Abstract
Extrachromosomal rDNA circles (ERCs) and recombinant origin-containing plasmids (ARS-plasmids) are thought to reduce replicative life span in the budding yeast Saccharomyces cerevisiae due to their accumulation in yeast cells by an asymmetric inheritance process known as mother cell bias. Most commonly used laboratory yeast strains contain the naturally occurring, high copy number 2-micron circle plasmid. 2-micron plasmids are known to exhibit stable mitotic inheritance, unlike ARS-plasmids and ERCs, but the fidelity of inheritance during replicative aging and cell senescence has not been studied. This raises the question: do 2-micron circles reduce replicative life span? To address this question we have used a convenient method to cure laboratory yeast strains of the 2-micron plasmid. We find no difference in the replicative life spans of otherwise isogenic cir+ and cir0 strains, with and without the 2-micron plasmid. Consistent with this, we find that 2-micron circles do not accumulate in old yeast cells. These findings indicate that naturally occurring levels of 2-micron plasmids do not adversely affect life span, and that accumulation due to asymmetric inheritance is required for reduction of replicative life span by DNA episomes.
Keywords: DNA episome, Life span, Asymmetric inheritance, Saccharomyces cerevisiae
1. Introduction
The budding yeast Saccharomyces cerevisiae undergoes asymmetric cell division that yields a larger mother cell and a smaller daughter cell. Mother cells divide a limited number of times to give rise to a limited number of daughter cells. The number of cell divisions completed by a mother cell determines its replicative life span. For yeast derived from the W303 strain background, mother cells typically give rise to an average of ~30 daughter cells and a maximum of ~40 daughter cells [1]. Changes in cell morphology and physiology take place as mother cells age and approach senescence (reviewed in [2]). Such changes reflect differences in cell regulation between mother and daughter cells, and are likely due, at least in part, to asymmetric inheritance of nucleic acids, proteins, and potentially other macromolecules [3,4].
We have shown that recombinant plasmids containing an origin of replication are inherited asymmetrically, accumulate in old cells, and reduce yeast life span [1]. Replication origins in yeast are known as autonomous replicating sequences (ARSs), and we refer to plasmids containing them as ARS-plasmids. Such recombinant plasmids are also known as yeast replicating plasmids (YRps). ARS-plasmids mimic extrachromosomal rDNA circles (ERCs) insofar as both are circular DNA episomes that contain ARSs and are inherited asymmetrically. ERCs arise due to recombination at the repetitive rDNA locus, and have been shown to accumulate in old mother cells and reduce life span [4]. Asymmetric inheritance of ARS-plasmids and ERCs and their accumulation during aging is thought to be due to mother cell bias inheritance. Mother cell bias is a well-known, but poorly understood, inheritance behavior of ARS-plasmids in yeast [5]. This relationship between reduction in life span and accumulation of plasmid DNAs in mother cells raises the question: do yeast 2-micron circle plasmids reduce replicative life span?
2-micron circles are naturally occurring DNA episomes in yeast named for the length of their circular DNA molecule [6]. The majority of laboratory strains of S. cerevisiae contain 2-micron circles (i.e., they are cir+). 2-micron circles encode four open reading frames (FLP1, RAF1, REP1, and REP2) and three well-known cis-acting sequences, ORI, FRT, and STB (reviewed in [6–8]). ORI is an origin of replication or ARS. FRT is the site of binding and recombination catalyzed by the Flp recombinase. STB is a “stability” element required for high fidelity transmission during cell division. The primary functions of these genetic elements are: (1) to maintain a copy number of ~60 copies per cell by a DNA replication and recombination-based mechanism, and; (2) to transmit 2-micron circles to daughter cells during cell division [6–8]. 2-micron circles have been shown to be lost from a population at a very low rate (10−4–10−5 per cell per generation in the LL20 strain background [9]). Transmission of 2-micron circles to daughter cells is an active process that requires proteins encoded by REP1 and REP2, as well as host cell factors, and is thought to involve attachment of 2-micron DNA to specific nuclear subregions associated with chromosomes during cell division [7,10,11]. Disruption of normal inheritance of 2-micron circles has been linked to lethality in certain yeast mutants. Clonal lethality has been reported in nib1(upl1) and mlp1/mlp2 mutant strains, in which abnormal partitioning of 2-micron circles leads to an increase their copy number [12–15]. Overexpression of Flp1p causes increases in 2-micron copy number, cells size, and cell death in a cir+ strain, but not a cir0 strain [16]. One possibility is that clonal lethality in such mutants is the result of 2-micron circle accumulation in yeast mother cells due to mother cell bias. The 2-micron circle partitioning machinery in wild type cells may function to prevent accumulation of 2-micron circles to lethal levels by counteracting mother cell bias.
There are no specific growth phenotypes associated with the presence or absence of 2-micron circles [6,8]. However, some studies have shown that cir0 strains have a competitive growth advantage of 1–3% over otherwise isogenic cir+ strains [9,17]. This raises the possibility that a cir0 strain may have a competitive advantage in terms of its replicative longevity over a cir+ counterpart. On the other hand, because 2-micron circle copy number and transmission is carefully regulated, 2-micron circles may not accumulate in old cells or reduce yeast life span. Unfortunately, little is known about inheritance of 2-micron circles over the yeast life span. For example, we do not know if the 2-micron plasmid segregation system continues to operate with high fidelity as yeast cells age and undergo senescence. To address these unknowns, we have compared the life spans and 2-micron circle levels in otherwise isogenic cir+ and cir0 strains. To construct cir0 strains, we used a convenient method to cure yeast strains of 2-micron circle plasmids and describe this method herein.
2. Materials and methods
2.1. Yeast strains and plasmids
Saccharomyces cerevisiae strain W303AR5 (MATa-leu2–3,112 his3–11,15 ura3–1 ade2–1 trp1–1 can1–100 rDNA∷ADE2,RAD5, cir+; see [4]) was used for all experiments described below. Plasmids pJPA113, pJPA114, pJPA116, and pJPA117 have been described [1], and are based on the shuttle vector pRS306 [18], which carries the URA3 selectable marker. Yeast were transformed with plasmid DNA using a standard lithium acetate method [19]. Untransformed strains were grown in YPD medium [20]. Transformed strains were grown on selective SD “drop in” medium [20]. Strains yAF7 and yAF8 were isolated as follows. Freshly prepared pJPA114 transformants were colony-purified on selective SD agar medium, and streaked on non-selective SD medium. Individual colonies were streaked on non-selective SD medium containing 1 mg/ml 5-fluoroorotic acid (5-FOA) to select against the URA3 marker on pJPA114. 5-FOA resistant isolates were tested for 2-micron circle DNA as described below.
2.2. DNA isolation, Southern blotting, cell sorting, and replicative life span determinations
DNA was extracted from freshly saturated overnight liquid cultures of transformed or untransformed cells grown as indicated above. Old cells were obtained by magnetic sorting as described [21], and corresponding young cells were obtained during the same experiment. DNA was extracted from yeast using a glass beads/phenol method, digested with the restriction enzyme PstI (New England Biolabs), and fragments were separated on 1% agarose gels, stained with ethidium bromide, and capillary transferred to positively-charged nylon membranes under alkaline conditions using standard methods as described [22]. 32P-labeled probe was generated using 2-micron or URA3 DNA as template and a random-primed labeling kit (New England Biolabs). Southern data were acquired with a Typhoon 9400 PhosphorImager and analyzed using ImageQuant software (Amersham Biosciences). Replicative life span determinations were done by microdissection on YPD agar medium as described [1,21]. Bud scars on young and old cells were stained with calcofluor as described [23] and counted with a Zeiss Axiophot microscope using a total magnification of approximately 2000×.
3. Results
3.1. 2-micron circle inheritance is affected by ARS-plasmids
During the course of previously published experiments [1], we observed that yeast transformed with ARS-plasmids were cured of 2-micron plasmids with a higher than expected frequency (data not shown). The majority of laboratory yeast strains, including the W303-derived strains we use, contain the naturally occurring 2-micron plasmid. To investigate this preliminary result further, we tested if ARS-plasmids exert an effect on inheritance of the 2-micron plasmid. For this, W303AR5 was transformed with a series of ARS-and ARS/CEN-plasmids, and individual transformants were analyzed for presence or absence of 2-micron circle DNA by Southern blotting. As expected, fewer plasmid transformants were found to contain 2-micron DNA than untransformed controls (Fig. 1A). Transformation resulted in curing from a cir+ to a cir0 genotype in 15–20% of the transformants we analyzed (Fig. 1A and B). The W303 strain background does not exhibit a high rate of spontaneous loss of 2-micron circles. Many individual untransformed W303 isolates did not exhibit loss of 2-micron circles (Fig. 1A and B). The effect of ARS-plasmids on 2-micron inheritance was observed with plasmids carrying ARS1 as well as the rDNA ARS (Fig. 1), indicating that this effect is observed with plasmids having both strong (ARS1) and weak (rDNA ARS) replication origins. Transformation with an ARS1/CEN4-plasmid did not affect 2-micron inheritance whereas transformation with an rDNA ARS/CEN4 plasmid did (Fig. 1A and B). The fact that transformation with the ARS1/CEN4-plasmid did not promote curing to cir0 demonstrates that the transformation method per se does not promote 2-micron loss. The observation than the rDNA ARS/CEN4 plasmid cures transformants to cir0 indicates that the presence of CEN4 does not preclude an effect on 2-micron transmission. We note that we have studied the effects of ARS-plasmids on 2-micron loss in a W303 strain, and it is possible that different results may be observed in strains derived from other genetic backgrounds.
Fig. 1.
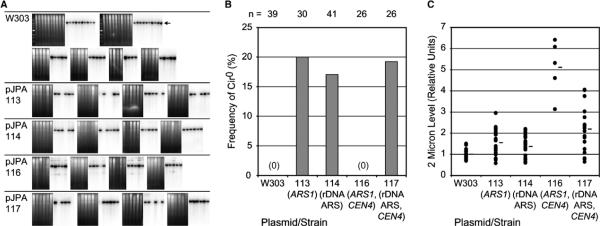
Loss of 2-micron circle DNA in ARS-plasmid transformants. (A) Southern blot analysis of untransformed W303AR5 (W303) and W303AR5 transformants bearing plasmids pJPA113, pJPA114, pJPA116, or pJPA117. DNAs from individual, colony-purified isolates were analyzed for the presence of 2-micron circle DNA. Companion ethidium bromide stained gels (on left) and Southern blots (on right) are shown. 2-micron circle DNA was linearized with PstI to yield a single 6.3 kb band (arrow). Long exposures of blots did not reveal faint 2-micron DNA bands (data not shown). (B) Summary of all Southern blot data (n, number of colonies analyzed). (C) Relative 2-micron DNA levels in untransformed and plasmid-transformed isolates. Southern blots were hybridized to URA3 probe to normalize 2-micron DNA levels to chromosomal DNA levels. In addition, relative levels of 2-micron DNA were normalized to the average level in the untransformed strain W303AR5. Units are arbitrary and are for comparison purposes only (i.e., units are not an accurate measure of 2-micron circle copy number per cell).
The results with the CEN4 plasmids pJPA116 (ARS1/CEN4) and pJPA117 (rDNA ARS/CEN4) appear contradictory and raise an interesting question: Why is pJPA116 associated with 2-micron loss whereas pJPA117 is not (Fig. 1A and B)? We have previously shown that pJPA116 and pJPA117 have similar per cell copy numbers: ~1.7 for pJPA116 and ~1.2 for pJPA117 [1]. However, the mitotic stabilities of pJPA116 and pJPA117 are different: ~88% for pJPA116 and ~34% for pJPA117 [1]. This suggests a correlation between low mitotic stability of plasmids and curing of 2-micron circles. Consistent with this, pJPA113 (ARS1) and pJPA114 (rDNA ARS) have mitotic stabilities of ~21% and ~1%, respectively [1]. Thus, CEN4 may suppress curing to cir0, but only in the context of a strong ARS, which supports a high mitotic stability.
There are a number of possible explanations for the reduced frequency of 2-micron circles in ARS-plasmid transformants. One possibility is that ARS-plasmids compete with 2-micron circle plasmids for replication factors, and thereby reduce 2-micron circle copy number and mitotic transmission. To test this, we analyzed the relative levels of 2-micron circle DNA in individual transformants using Southern blotting. In our analyses, the 2-micron circle DNA level in each sample was normalized to the amount of genomic DNA using the URA3 locus as an internal standard. This yields a relative measure of 2-micron DNA levels that is expressed in arbitrary units. In contrast to our expectations, the relative levels of 2-micron circle DNA were found to be elevated up to approximately sixfold in transformed cells compared to untransformed controls (Fig. 1C). In addition, a wide range of 2-micron DNA levels was observed in transformants compared to untransformed cells (Fig. 1C). 2-micron DNA levels in untransformed W303AR5 varied over a twofold range (i.e., from ~0.75–1.5), whereas pJPA117 (rDNA ARS/CEN4) transformants contained levels of 2-micron DNA that varied over a sixfold range (i.e., from ~0.65–4.1) (Fig. 1C). These findings reveal that ARS-plasmids influence 2-micron DNA levels, which is consistent with an effect of ARS-plasmids on 2-micron circle inheritance. However, increased levels of 2-micron circle DNA do not correlate with curing to cir0. pJPA116 (ARS1/CEN4) exhibits the highest levels of 2-micron circles, but is not associated with curing to cir0 (Fig. 1C). Also, the observation of increased 2-micron DNA levels and impaired inheritance in transformants is somewhat paradoxical. One possibility is that there is a reduced likelihood that ARS-plasmids and 2-micron circles are inherited together, and that progeny cells, most of which do not contain an ARS-plasmid, experience an increase in 2-micron circles. Although the mechanism remains unclear at this point, our interpretation is that ARS-plasmids interfere with the normal pathways that regulate the copy number and inheritance of 2-micron circles.
3.2. 2-micron circles do not accumulate in old cells and do not reduce life span
Realizing that ARS-plasmids could be used to conveniently cure strains of 2-micron circles, we wished to compare replicative life spans of cir+ and cir0 strains. For this, we transformed W303AR5 with pJPA114 and streaked transformants on minimal medium containing 5-fluoroorotic acid to select against the URA3 nutritional marker on pJPA114. Because pJPA114 has very a low mitotic stability (~1%, [1]), it is readily lost from transformants grown in the absence of selection. Two strains prepared in this manner, yAF7 and yAF8, were shown by Southern blotting not to possess 2-micron circle DNA (Fig. 2A). To determine if the presence of 2-micron plasmids influenced replicative life span, microdissection-based life span determinations were done as described [1,21]. The two cir0 strains yAF7 and yAF8 were compared to the parental strain W303AR5. No apparent differences in replicative life spans were observed (Fig. 2B). The three life span curves are indistinguishable by Wilcoxon two-sample paired signed rank tests (p < 0.04). The average replicative life spans for yAF7, yAF8, and W303AR5 were 22.7 (±6.2), 21.9 (±7.3), and 23.5 (±5.9) generations, respectively. This is comparable to life spans of W303-derived strains determined by us and others in the past [1,4]. Thus, the presence of 2-micron circles does not reduce replicative life span.
Fig. 2.
Comparison of otherwise isogenic cir+ and cir0 yeast strains. (A) Gel and Southern blot of DNA from parental cir+ (W303AR5) and cir0 isolates yAF7 and yAF8 probed to detect 2-micron DNA. (B) Life span analysis. Number of daughter cells (generations) produced per mother cell is plotted as a function of mother cell viability. The number of mother cells (n) analyzed for each curve equals 55, 55, and 56 for W303AR5, yAF7, and yAF8, respectively. (C) 2-micron plasmid levels in young and old cells. Young and old cells were isolated, and total cellular DNAs were analyzed by Southern blotting using 2-micron and URA3 probes (see Section 2). Average bud scar count equals mean age. Size markers (M) are shown (in kb). Arrowhead denotes 6.3 kb 2-micron circle DNA band.
We also determined the replicative life spans of W303AR5 and yAF8 transformed with the ARS-plasmid pJPA113 (data not shown). The purpose of this experiment was to address the possibility that reduction in life span by ARS-plasmids is dependent on 2-micron circles. The observation that ARS-plasmids increase 2-micron circle copy number raised the possibility that ARS-plasmids reduce replicative life span due to an increase in 2-micron circle copy number. Clonal lethality has been reported in strains carrying nib1 (upl1) and mlp1/mlp2 mutations, which disrupt normal partitioning of 2-micron circles and increase their copy number [12–15]. Although clonal lethality is not observed in ARS-plasmid transformants grown under standard lab conditions (e.g., colony formation on agar media), it is conceivable that the situation may be different in old cells. If ARS-plasmids cause 2-micron levels to continue to rise over the life span, this may be the basis of reduced life span in ARS-plasmid transformants. To address this possibility, we determined the replicative life spans of W303AR5 and yAF8 transformants carrying pJPA113. We found that the life spans were statistically indistinguishable (data not shown). Thus, reduction in replicative life span by ARS-plasmids, which we have reported previously [1], is not dependent on the presence of 2-micron circles.
The fact that 2-micron plasmids do not reduce replicative life span begs the question as to whether or not 2-micron circles accumulate during the aging process in yeast. Because 2-micron circles did not reduce life span, we predicted that 2-micron circles would not accumulate during replicative aging. To test this prediction, ~6-generation old yeast cells were prepared by magnetic sorting (see Section 2) and 2-micron circle DNA levels were analyzed by Southern blotting. Amounts of 2-micron DNA in young and old cell samples were normalized to the amounts of chromosomal DNA present, using the URA3 gene as an internal standard. This yields a relative measure of 2-micron DNA levels. We found that the levels of 2-micron DNA in young and old cells were very similar: 2-micron/URA3 ratios of 3.41 and 3.62 were obtained from populations with average ages of 1.1 and 6.2, respectively (Fig. 2C). The absolute values of these ratios are meaningless in terms of the copy number of 2-micron circles per genome. However, these ratios can be used as a basis of comparison between the young and old cell preparations. Importantly, the difference between normalized 2-micron DNA levels in young and old cells was very small. Our previous studies have documented substantial accumulations of ERCs (5- to 50-fold) and recombinant ARS-plasmids (5- to 25-fold) in 7-generation old yeast cells [1]. No similar accumulation was observed with 2-micron circle plasmids after six generations. We conclude that 2-micron plasmids are unlike ERCs and ARS-plasmids, and do not accumulate in old cells.
4. Discussion
2-micron circles are naturally occurring circular DNA episomes that exist at an average copy number of ~60 per cell, depending on the genetic background of the strain, and are partitioned during mitosis by an active process. For the first time to our knowledge, we show that 2-micron circles do not accumulate in old cells and do not reduce replicative life span. These findings agree with previous results of ours and others that link accumulation of episomal DNAs in mother cells with reduced life span [1,4]. Specifically, our findings support the conclusion that plasmids that are actively partitioned during mitosis, and contravene mother cell bias, do not accumulate to suffcient levels to adversely affect life span. Taken together with our studies of recombinant ARS-plasmids [1], these studies provide strong support for an association between plasmid accumulation due to mother cell bias and replicative senescence in yeast.
The fact that 2-micron circles do not adversely affect life span indicates that certain levels of DNA episomes are tolerated by yeast cells of different ages over the course of the life span. Furthermore, this suggests that higher levels of episomal DNAs are required to reduce life span in yeast. Consistent with this, we demonstrated in a previous report [1] that ARS-plasmids that reduce life span achieve per cell copy numbers in the range of 200–300 in several-generation old cells, which is significantly above levels known for 2-micron circles [6]. Perhaps yeast cells can support a per cell copy number of up to ~100 without incurring an adverse effect on life span. It is important to note that 2-micron circles are capable of reducing life span in certain mutant strains. Mutations that lead to elevated levels of 2-micron plasmids are known to cause clonal lethality. This is observed in nib1 (upl1) single, and mlp1/mlp2 double, mutants [12–15]. However, such 2-micron circle dependent lethality appears unrelated to reduction in life span by ARS-plasmids. Our results show that 2-micron circles are not required for reduction in life span by ARS-plasmids.
Our findings raise another interesting issue relating to plasmid accumulation and life span. We have previously shown that recombinant plasmids containing a 2-micron origin of replication accumulate in yeast mother cells and reduce replicative life span. The recombinant 2-micron origin plasmids that we have studied contain a 1346-bp region of 2-micron DNA that carries one copy of the following cis-acting elements: STB, ORI, FRT and a 599-bp repeat region extending across it [1]. This is typical for this class of yeast episomal plasmid (i.e., YEp) vectors. Endogenous 2-micron circle plasmids, however, contain two FRT sites within two inverted repeats, as well as four open reading frames. These differences between recombinant 2-micron origin-containing plasmids and 2-micron circles most likely account for differences in mitotic stability and effects on life span (i.e., 2-micron circles have higher mitotic stability and no effect on life span whereas recombinant 2-micron origin plasmids have a lower mitotic stability and reduce replicative life span). Beyond this, however, we cannot explain differences between 2-micron circles and recombinant 2-micron origin plasmids at a mechanistic level. Many aspects of the mechanisms that regulate copy number and segregation of 2-micron circles remain unknown [6–8]. Even less is known about how recombinant 2-micron origin-containing recombinant plasmids regulate copy number.
Finally, our studies reveal an effect of ARS-plasmids on the inheritance of 2-micron circles. ARS-plasmids interfere with normal inheritance of 2-micron circles, leading to curing of strains from a cir+ to a cir0 genotype. This effect is unexpected because these two types of yeast plasmids are thought to be transmitted to daughter cells via different mechanisms. At this point, we cannot explain this effect on 2-micron circle inheritance. Despite this, the effect of ARS-plasmids on 2-micron circle inheritance provides a simple-to-use method for curing a cir+ strain to cir0 status. Previously described methods for curing strains of 2-micron plasmids are more time-consuming and less convenient [24,25]. The facility of curing yeast strains of 2-micron circles has led to two important results that are relevant to our aging studies, as discussed above. This method is likely to be generally useful in studying aspects of 2-micron circle biology that require curing to the cir0 genotype. Furthermore, examination of potential “interactions” between ARS-plasmids and 2-micron circles may shed light on mechanisms of inheritance of these two episomes in yeast.
Acknowledgements
These studies were supported by the Ellison Medical Foundation and the National Institutes of Health (Grant AG-23719).
References
- [1].Falcón AA, Aris JP. Plasmid accumulation reduces life span in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:41607–41617. doi: 10.1074/jbc.M307025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- [4].Sinclair DA, Guarente L. Extrachromosomal rDNA circles – a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- [5].Murray AW, Szostak JW. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- [6].Broach JR, Volkert FC. Circular DNA plasmids of yeasts. In: Broach JR, Jones EW, Pringle JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 1. Cold Spring Harbor Press; 1991. pp. 297–331. [Google Scholar]
- [7].Jayaram M, Mehta S, Uzri D, Voziyanov Y, Velmurugan S. Site-specific recombination and partitioning systems in the stable high copy propagation of the 2-micron yeast plasmid. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:127–172. doi: 10.1016/S0079-6603(04)77004-X. [DOI] [PubMed] [Google Scholar]
- [8].Futcher AB. The 2 micron circle plasmid of Saccharomyces cerevisiae. Yeast. 1988;4:27–40. doi: 10.1002/yea.320040104. [DOI] [PubMed] [Google Scholar]
- [9].Futcher AB, Cox BS. Maintenance of the 2 μm circle plasmid in populations of Saccharomyces cerevisiae. J. Bacteriol. 1983;154:612–622. doi: 10.1128/jb.154.2.612-622.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Scott-Drew S, Wong CM, Murray JA. DNA plasmid transmission in yeast is associated with specific sub-nuclear localisation during cell division. Cell Biol. Int. 2002;26:393–405. doi: 10.1006/cbir.2002.0867. [DOI] [PubMed] [Google Scholar]
- [11].Mehta S, Yang XM, Chan CS, Dobson MJ, Jayaram M, Velmurugan S. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 2002;158:625–637. doi: 10.1083/jcb.200204136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Holm C. Clonal lethality caused by the yeast plasmid 2 mu DNA. Cell. 1982;29:585–594. doi: 10.1016/0092-8674(82)90174-x. [DOI] [PubMed] [Google Scholar]
- [13].Holm C. Sensitivity to the yeast plasmid 2mu DNA is conferred by the nuclear allele nibl. Mol. Cell. Biol. 1982;2:985–992. doi: 10.1128/mcb.2.8.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dobson MJ, Pickett AJ, Velmurugan S, Pinder JB, Barrett LA, Jayaram M, Chew JS. The 2 microm plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol. Cell. Biol. 2005;25:4299–4310. doi: 10.1128/MCB.25.10.4299-4310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhao X, Wu CY, Blobel G. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 2004;167:605–611. doi: 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reynolds AE, Murray AW, Szostak JW. Roles of the 2 microns gene products in stable maintenance of the 2 microns plasmid of Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mead DJ, Gardner DC, Oliver SG. The yeast 2 micron plasmid: strategies for the survival of a selfish DNA. Mol. Gen. Genet. 1986;205:417–421. doi: 10.1007/BF00338076. [DOI] [PubMed] [Google Scholar]
- [18].Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- [20].Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- [21].Park PU, McVey M, Guarente L. Separation of mother and daughter cells. Methods Enzymol. 2002;351:468–477. doi: 10.1016/s0076-6879(02)51865-6. [DOI] [PubMed] [Google Scholar]
- [22].Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Greene Publishing and Wiley-Interscience; New York; 2005. [Google Scholar]
- [23].Pringle JR. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- [24].Rose AB, Broach JR. Propagation and expression of cloned genes in yeast: 2-micron circle-based vectors. Methods Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
- [25].Tsalik EL, Gartenberg MR. Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast. 1998;14:847–852. doi: 10.1002/(SICI)1097-0061(19980630)14:9<847::AID-YEA285>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]



