is possible that different injury types introduce distinct outcomes of myocardial regeneration. Recent studies have reported new approaches to injure the zebrafish heart. In one of these studies, Wang and colleagues generated a transgenic system to facilitate cell type-specific ablation in zebrafish (Wang et al 2011). This system employs two transgenes: 1) a 4-hydroxytamoxifen (4-HT)-inducible Cre recombinase (CreER) restricted to cardiomyocytes by the regulatory sequences of cardiac myosin light chain 2 (cmlc2); and 2) a cytotoxic DTA (diphtheria toxin A chain) gene that can be inducibly targeted to CreER-expressing cells. In these transgenic fish, referred to as Z-CAT (zebrafish cardiomyocyte ablation transgenes), a single injection of 4-HT could eliminate more than 60% of cardiomyocytes throughout the heart. While such massive loss of myocardium did not normally affect survival, it caused lethargy, a gasping phenotype, and reduced exercise capacity, classic indicators of heart failure that are not seen after resection injury. Interestingly, these signs of heart failure reversed within several days, a recovery that correlated with massive cardiomyocyte proliferation detected throughout the ventricle. By 30 days after injury, the ventricle was filled with new muscle and displayed little or no scar tissue.
It is important to test whether regenerated cardiomyocytes incorporate functionally with existing cardiac muscle and do not generate arrhythmias. This was investigated using optical voltage mapping of surface myocardium at various stages of regeneration. At 7 days after injury, when cardiomyocytes begin to proliferate, muscle at the regenerating apex was uncoupled. A week later, coupling was evident, and by 30 dpa, electrical conduction through the apex occurred at normal velocities (Kikuchi et al 2010). Loss and recovery of conduction velocities was also evident after genetic ablation, and then regeneration, of cardiomyocytes in the Z-CAT model (Wang et al 2011). These data indicated that the newly created cardiomyocytes show evidence of functional integration in the regenerated zebrafish heart.
MI is caused by ischemic injury, and coronary artery occlusion is routinely used as an injury model in small and large mammalian model systems. However, the zebrafish ventricle is diminutive (~1 mm3), and the coronary vascular network perfuses a relatively small proportion of ventricular muscle, making this type of injury a difficult task. Cryocauterization has occasionally been used as an alternative model to coronary artery ligation in mouse (de Crom & Duncker 2005). In this model, a thin metal filament cooled by liquid nitrogen is delivered to the surface of the heart to introduce a rapid temperature change, resulting in rupture of cells and massive tissue necrosis. Recently, this injury model was applied to zebrafish (Chablais et al 2011, González-Rosa et al 2011, Schnabel et al 2011). Within a day after cryoinjury to the ventricular apex, a target of approximately 25% of the ventricular tissue underwent necrosis. In the initial report of this injury model, collagen deposits formed during the 3 weeks following injury, yet were subsequently replaced with new cardiac muscle by 130 days post-injury (González-Rosa et al 2011). Although cardiac muscle and coronary vasculature were recovered at the cryoinjured area, perfect ventricular shape was not restored, as also observed in the resection model. Whereas the amount of cauterized ventricular tissue is similar to that removed in the resection model, dynamics of cardiac regeneration differed with injury type. This delay likely reflects the need to remove necrotic tissue after cryoinjury for regeneration in the damaged area to take place. A recent study has shown the capacity of heart regeneration after an analogous necrotic injury in the giant danio, a teleost fish closely related to zebrafish (Lafontant et al 2011).
These 3 quite different injury models each stimulate robust myocardial regeneration in zebrafish, although at different rates and likely with some differences in mechanism that may turn out to be informative.
What limits the regenerative capacity of the mammalian heart?
Explaining these clear differences in cardiac regenerative capacity among vertebrate species is a central pursuit of the field. One possible reason for this may be intrinsic differences in cardiomyocytes. Lower vertebrate cardiomyocytes tend to be mononucleated, smaller in size, and containing fewer myofibrils, as compared to those of adult mammals. In fact, these characters are typical of cardiomyocytes in young mammals, and might facilitate cell cycle reentry after injury (Figure 1) (Rumyantsev 1977).
This correlation, in addition to detection of apparent age-dependent rates of cardiomyocyte turnover in humans mentioned earlier, prompt the question of whether regenerative capacity remains constant throughout life. By way of a conditional cardiomyocyte-lethal mutant in an X-linked gene, a recent study assessed the effects of ablating approximately one-half of the cardiomyocytes of female mouse embryos at 12.5 days post-coitum. In these experiments, fetal hearts compensated for cardiac muscle loss by increased representation of cardiomyocytes with a non-inactivated wild-type allele, indicating regenerative potential of the fetal heart (Drenckhahn et al 2008).
More recently, Sadek and colleagues have applied a resection injury model to the neonatal mouse heart (Porrello et al 2011b). In this study, approximately 15% of the muscle was removed from the left ventricular apex of one day-old mice. At this age, mice are in the process of major growth, as are their cardiac chambers. Similar to the zebrafish and salamander models, a large blood clot quickly sealed the wound after injury. After surgery and sutures, pups were cared for by mothers until weaning. Strikingly, during this 3-week period, the ventricles fully healed without major scarring. Cardiomyocyte proliferation indices were boosted both near to and away from the resection plane to levels even higher than normally seen in growing hearts. By contrast, resection injuries performed at 7 days after birth led to the formation of a fibrotic scar. Thus, the capacity of myocardial regeneration is transiently present in the neonatal mouse heart, but is quickly lost by 7 days after birth.
Postnatal switches in cardiomyocyte proliferation and regenerative capacity coincide with changes of the expression of cell cycle regulator genes (Walsh et al 2010), and a recent study suggested the role of microRNAs (miRNAs) in this regulation (Porrello et al 2011a). In this study, microarray analysis was carried out to identify subsets of miRNAs of which expression is changed in murine cardiac ventricles between 1 and 10 days after birth. The analysis identified miR-195, a member of the miR-15 family that is reported to regulate B cell proliferation and contribute to leukemogenesis, as being highly upregulated during the postnatal period. Myocardial specific overexpression as well as knockdown experiments indicated that miR-195 negatively regulates cardiomyocyte mitosis. Not surprisingly, a number of cell cycle-associated genes were identified as potential targets through gene expression analysis of miR-195-overexpressing hearts. For example, Checkpoint kinase 1 (Check1) was shown to be directly regulated through a binding site in its 3′UTR.
While cardiomyocyte characteristics would appear to have a primary role in regenerative capacity, another basis for the poor regenerative potential of the mammalian heart may be the activity of non-myocardial cardiac cells in response to injury. For instance, fibroblasts make up a high percentage of adult mammalian cardiac cells, and a much lower percentage of fetal mammalian or adult non-mammalian vertebrate hearts. These fibroblasts not only have the capacity to form scar tissue, but also appear to impact the proliferative capacity of cardiomyocytes. To this point, a recent study found that adult cardiac fibroblasts co-cultured with neonatal cardiomyocytes inhibited their proliferation, while embryonic cardiac fibroblasts had no such effect (Ieda et al 2009). Thus, age-related changes in fibroblast characters might modify cardiac regenerative capacity.
Additionally, the hearts of lower vertebrates such as zebrafish or axolotl have long, exaggerated trabeculae that protrude into the ventricular lumen. These trabeculae are lined by a large total surface area of endocardial cells. The mammalian cardiac chambers briefly display similar anatomy in their fetal form. During maturation, mammalian ventricles then acquire a thick, vascularized wall with limited trabeculation and low relative endocardial surface area. Recent evidence suggests that this different endocardial presence might be important for regeneration. Notably, the zebrafish endocardium quickly responds to injury and induces a signal(s) that is required for myocardial proliferation, while the endocardium of the adult mouse heart does not appear to mount an analogous response (Kikuchi et al 2011b). The important roles of non-myocardial cardiac cells during regeneration will be discussed again in the later section of this review, and may thus be contributors to regenerative capacity.
CELLULAR SOURCES OF CARDIAC MUSCLE REGENERATION
Identification of endogenous cardiac stem and progenitor cells in the postnatal mammalian heart
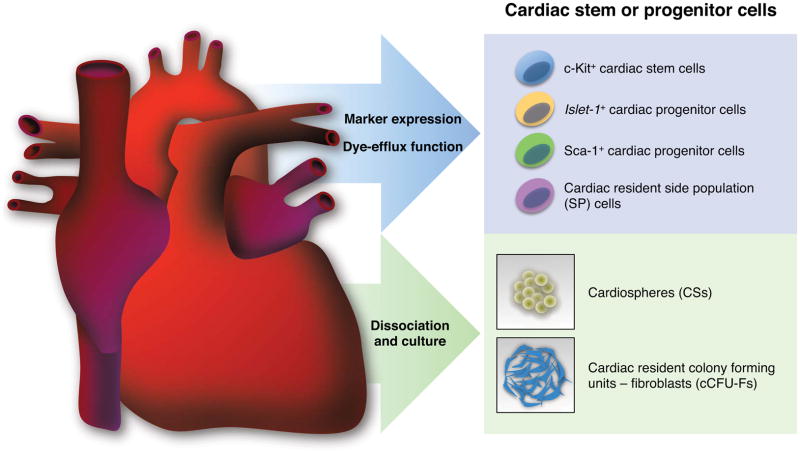
As mentioned, adult mammalian cardiomyocytes have low proliferative character, and thus it makes good sense for biologists to search for undifferentiated progenitor cells that have the potential to mature into contractile cells. Several cell types with the potential to create cardiomyocytes postnatally have been described, expressing either the pan-stem cell marker c-Kit (Bearzi et al 2007, Beltrami et al 2003), the transcription factor Islet1 (Laugwitz et al 2005), or the cell surface marker stem cell antigen 1 (Sca-1) (Matsuura et al 2004, Oh 2003). Other candidate cardiac progenitor cells include “side population” cells that possess physiological properties to efflux fluorescent dye (Hierlihy et al 2002, Pfister et al 2010), or to form multicellular clusters, referred to as cardiospheres, in culture (Figure 2) (Messina et al 2004, Smith et al 2007, Ye et al 2012).
Figure 2.
Endogenous cardiac stem or progenitor cells in the postnatal mammalian heart. These cell populations have been directly identified based on cell surface marker expression (c-Kit+ and Sca-1+ cells), genetic marker expression (Islet1+ cells), and dye-efflux function (SP cells), or indirectly identified as cell clusters derived from the culture of dissociated cardiac tissues (CSs and cCFU-Fs).
In another recent study, Harvey and colleagues reported colony-forming cells (cardiac resident colony forming units - fibroblasts, cCFU-Fs) in the adult mouse heart that have long-term growth potential in culture (Figure 2) (Chong et al 2011). Clonally derived cCFU-Fs were shown to give rise to multiple mesodermal lineages in vitro including cardiomyocytes, endothelial cells, smooth muscles, adipocytes, cartilage and bone. Injection of GFP-tagged cCFU-Fs into the infarcted heart demonstrated that those cells have the capacity to create cardiomyocytes, endothelial cells, and smooth muscle cells in vivo. Surprisingly, differentiation of endodermal and ectodermal cell types, such as hepatocytes, neurons, and oligodendrocytes, was also detected in vitro, suggesting trans-germ layer plasticity of this population. Gene expression profiles and marker expression, as well as perivascular localization of cCFU-Fs, were analogous to those of mesenchymal stem cells derived from bone marrow. However, transplantation assays and Cre-based genetic fate-mapping indicated that cCFU-Fs are likely to derive from the epicardium, a finding of interest given other findings that suggest epicardial transdifferentiation capacity (see below).
Establishment of cell therapy with endogenous stem cells is one of the major goals in regenerative medicine. Several stem cell populations, such as c-Kit+ cells (Bolli et al 2011) and bone marrow mononucleated cells (Janssens et al 2006), have been subjects of clinical trials in patients with heart disease. Recently, Marbán and colleagues reported findings from a randomized phase 1 trial with cardiosphere-derived stem cells (Makkar et al 2012). In this trial, cardiospheres were grown from explant culture biopsies of MI patients suffering from left ventricular dysfunction. Autologous cardiosphere-derived cells (CDCs) were then infused into the artery associated with the infarct. Functional examinations of CDC-treated hearts showed that although there were some beneficial effects, overall ejection fraction of left ventricle was not significantly recovered. Nevertheless, MRI examinations revealed that mean scar mass was significantly reduced in CDC groups, suggesting recovery of myocardium. This change was likely induced by indirect mechanisms, as human CDCs exert beneficial effects through paracrine mechanisms when injected into immunocompromised mice after MI (Chimenti et al 2010). This study provides an optimistic projection for future cell therapy of diseased hearts.
Given findings over the years that many adult mammalian organs maintain one or more progenitor cell types, the identification of multiple cardiac stem/progenitor pools itself may not be surprising. Important perspective may come from particularly well-examined stem cell-based tissues like the intestinal epithelium, which is rapidly renewed in adult mammals. Previous genetic fate-mapping studies revealed that all cell types of the small intestinal epithelium are generated by at least two distinct intestinal stem cell populations: Lgr5+ cells at the crypt base (Barker et al 2007) and Bmi1+ cells at the +4 position relative to the crypt base (Sangiorgi & Capecchi 2008). Further genetic lineage tracing experiments showed that Bmi1+ cells give rise to Lgr5+ cells, and the intestinal epithelium can renew without difficulty when Lgr5+ cells are genetically ablated (Tian et al 2011). Interestingly, a recent fate-mapping study using Hopx, a novel marker for +4 cells, showed that Hopx+ cells generate Lgr5+ cells, but conversely, Lgr5+ cells can also form Hopx+ cells (Takeda et al 2011). Thus, each stem cell population in intestine is interrelated, and the hierarchy seems bidirectional. Similarly, cardiac stem/progenitor populations are likely to be interrelated during development or homeostasis. Genetic fate-mapping experiments using multiple specific regulatory sequences should enable this issue to be addressed directly.
Overall, evidence is accumulating that the mammalian heart possesses stem or progenitor cell populations that differentiate into cardiomyocytes in vitro or in vivo after transplantation. The mystery, of course, is why the damaged mammalian heart fails to efficiently harness the potential of progenitor cells to create a significant amount of new cardiac muscle after injury.
Contributions of cardiomyocytes to regenerated myocardium
Zebrafish provide a model to directly assess the cellular source(s) of naturally regenerated heart muscle. Early studies performed prior to the introduction of Cre-based genetic fate-mapping tools to this animal suggested a model in which undifferentiated progenitor cells are a major source of proliferating cardiomyocytes. Key evidence for this model came from assessment of fast- (EGFP) and slow-folding (nuclear DsRed2) reporter proteins in transgenic expression cassettes driven by regulatory sequences of the contractile gene cmlc2. In this developmental timing assay, many EGFP+nucDsRed2− cardiomyocytes were detected at apical edge of the wound in the regenerating adult ventricle (Lepilina et al 2006). This expression phenotype is identical to that observed in de novo cardiomyocytes differentiating from heart fields in early zebrafish embryos (de Pater et al 2009), and suggested fresh maturation of Cmlc2− progenitor cells into proliferative cardiomyocytes (Kikuchi et al 2010).
Recently, this assay was re-explored by substituting a cytosolic DsRed2 reporter of cmlc2 promoter activity, allowing visualization of the two reporters within the same subcellular compartment and removing the element of nuclear localization. Here, regenerates contained EGFP+DsRed2+ myocytes with each reporter fluorescing at lower intensities than in non-regenerating muscle (Kikuchi et al 2010). Taken together, the most likely interpretation of the developmental timing assays is that cardiomyocytes reduce contractile gene expression, but do not fully lose the cardiomyocyte phenotype, as they participate in regeneration. A similar reduction in the expression of contractile protein was also reported in the newt ventricle upon injury (Laube et al 2006).
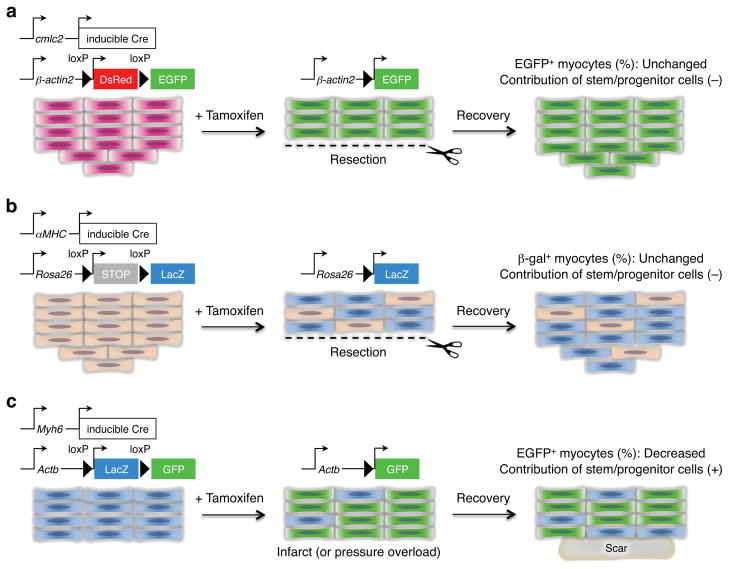
This non-implication of resident progenitor cells during instances of natural heart regeneration is supported by two recent studies, each of which directly assessed the contribution of cardiomyocytes to zebrafish heart regeneration by inducible genetic fate-mapping techniques (Buckingham & Meilhac 2011) (Figure 3). In each case, transgenic lines were generated with CreER driven by the regulatory sequences of cmlc2, as well as an indicator line that would permit visualization of cardiomyocyte EGFP fluorescence after excision of loxP-flanked stop sequences. In these experiments, 4-HT was used to pre-label cardiomyocytes with EGFP fluorescence prior to tests of regeneration. There was no significant difference in the proportion of EGFP cardiomyocytes in regenerated tissue compared to uninjured ventricles, indicating that the vast majority of new cardiomyocytes derive from cells expressing cmlc2 before injury (Jopling et al 2010, Kikuchi et al 2010).
Figure 3.
Cellular origins of regenerating cardiomyocytes. (a) Genetic fate-mapping to determine the source of new cardiomyocytes during zebrafish heart regeneration. Treatment with tamoxifen leads to marking of nearly all cardiomyocytes with EGFP. Following resection, new cardiomyocytes expressed EGFP, indicating derivation from existing cardiomyocytes rather than a different unmarked cell source (Kikuchi et al 2010). (b) A similar fate-mapping experiment was performed in the neonatal mouse heart, in this case with an inducible LacZ reporter. Although the labeling of cardiomyocytes with this system was ~60%, the ratio of labeled cells was maintained between the regenerate and uninjured myocardium. This result indicates that the new cardiomyocytes are derived from existing cardiomyocytes rather than a progenitor cell (Porrello et al 2011b). (c) Fate-mapping experiments were performed in the adult mouse heart with an inducible GFP reporter. The labeling of cardiomyocytes with this system was ~83%, and the ratio of labeled cells was decreased to ~68% in areas bordering the MI, and decreased to ~76% in hearts subjected to pressure overload. Thus, stem or progenitor cells might refresh cardiomyocytes in hearts after pressure overload or myocardial infarction (Hsieh et al 2007).
Furthermore, most regenerating myocytes were found to activate regulatory sequences of the transcription factor gata4, a gene required for embryonic heart development (Kikuchi et al 2010). Regenerating cardiomyocytes maintained this signature throughout the process, suggesting that the tissue had activated an embryonic program. This is consistent with results of transmission electron microscopy and sarcomere stains, which indicated that regenerating cardiomyocytes acquire a less organized sarcomeric structure during regeneration (Jopling et al 2010, Kikuchi et al 2010, Wang et al 2011). Earlier supporting data included detection of increased expression of other embryonic cardiogenesis genes during zebrafish heart regeneration (Lepilina et al 2006). The most likely model to synthesize all results is that existing differentiated cardiomyocytes reduce their contractile state to acquire a more embryonic form, in which cell division is facilitated. Thus, the current thinking in the field is that dedifferentiation of existing cardiomyocytes is the dominant source mechanism for heart regeneration in zebrafish.
A similar fate-mapping analysis was performed in the context of neonatal heart regeneration in mice, in this case employing the cardiomyocyte-specific α-myosin heavy chain (αMHC or Myh6) promoter to drive a tamoxifen-inducible Cre recombinase in combination with a loxP-based reporter strain (Porrello et al 2011b) (Figure 3). Neonatal mice were given a single subcutaneous dose of tamoxifen at birth to label the cardiomyocyte population prior to resection injury. Histological analysis revealed that the majority of cardiomyocytes within the newly formed apex (3 weeks post-resection) were lineage-labeled, indicating that these myocytes arose from an αMHC-positive cardiac lineage. Echocardiograms of these hearts suggested the recovery of contractile function after regeneration. Thus, the primary source for regeneration of functional myocardium in robust systems appears to be resident cardiomyocytes.
Importantly, a similar experiment has been performed using adult mice, with intriguing results that keep stem cells on the radar. Here, a high percentage of cardiomyocytes were pre-labeled with the reagents described above, and then assessed for cardiomyocyte labeling during normal aging, or after an MI injury (Figure 3). This approach detected no changes in the percentage of labeled cardiomyocytes during aging, suggesting that a non-muscle cell could not be responsible for new cardiomyocyte addition during cardiac homeostasis. However, in mice subjected to MI, percentages of labeled cardiomyocytes were reduced in the peri-infarct zone, suggesting the contribution of undifferentiated progenitor cells (Hsieh et al 2007).
In a more recent study that followed up this work, the investigators examined effects of injecting bone marrow-derived c-Kit+ cells into the infarcted heart (Loffredo et al 2011). Upon cell injection, the percentage of labeled cardiomyocytes was further decreased compared to sham or injections with bone marrow-derived mesenchymal stem cells. These results suggest that c-Kit+ cells somehow stimulate some degree of cardiomyocyte regeneration from an endogenous progenitor or unlabeled cardiomyocyte source. Importantly, by use of a second cell-marking tag as well as sex-mismatched wild-type recipients, Loffredo and colleagues could argue against transdifferentiation or cell fusion with a high degree of certainty. Together, the results point to paracrine effects by a still unidentified signal(s) released from the transplanted cells, a signal that can enable de novo cardiomyocyte creation from a still unidentified source.
Does heart regeneration occur from a subset of “elite” cardiomyocytes?
Fate-mapping experiments described above do not address the possibility that cardiomyocytes are a heterogeneous population with respect to their regenerative capacity, containing certain muscle cells that may be better suited for division after injury. That is, myocardial regeneration might depend on such “elite cardiomyocytes”, perhaps even identifiable by a specific gene expression signature. This seems logical, given that the heart is initially contributed by two recognized cardiac fields (Buckingham et al 2005, Cai et al 2003, Farrell et al 1999, Musunuru et al 2010, Wu et al 2008), and that various cardiomyocytes have different physiologic and/or functional properties (Domian et al 2009). As mentioned above, regeneration of the zebrafish ventricular apex involves activation of gata4 regulatory sequences in a subpopulation of cardiomyocytes within the wall near the injury. Moreover, fate-mapping experiments indicated that those myocytes inducing gata4 make major contributions to regeneration after resection injury (Kikuchi et al 2010). Interestingly, newt cardiomyocytes isolated from the adult ventricle showed heterogenous proliferation in cell culture. Only one-third of these cells progressed through mitosis and underwent successive cell divisions (Bettencourt-Dias et al 2003). It is thus of interest to look further at regenerative cardiomyocytes in adult zebrafish or neonatal mice for heterogeneity and potential underlying factors.
In adult mammals, a recent study by Bersell and colleagues reported that Neuregulin1 (NRG1) promotes proliferation of differentiated adult mouse cardiomyocytes in cell culture and when introduced in vivo (Bersell et al 2009). These treatments appeared to predominantly affect a subpopulation of mononucleated (versus binucleated) cardiomyocytes, a finding consistent with the idea that some cardiomyocytes are more receptive to regeneration signals.
Transdifferentiation and cardiac muscle regeneration
Transdifferentiation is a regenerative phenomenon in which one cell type converts to another, sometimes using an undifferentiated intermediate. A classic example of transdifferentiation occurs after removal of the lens from an adult next. Here, a new lens emerges from the dorsal, but not the ventral, pigmented iris tissue (Eguchi et al 1974, Grogg et al 2005). This is an intriguing model, but it is unclear whether events like these are common during organ regeneration. A recent study used transgenic axolotls that ubiquitously express EGFP to examine whether transdifferentiation drives salamander limb regeneration. Using elegant tissue transplantation experiments, Tanaka and colleagues demonstrated that while dermal cells might show multipotency, most cells in the limb stump are restricted to contributing their own tissue type during regeneration (Kragl et al 2009). Similar lineage and germ layer restriction properties were shown by a series of genetic fate-mapping experiments on regenerating digit tips in mice (Rinkevich et al 2011).
In the field of cardiac regeneration, there is a considerable interest in whether transdifferentiation events might create new cardiomyocytes. Bone marrow-derived cells like hematopoietic stem cells (Orlic et al 2001) and mesenchymal stem cells (Toma et al 2002) were thought to differentiate to cardiac muscle and contribute to functional recovery after MI. However, results from subsequent studies indicate that these cell types may contribute to cardiac muscle survival/repair by indirect paracrine mechanisms, as opposed to direct differentiation into myocardium (Balsam et al 2004, Mirotsou et al 2007, Murry et al 2004).
On the surface, it seems a rare and difficult task for differentiated cells to switch a determined lineage under natural conditions. However, cumulative evidence demonstrates that experimental manipulations can overcome this hurdle. Recent findings indicate that forced expression of fate-determining transcription factors can eventually wrest control of the developmental program of a cell type that has previously been committed to a specific lineage. Notable examples are the derivation of induced pluripotent stem cells from adult somatic cells (Takahashi & Yamanaka 2006), the direct reprogramming of pancreatic β-cells from exocrine cells (Zhou et al 2008b), and the conversion of fibroblasts into neurons (Ambasudhan et al 2011, Caiazzo et al 2011, Kim et al 2011, Son et al 2011, Vierbuchen et al 2010). Relevant to cardiac cells, direct differentiation of non-cardiogenic mesoderm into beating cardiomyocytes (Takeuchi & Bruneau 2009) and direct reprogramming of cardiac or dermal fibroblasts to cardiac muscle cells (Ieda et al 2010) have been demonstrated. Although the underlying mechanisms remain incompletely understood, reprogramming is an exciting methodology for generating patient-derived cardiomyocytes. Directed in vitro differentiation of ES cells or reprogrammed cells into cardiomyocytes is becoming efficient (Kattman et al 2011, Yang et al 2008). Yet, an ideal scenario would involve targeted reprogramming of an abundant resident source in the heart, one that could transdifferentiate to the myocardial lineage with minimal steps.
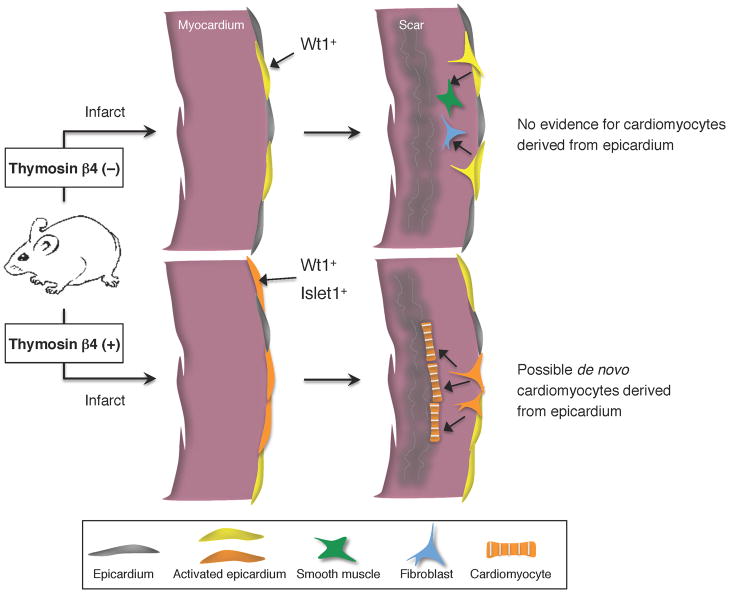
In this regard, the epicardial covering of the heart is an interesting candidate; indeed, this tissue has been implicated as a source for cardiomyocytes during mouse cardiac development (Cai et al 2008, Zhou et al 2008a). In the infarcted adult mouse heart, lineage tracing experiments indicated that the epicardium does not differentiate into cardiac muscle; instead, epicardial cells contribute to the canonical epicardial lineage (epicardium, fibroblasts, smooth muscle, perivascular cells) (Figure 4) (Zhou et al 2011). The fate of the epicardium has also been examined during robust cardiac regeneration in zebrafish (Kikuchi et al 2011a). In this system, lineage-traced descendants of epicardial cells were detected in perivascular cells but not cardiomyocytes during either cardiac development or regeneration.
Figure 4.
Potential role of Thymosin β4 in cardiomyocyte differentiation from non- myocytes. In adult mouse hearts, Wt1 is absent or expressed at low levels in epicardial cells. Epicardial cells re-express Wt1 after infarction, proliferate, and give rise to mesenchymal cells expressing smooth muscle and fibroblast cell markers, but never differentiate into cardiomyocytes (Zhou et al 2011). With Thymosin β4 injections prior to the injury, more epicardial cells induce Wt1 expression after MI, some of which coexpress cardiac progenitor markers such as Islet1. Evidence suggests that some of these activated epicardial cells transdifferentiate to cardiomyocytes (Smart et al 2011).
While data indicate that epicardial cells lack natural myogenic potential under most contexts (Christoffels et al 2009, Kikuchi et al 2011a, Zhou et al 2011), a recent study suggests that this restriction can be modulated. Thymosin β4 (Tβ4) is a peptide that has been shown to enhance vascular potential to adult epicardial- derived cells (EPDCs) and improve responses to MI (Bock-Marquette et al 2004, Smart et al 2006). When Tβ4 was injected into mice prior to infarction, epicardial cells induced the expression of the embryonic epicardial gene Wt1 and cardiac progenitor markers. Genetic fate-mapping analysis combined with transplantation assays with purified epicardial cells provided evidence that EPDCs near the infarcted area turn into functional cardiomyocytes at low frequency (Smart et al 2011) (Figure 4). Although the low reprogramming efficiency and the preconditioning with Tβ4 injections may not be realistic for therapy, this study provided rationale for considering the adult epicardium as a source for creating new myocardium. It is a mystery how Tβ4, an intracellular actin binding molecule, might induce myocardial differentiation from the epicardium, and resolution of this mechanism may provide novel insight for regenerative mechanisms.
MECHANISMS STIMULATING MYOCARDIAL REGENERATION
Organ-wide injury responses
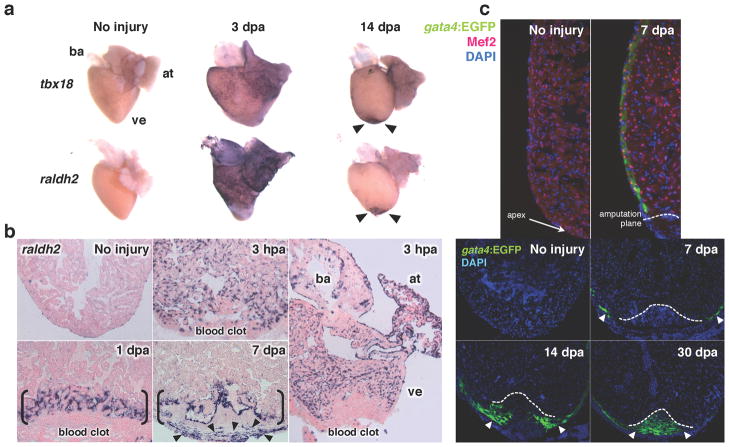
A hallmark of zebrafish heart regeneration is the presence of injury responses that occur not only near trauma, but also in an organ-wide manner. Studies thus far have found that all major cardiac tissues - epicardium, endocardium and myocardium -employ this strategy in response to injury (Kikuchi et al 2011b, Kikuchi et al 2010, Lepilina et al 2006) (Figure 5). The endocardium stands out among these tissues, as it shows the earliest responses yet seen after cardiac injury. Within an hour of local injury, endocardial cells throughout the heart take on a rounded morphology and show detachment from underlying myofibers. Concomitant with these morphological changes, endocardial cells induce the expression of developmental marker genes, raldh2 and heg, in an organ-wide manner by 3 hours post-injury (Kikuchi et al 2011b) (Figure 5). Interestingly, this activation does not occur in the vascular endothelium, suggesting a distinct role of the endocardial endothelium in this response. Similarly, embryonic epicardial markers tbx18 and raldh2 are induced in adult epicardial cells as early as 1 day after injury, and become detectable around the periphery of the entire heart by 3 days post trauma (Lepilina et al 2006) (Figure 5). In the myocardium, gata4 regulatory sequences are activated in ventricular cardiomyocytes located in the subepicardial compact layer of the entire ventricle by 7 days post-injury, before this signature localizes to regenerating cardiomyocytes (Kikuchi et al 2010) (Figure 5). At different time courses depending on the cell type, these injury-activated expression signatures disappear globally and localize to the injury site, where they aid or indicate cardiac muscle regeneration, as described later.
Figure 5.
Injury responses of the three major cardiac cell types in zebrafish. (a) Ostensibly the entire epicardium activates developmental genes such as tbx18 and raldh2 (violet) within several days following injury. Enhanced gene expression localizes to the wound area by two weeks post-injury (arrowheads). (b) Endocardial cells in the entire heart upregulate raldh2 (violet) within hours of amputation, before localization to the injury site. (c) Myocardial activation, represented by induction of a gata4:EGFP reporter, is first detected throughout the compact layer of the myocardium by 7 days post-amputation (dpa) and becomes localized to the regenerating myocardium by 14–30 dpa (arrowheads). Cardiomyocyte nuclei are visualized by Mef2. ba, bulbus arteriosus; at, atrium; ve, ventricle. Dotted lines indicate approximate amputation planes. Modified from previous studies (Kikuchi et al 2011b, Kikuchi et al 2010, Lepilina et al 2006).
The organ-wide response is not unique to the adult zebrafish heart. When neonatal mouse ventricles are injured by resection, cardiomyocyte mitoses and sarcomere disassembly are increased not only near the injury but also in areas distant from the injury (Porrello et al 2011b). Like the zebrafish, the neonatal mouse activates this organ-wide response quickly; indices are boosted a day after injury and peak at 7 days after apical resection of the ventricle. Thus, local injury can induce global cardiomyocyte morphology changes and proliferation in the neonatal mouse heart.
Mechanisms by which these organ-wide responses are activated remain unclear. Results from various injury models suggest that the activation process in zebrafish does not require tissue removal or direct injury to the endocardium and epicardial tissue, and is not maintained by circulating systemic factors (Kikuchi et al 2011b). Interestingly, when zebrafish are intraperitoneally injected with Lipopolysaccharide (LPS), an agent that can induce systemic inflammation, the expression of the retinoic acid (RA) synthesizing enzyme raldh2 is induced in the entire endocardium and epicardium of the uninjured heart (Kikuchi et al 2011b). During mammalian liver regeneration, partial hepatectomy is known to affect tissue distant from trauma and activates compensatory hepatocyte proliferation in spare lobes, partly through inflammatory factors such as interleukin-6 and TNFα (Taub 2004). Similarly, factors released during cardiac inflammation may help to trigger organ-wide injury responses during heart regeneration. It seems natural to imagine that in tissues that are competent for regeneration, local signals provoked by injury target regenerative events. However, cumulative examples of natural heart regeneration indicate that, instead, injury responses are initially activated throughout the entire chamber or organ, a property that might be key to regenerative success.
Regulation by non-myocardial cells
The epicardium and the endocardium appear to play important signaling and structural roles during heart regeneration in zebrafish. As indicated earlier, morphological changes in endocardial cells start organ-wide but become localized to the wound area by around 1 dpa. By 7–14 dpa, epicardial cells that have amplified in response to injury accumulate in the wound site (Lepilina et al 2006). Together, endocardial cells near the injury site and epicardial cells integrated into the wound maintain high expression of raldh2 while regeneration continues (Figure 5). Recent transgenic experiments involving overexpression of a dominant-negative form of RA receptor alpha, or an RA-degrading enzyme, Cyp26a1, indicated that RA produced by activated endocardial and epicardial cells is essential to maintain myocardial proliferation at the injury site. Introduction of RA or analogues to uninjured animals failed to induce myocardial proliferation, suggesting a permissive role of endocardial and epicardial RA during regeneration (Kikuchi et al 2011b).
Establishing new vasculature is critical for tissue regeneration. Multiple developmental signaling pathways have been functionally implicated in this process during zebrafish heart regeneration. Here, as during embryonic heart development in higher vertebrates (Gittenberger-de Groot et al 2000, Mikawa & Fischman 1992, Mikawa & Gourdie 1996, Perez-Pomares et al 2002, Tevosian et al 2000), the creation of new vascular components appears to be facilitated by epicardial cells. As mentioned above, genetic fate-mapping of epicardial cells using the marker tcf21 identified contributions to perivascular cell types – thus, epicardial cells are ostensibly recruited into the regenerate to facilitate growth (Kikuchi et al 2011a). Members of the Fgf signaling pathway are upregulated after resection injury and may serve this purpose. Expression of the ligand fgf17b is activated in injured myocardium, corresponding with upregulation of the receptors fgfr2 and fgfr4 in epicardial cells within the regenerate. Inhibition of Fgf signaling by transgenic overexpression of a dominant-negative Fgfr inhibits epicardial cell integration into the wound area, and also blocks neovascularization of regenerating myocardium. This manipulation arrested muscle regeneration and caused scar formation (Lepilina et al 2006).
Platelet-derived growth factor (Pdgf) may have similar roles as Fgfs during regeneration. Lien and colleagues performed a microarray to compare gene expression after resection injury in zebrafish and identified two ligand members with increased expression. Initially, Pdgf was implicated in directly activating cardiomyocyte proliferation (Lien et al 2006). Follow-up studies found that expression of a receptor for Pdgf, pdgfrβ, is induced during heart regeneration, and that pharmacological inhibition of Pdgf receptors inhibits proliferation in epicardial cells and coronary vasculature formation during regeneration (Kim et al 2010). Thus, both Fgf and Pdgf signaling both appear to reactivate vascular development during myocardial regeneration.
Cardiomyocyte dedifferentiation
Proliferation by resident cardiomyocytes is the primary source mechanism for regeneration of the adult zebrafish or neonatal mouse heart. While the non-muscle injury environment likely contributes, intrinsic mechanisms must also underlie their regenerative capacity. As mentioned earlier, cardiomyocyte dedifferentiation is typically characterized by reduction of sarcomere structures and expression of fetal gene markers, and appears to be a shared mechanism associated with cardiac muscle regeneration. Thus, a focus of the field will be to understand how dedifferentiation is initiated.
Interestingly, dedifferentiation characters are detected in the adult mammalian heart under some pathologic situations (Dispersyn et al 2002, Driesen et al 2009, Rücker-Martin et al 2002, Sharov et al 1997). The link between such phenotypes to physiological regeneration is unclear, as the innate regenerative ability of the mammalian heart is extremely limited. However, understanding mechanisms that underlie these changes may provide a clue to identifying molecule(s) that induce cardiomyocyte dedifferentiation. Braun and colleagues recently investigated heart tissue samples from chronic dilated cardiomyopathy (DCM) patients, in an effort to discover factors that cause dedifferentiated phenotypes in human cardiomyocytes (Kubin et al 2011).
By using proteomics and biochemical approaches, Oncostatin M (OSM) was found to be highly expressed in DCM hearts but not healthy hearts. OSM is a cytokine that has pleiotropic functions and transduces signals through a heterodimeric receptor composed of gp130, a co-receptor shared with many other cytokines, and OSM receptor (Oβ) or LIF receptor (Heinrich et al 2003, Tanaka & Miyajima 2003). The authors found that OSM induced loss of sarcomeric structures and re-expression of embryonic markers in rat adult cardiomyocytes in vitro and in vivo, through signals mediated by Oβ. It could also enhance cell-cycle entry in neonatal cardiomyocytes in vitro, and Oβ was required for dedifferentiation phenotypes in cardiomyocytes at the border zone in mouse MI models. These results suggest a potential target for manipulating dedifferentiation, and possibly cardiomyocyte proliferation, in the injured heart.
SUMMARY AND PROSPECTS
The adult mammalian heart was once believed to be a post-mitotic organ without any capacity for regeneration, but recent findings have challenged this dogma. A modified view assigns to the mammalian heart a measurable capacity for regeneration throughout life. The ultimate goals of the cardiac regeneration field have been pursued by multiple strategies, including understanding the developmental biology of cardiomyocytes and cardiac stem/progenitor cells, applying chemical genetics, and engineering biomaterials and delivery methods that facilitate cell transplantation.
Successful stimulation of endogenous regenerative capacity in injured adult mammalian hearts can benefit from studies of natural cardiac regeneration. From lineage-tracing studies in zebrafish and neonatal mice, we recognize that a major cellular source for regenerating muscle is resident cardiomyocytes. Given this identified source, groups can more efficiently search for intrinsic and extrinsic signals that control cardiac regeneration. Such signals will be generated from tissues like epicardium and endocardium, and will affect regenerative proliferation of cardiomyocytes directly, and indirectly through events like neovascularization. New sources of cardiomyocytes might be identified by further lineage tracing results, or by methodology to stimulate transdifferentiation, dedifferentiation, or direct reprogramming by defined factors. These approaches together provide growing perspective for understanding how to achieve heart regeneration.
SUMMARY POINTS.
Mammalian hearts maintain a measurable capacity for cardiomyocyte turnover throughout life, providing a rationale for the development of manipulations that might enhance myocardial regeneration.
Postnatal mammalian hearts contain endogenous stem or progenitor cells with potential to differentiate into cardiomyocytes in vitro or in vivo after transplantation.
Resident cardiomyocytes are the primary source for myocardial regeneration in robust regeneration models, such as the adult zebrafish heart or the neonatal mouse heart.
Highly regenerative hearts initially respond to injury in an organ-wide manner before localization of regenerative activity to trauma.
Endocardial and epicardial cells in the zebrafish heart release factors like retinoic acid at the injury site that contribute to local regenerative proliferation of cardiomyocytes.
Epicardial cells do not normally give rise to cardiomyocytes; however, it is possible that exogenous factors may induce their transdifferentiation in mammalian hearts after MI.
FUTURE ISSUES.
Which signaling pathways, factors, and small molecules can enhance the regenerative ability of cardiomyocytes or endogenous stem/progenitor cells in adult mammalian hearts?
What molecular and cellular mechanisms induce cardiomyocyte dedifferentiation?
Can subpopulations of cardiomyocytes with high regenerative capacity be identified and activated?
What underlies differences in cardiac regenerative ability between lower and higher vertebrates?
What are the evolutionary factors that have selected for or against cardiac regenerative capacity?
Acknowledgments
We apologize to colleagues whose work was not discussed due to space limitations. K.D.P. is an Early Career Scientist of the Howard Hughes Medical Institute and acknowledges grant support for heart regeneration research in his laboratory from the National Heart, Lung, and Blood Institute, American Heart Association, and American Federation for Aging Research.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial, holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, et al. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–18. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader D, Oberpriller JO. Repair and reorganization of minced cardiac muscle in the adult newt (Notophthalmus viridescens) Journal of Morphology. 1978;155:349–57. doi: 10.1002/jmor.1051550307. [DOI] [PubMed] [Google Scholar]
- Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL, Robbins RC. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature. 2004;428:668–73. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–07. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Bearzi C, Rota M, Hosoda T, Tillmanns J, Nascimbene A, et al. Human cardiac stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14068–73. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. The Journal of Comparative Neurology. 1997;377:577–95. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–76. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, et al. Evidence for Cardiomyocyte Renewal in Humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 Signaling Induces Cardiomyocyte Proliferation and Repair of Heart Injury. Cell. 2009;138:257–70. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Mittnacht S, Brockes JP. Heterogeneous proliferative potential in regenerative adult newt cardiomyocytes. Journal of Cell Science. 2003;116:4001–09. doi: 10.1242/jcs.00698. [DOI] [PubMed] [Google Scholar]
- Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432:466–72. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–57. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–23. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nature Reviews Genetics. 2005;6:826–35. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Buckingham ME, Meilhac SM. Tracing cells for tracking cell lineage and clonal behavior. Developmental Cell. 2011;21:394–409. doi: 10.1016/j.devcel.2011.07.019. [DOI] [PubMed] [Google Scholar]
- Cai C-L, Liang X, Shi Y, Chu P-H, Pfaff SL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Developmental Cell. 2003;5:877–89. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C-L, Martin JC, Sun Y, Cui L, Wang L, et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–08. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–27. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Chablais F, Veit J, Rainer G, JaŸwińska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Developmental Biology. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimenti I, Smith RR, Li TS, Gerstenblith G, Messina E, et al. Relative roles of direct regeneration versus paracrine effects of human cardiosphere-derived cells transplanted into infarcted mice. Circulation Research. 2010;106:971–80. doi: 10.1161/CIRCRESAHA.109.210682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JJH, Chandrakanthan V, Xaymardan M, Asli NS, Li J, et al. Adult Cardiac-Resident MSC-like Stem Cells with a Proepicardial Origin. Cell Stem Cell. 2011;9:527–40. doi: 10.1016/j.stem.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Grieskamp T, Norden J, Mommersteeg MTM, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458:E8–E9. doi: 10.1038/nature07916. [DOI] [PubMed] [Google Scholar]
- de Crom R, Duncker D. A novel model of cryoinjury-induced myocardial infarction in the mouse: a comparison with coronary artery ligation. American Journal of Physiology Heart and Circulatory Physiology. 2005;289:H1291–300. doi: 10.1152/ajpheart.00111.2005. [DOI] [PubMed] [Google Scholar]
- de Pater E, Clijsters L, Marques SR, Lin Y-F, Garavito-Aguilar ZV, et al. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–41. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dispersyn GD, Mesotten L, Meuris B, Maes A, Mortelmans L, et al. Dissociation of cardiomyocyte apoptosis and dedifferentiation in infarct border zones. European Heart Journal. 2002;23:849–57. doi: 10.1053/euhj.2001.2963. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Chiravuri M, van der Meer P, Feinberg AW, Shi X, et al. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326:426–9. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenckhahn J-D, Schwarz QP, Gray S, Laskowski A, Kiriazis H, et al. Compensatory Growth of Healthy Cardiac Cells in the Presence of Diseased Cells Restores Tissue Homeostasis during Heart Development. Developmental Cell. 2008;15:521–33. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Driesen RB, Verheyen FK, Debie W, Blaauw E, Babiker FA, et al. Re-expression of alpha skeletal actin as a marker for dedifferentiation in cardiac pathologies. Journal of Cellular and Molecular Medicine. 2009;13:896–908. doi: 10.1111/j.1582-4934.2008.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi G. Cellular and molecular background of Wolffian lens regeneration. Cell Differentiation and Development. 1988;25:147–58. doi: 10.1016/0922-3371(88)90111-6. [DOI] [PubMed] [Google Scholar]
- Eguchi G, Abe SI, Watanabe K. Differentiation of lens-like structures from newt iris epithelial cells in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:5052–56. doi: 10.1073/pnas.71.12.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell M, Waldo K, Li YX, Kirby ML. A novel role for cardiac neural crest in heart development. Trends in Cardiovascular Medicine. 1999;9:214–20. doi: 10.1016/s1050-1738(00)00023-2. [DOI] [PubMed] [Google Scholar]
- Flink IL. Cell cycle reentry of ventricular and atrial cardiomyocytes and cells within the epicardium following amputation of the ventricular apex in the axolotl, Amblystoma mexicanum: confocal microscopic immunofluorescent image analysis of bromodeoxyuridine-labeled nuclei. Anatomy and Embryology. 2002;205:235–44. doi: 10.1007/s00429-002-0249-6. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Thorogood P, Ferretti P. Regenerative capability of upper and lower jaws in the newt. The International Journal of Developmental Biology. 1994;38:479–90. [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Bergwerff M, Mentink MM, Poelmann RE. Epicardial outgrowth inhibition leads to compensatory mesothelial outflow tract collar and abnormal cardiac septation and coronary formation. Circulation Research. 2000;87:969–71. doi: 10.1161/01.res.87.11.969. [DOI] [PubMed] [Google Scholar]
- González-Rosa JM, Martín V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–74. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- Grogg MW, Call MK, Okamoto M, Vergara MN, Del Rio-Tsonis K, Tsonis PA. BMP inhibition-driven regulation of six-3 underlies induction of newt lens regeneration. Nature. 2005;438:858–62. doi: 10.1038/nature04175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochemical Journal. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierlihy AM, Seale P, Lobe CG, Rudnicki MA, Megeney LA. The post-natal heart contains a myocardial stem cell population. FEBS letters. 2002;530:239–43. doi: 10.1016/s0014-5793(02)03477-4. [DOI] [PubMed] [Google Scholar]
- Hsieh PCH, Segers VFM, Davis ME, MacGillivray C, Gannon J, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nature Medicine. 2007;13:970–74. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, et al. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell. 2010;142:375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong T-T, et al. Cardiac Fibroblasts Regulate Myocardial Proliferation through β1 Integrin Signaling. Developmental Cell. 2009;16:233–44. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Dubois C, Bogaert J, Theunissen K, Deroose C, et al. Autologous bone marrow-derived stem-cell transfer in patients with ST-segment elevation myocardial infarction: double-blind, randomised controlled trial. Lancet. 2006;367:113–21. doi: 10.1016/S0140-6736(05)67861-0. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Martí M, Raya A, Belmonte JCI. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–09. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajstura J, Urbanek K, Perl S, Hosoda T, Zheng H, et al. Cardiomyogenesis in the adult human heart. Circulation Research. 2010;107:305–15. doi: 10.1161/CIRCRESAHA.110.223024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–40. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Keefe JR. An analysis of urodelian retinal regeneration: II. Ultrastructural features of retinal regeneration inNotophthalmus viridescens. The Journal of Experimental Zoology. 1973;184:207–31. doi: 10.1002/jez.1401840207. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011a;138:2895–902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, et al. Retinoic Acid Production by Endocardium and Epicardium Is an Injury Response Essential for Zebrafish Heart Regeneration. Developmental Cell. 2011b;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature. 2010;464:601–05. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Su SC, Wang H, Cheng AW, Cassady JP, et al. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9:413–19. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proceedings of the National Academy of Sciences. 2010;107:17206–10. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138:4831–41. doi: 10.1242/dev.072587. [DOI] [PubMed] [Google Scholar]
- Kubin T, Pöling J, Kostin S, Gajawada P, Hein S, et al. Oncostatin M Is a Major Mediator of Cardiomyocyte Dedifferentiation and Remodeling. Stem Cell. 2011;9:420–32. doi: 10.1016/j.stem.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–35. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontant PJ, Burns AR, Grivas JA, Lesch MA, Lala TD, et al. The Giant Danio (D. Aequipinnatus) as A Model of Cardiac Remodeling and Regeneration. The Anatomical Record. 2011;295:234–48. doi: 10.1002/ar.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube F, Heister M, Scholz C, Borchardt T, Braun T. Re-programming of newt cardiomyocytes is induced by tissue regeneration. Journal of Cell Science. 2006;119:4719–29. doi: 10.1242/jcs.03252. [DOI] [PubMed] [Google Scholar]
- Laugwitz K-L, Moretti A, Lam J, Gruber P, Chen Y, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–53. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, et al. A Dynamic Epicardial Injury Response Supports Progenitor Cell Activity during Zebrafish Heart Regeneration. Cell. 2006;127:607–19. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Leri A, Kajstura J, Anversa P. Role of cardiac stem cells in cardiac pathophysiology: a paradigm shift in human myocardial biology. Circulation Research. 2011;109:941–61. doi: 10.1161/CIRCRESAHA.111.243154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lien C-L, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biology. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo FS, Steinhauser ML, Gannon J, Lee RT. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell. 2011;8:389–98. doi: 10.1016/j.stem.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LEJ, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012 doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, et al. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. The Journal of Biological Chemistry. 2004;279:11384–91. doi: 10.1074/jbc.M310822200. [DOI] [PubMed] [Google Scholar]
- Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circulation Research. 2004;95:911–21. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:9504–8. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Developmental Biology. 1996;174:221–32. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Minelli G, Franceschini V, Del Grande P, Ciani F. Newly-formed neurons in the regenerating optic tectum of Triturus cristatus carnifex. Basic and Applied Histochemistry. 1987;31:43–52. [PubMed] [Google Scholar]
- Mirotsou M, Zhang Z, Deb A, Zhang L, Gnecchi M, et al. Secreted frizzled related protein 2 (Sfrp2) is the key Akt-mesenchymal stem cell-released paracrine factor mediating myocardial survival and repair. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1643–48. doi: 10.1073/pnas.0610024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Soonpaa MH, Reinecke H, Nakajima H, Nakajima HO, et al. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004;428:664–68. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Domian IJ, Chien KR. Stem cell models of cardiac development and disease. Annual Review of Cell and Developmental Biology. 2010;26:667–87. doi: 10.1146/annurev-cellbio-100109-103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Steen WK, Walker BE. Radioautographic studies of regeneration in the common newt III. Regeneration and repair of the intestine. The Anatomical Record. 1962;142:179–87. doi: 10.1002/ar.1091420210. [DOI] [PubMed] [Google Scholar]
- Oberpriller JOJ. Mitosis in adult newt ventricle. The Journal of Cell Biology. 1971;49:560–63. doi: 10.1083/jcb.49.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. The Journal of Experimental Zoology. 1974;187:249–53. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- Oh H. Cardiac progenitor cells from adult myocardium: Homing, differentiation, and fusion after infarction. Proceedings of the National Academy of Sciences. 2003;100:12313–18. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlic D, Kajstura J, Chimenti S, Jakoniuk I, Anderson SM, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701–05. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Carmona R, Gonzalez-Iriarte M, Atencia G, Wessels A, Munoz-Chapuli R. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. The International Journal of Developmental Biology. 2002;46:1005–13. [PubMed] [Google Scholar]
- Pfister O, Oikonomopoulos A, Sereti K-I, Liao R. Isolation of resident cardiac progenitor cells by Hoechst 33342 staining. Methods in Molecular Biology. 2010;660:53–63. doi: 10.1007/978-1-60761-705-1_4. [DOI] [PubMed] [Google Scholar]
- Piatt J. Regeneration of the spinal cord in the salamander. The Journal of Experimental Zoology. 1955;129:177–207. [Google Scholar]
- Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam Y-J, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circulation Research. 2011a;109:670–79. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011b;331:1078–80. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nature Reviews Genetics. 2010;11:710–22. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Developmental Dynamics. 2003;226:202–10. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–49. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- Rinkevich Y, Lindau P, Ueno H, Longaker MT, Weissman IL. Germ-layer and lineage-restricted stem/progenitors regenerate the mouse digit tip. Nature. 2011;476:409–13. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubart M, Field LJ. Cardiac regeneration: repopulating the heart. Annual Review of Physiology. 2006;68:29–49. doi: 10.1146/annurev.physiol.68.040104.124530. [DOI] [PubMed] [Google Scholar]
- Rücker-Martin C, Pecker F, Godreau D, Hatem SN. Dedifferentiation of atrial myocytes during atrial fibrillation: role of fibroblast proliferation in vitro. Cardiovascular Research. 2002;55:38–52. doi: 10.1016/s0008-6363(02)00338-3. [DOI] [PubMed] [Google Scholar]
- Rumyantsev PP. Post-injury DNA synthesis, mitosis and ultrastructural reorganization of adult frog cardiac myocytes. Cell and Tissue Research. 1973;139:431–50. doi: 10.1007/BF00306596. [DOI] [PubMed] [Google Scholar]
- Rumyantsev PP. Interrelations of the proliferation and differentiation processes during cardiact myogenesis and regeneration. International Review of Cytology. 1977;51:186–273. [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nature Genetics. 2008;40:915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel K, Wu C-C, Kurth T, Weidinger G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS ONE. 2011;6:e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov VG, Sabbah HN, Ali AS, Shimoyama H, Lesch M, Goldstein S. Abnormalities of cardiocytes in regions bordering fibrous scars of dogs with heart failure. International Journal of Cardiology. 1997;60:273–79. doi: 10.1016/s0167-5273(97)00117-4. [DOI] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dubé KN, Vieira JM, Zhou B, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011 doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Risebro CA, Melville AAD, Moses K, Schwartz RJ, et al. Thymosin β4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2006;445:177–82. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, et al. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, et al. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–18. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. The American Journal of Physiology. 1997;272:H220–6. doi: 10.1152/ajpheart.1997.272.1.H220. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–24. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–11. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Miyajima A. Oncostatin M, a multifunctional cytokine. Reviews of Physiology, Biochemistry and Pharmacology. 2003;149:39–52. doi: 10.1007/s10254-003-0013-1. [DOI] [PubMed] [Google Scholar]
- Taub R. Liver regeneration: from myth to mechanism. Nature Reviews Molecular Cell Biology. 2004;5:836–47. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, et al. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000;101:729–39. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–59. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma C, Pittenger MF, Cahill KS, Byrne BJ, Kessler PD. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93–98. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. Journal of Neurobiology. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Walsh S, Pontén A, Fleischmann BK, Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo--an analysis based on cardiomyocyte nuclei. Cardiovascular Research. 2010;86:365–73. doi: 10.1093/cvr/cvq005. [DOI] [PubMed] [Google Scholar]
- Wang J, Panáková D, Kikuchi K, Holdway JE, Gemberling M, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–30. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AA, Holdway JE, Major RJ, Poss KD. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development. 2007;135:183–92. doi: 10.1242/dev.010363. [DOI] [PubMed] [Google Scholar]
- Witman N, Murtuza B, Davis B, Arner A, Morrison JI. Recapitulation of developmental cardiogenesis governs the morphological and functional regeneration of adult newt hearts following injury. Developmental Biology. 2011;354:67–76. doi: 10.1016/j.ydbio.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–43. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–28. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Ye J, Boyle A, Shih H, Sievers RE, Zhang Y, et al. Sca-1 cardiosphere-derived cells are enriched for isl1-expressing cardiac precursors and improve cardiac function after myocardial injury. PLoS ONE. 2012;7:e30329. doi: 10.1371/journal.pone.0030329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi BA, Wernet O, Chien KR. Pregenerative medicine: developmental paradigms in the biology of cardiovascular regeneration. The Journal of Clinical Investigation. 2010;120:20–28. doi: 10.1172/JCI40820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh J-H, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. The Journal of Clinical Investigation. 2011;121:1894–904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008a;454:109–13. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature. 2008b;455:627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]