Abstract
The quantitative analysis of complex biological samples has emerged as a key research area in the field of proteomics. Although quantitative proteomic experiments remain challenging, these strategies have been greatly facilitated by the development of newer high-performance mass spectrometers. In this work, we have evaluated the use of the LTQ-Orbitrap, a hybrid mass spectrometer in which a linear ion trap is coupled to an Orbitrap mass analyzer, for quantitative analyses. By analyzing a range of yeast protein standards, we found that the high mass accuracy, high resolution, large ion capacity, and large dynamic range of the LTQ-Orbitrap led to a significant improvement in the number and quality of the peptide ratio measurements compared to similar analyses done on the LTQ. We also successfully quantified protein expression differences that occur in metabolically labeled rat synapses during brain development to further demonstrate the suitability of the LTQ-Orbitrap for the comparative analysis of complex tissue samples.
Introduction
The analysis of complex biological mixtures using mass spectrometry has become a major tool for dissecting biological processes. A key component of this approach has been the development of quantitative methodologies that are capable of accurately measuring the abundances of the protein constituents of these samples 1, 2. These strategies rely primarily on the introduction of an isotopically labeled internal standard into the sample followed by mass spectrometric analysis to measure the relative abundances of the labeled and unlabeled peptide pair and have been successfully used for comparative proteomic analyses in a variety of different experimental systems. Despite the successes of these methods, quantitative proteomic studies remain technically challenging and are often limited by problems of dynamic range and the inability to accurately quantitate low signal-to-noise peptide measurements 3, 4.
The myriad of bioanalytical challenges associated with proteomics has been a major driving force in the development of new high performance instruments that are capable of meeting the needs of the scientific community. One such high performance instrument that has recently become commercially available is a hybrid tandem mass spectrometer called the LTQ-Orbitrap in which a linear ion trap (LTQ) is coupled to an Orbitrap mass analyzer via a C-shaped ion storage trap 5, 6. Early studies using this hybrid instrument have demonstrated a number of performance advantages including high mass accuracy (< 2ppm), high resolution (up to 100,000), large space charge capacity, and high dynamic range 7, 8. Although these features should make the LTQ-Orbitrap useful for a wide-range of biological applications, its direct application in different areas of proteomics such as quantitation and the identification of post-translational modifications has not yet been reported.
To assess the suitability of the LTQ-Orbitrap for the quantitative analysis of complex protein mixtures, samples containing different ratios of unlabeled and 15N-labeled proteins isolated from Saccharomyces cerevisiae were analyzed by data-dependent μLC/MS/MS on a ThermoElectron LTQ-Orbitrap. The relative abundances of each peptide identified in the mixture were determined using the Census software platform and the quality of the measurements derived from Orbitrap spectra were compared to measurements derived from LTQ spectra. The measurements from the Orbitrap dataset were generally more accurate and of higher quality compared to the LTQ measurements and convincingly demonstrated the performance advantages of the Orbitrap versus the LTQ for quantitative analyses. To further demonstrate the improved quantitative analysis capabilities of the Orbitrap, we also quantitatively analyzed changes in protein expression in 15N metabolically labeled rat-derived synaptosome samples during the course of rat brain development and successfully identified 149 synaptic proteins whose abundance changes during development.
Experimental
Materials
Angiotension I (Homo sapiens) and [Glu1]-fibrinopeptide B (Homo sapiens) were obtained from Sigma Chemical Co. (St. Louis, MO). Peptide stock solutions were prepared by dilution of standards with 3% formic acid (J.T.Baker) to a final concentration of 100 ρmol/μL.
Preparation of Samples
Metabolic 15N Labeling and Preparation of S.cerevisiae Samples
Yeast were grown and labeled using a previously published protocol 9, 10. Unlabeled and 15N-labeled yeast were mixed in known ratios (i.e., 1:1, 5:1, 10:1, 50:1 and 100:1) as determined by OD600/ml. Mixtures of yeast cells were collected by centrifugation at 1000g, after which yeast were lysed. Lysates were subjected to trichloroacetic acid precipitation followed by denaturation, reduction and alkylation. Proteins were then digested overnight at 37 °C with 5 μg of endonuclease Lys-C (Roche). After dilution to 2 M urea with 100 mM tris pH 8.5, 1 μg of modified trypsin (Promega) was added and the mixture was incubated over night at 37 °C. The resulting peptide mixture was acidified with formic acid (5% final).
Metabolic 15N Labeling of Rat brains
Sprague-Dawley rats were labeled with 15N as previously described 10. Briefly, a female rat was fed a 15N diet starting after weaning, remaining on the 15N diet through its pregnancy, and while weaning its pups. On postnatal day 45 (p45), the pups were subjected to halothane by inhalation until unresponsive and the brains were quickly removed. The brains were homogenized in 0.32M sucrose, 4mM HEPES, and protease inhibitors, and the homogenates were stored at −80°C. The 15N enrichment was determined to be 94% using a previously described protocol 11.
Isolation and Digestion of Synaptosomes
The brain homogenate from Sprague-Dawley rats at four postnatal development time points (p1, p10, p20, and p45,) were prepared in an identical manner as the 15N label brains. Each developmental brain homogenate was mixed in a 1:1 ratio with 15N labeled rat brain homogenate and synaptosomes were isolated as previously described12. The synaptosome enriched samples was digested with proteinase K as previously described 13.
Methods
Flow Injection Analysis
Flow injection was performed using a Surveyor HPLC pump (ThermoElectron) and an LTQ-Orbitrap hybrid mass spectrometer (ThermoElectron) with an ESI source. For all experiments, the pump was operated at a flow rate of 5 μL/min, and the switching valve was employed with a 2 μL sample injection loop. Samples were loaded manually into the injection loop using a full loop injection, and injected ~0.1 min into the analysis. MS scans (760–825 m/z) were acquired in both the LTQ-Orbitrap (R=60,000) and LTQ in alternating fashion. Instrumental parameters included a spray voltage of 4kV, a maximum injection time of 100 ms, LTQ AGC target for MS of 50,000 ions and an FTMS AGC target for MS of 1,000,000 ions.
Multidimensional Liquid Chromatography Tandem Mass Spectrometry of Peptide Mixtures (MudPIT)
This approach has been described in detail by several authors 14–16, so it will only briefly be detailed here. A three phase microcapillary column was constructed and equilibrated with 5% acetonitrile/0.1% formic acid for ~30 min before the peptide mixture was loaded. For analysis, the microcolumn was positioned in-line with an Eksigent nano-flow HPLC pump (Eksigent) directly in front of the heated capillary opening of an LTQ-Orbitrap hybrid mass spectrometer (ThermoElectron). Peptide mixtures were analyzed using a six step separation procedure. 3, 17, 18 As peptides were eluted and ionized into the mass spectrometer, acquisition of tandem mass spectra was repeated continuously during the course of the analysis. All tandem mass spectra were collected using a normalized collision energy of 35%, an isolation window of 3 m/z, and 1 μ scan. Data-dependent acquisition was employed and consisted of two full scan mass spectra (i.e., one in the Orbitrap, and one in the LTQ) followed by 4 tandem mass spectra in the LTQ. The master scan was collected in the Orbitrap with R=60,000, and a second MS scan was collected in the LTQ for comparison purposes. Other instrumental parameters included a spray voltage of 2.4kV, a maximum injection time of 100 ms, LTQ AGC target for MS/MS of 10,000 ions and an FTMS AGC target for MS of 500,000 ions.
Software
Interpretation of Tandem mass spectra
Both MS and tandem mass spectra were extracted from the Xcalibur data system format (.RAW) into MS1 and MS2 formats19 using in house software (RAW_Xtractor). Tandem mass spectra were interpreted by SEQUEST20, which was parallelized on a Beowulf cluster of ~150 computers21 and results were filtered, sorted, and displayed using the DTASelect2 program utilizing a decoy database strategy, that sets scoring threshold values based on a specific false positive rate (user supplied). Searches were performed against the yeast database from Saccharomyces Genome Database (SGD, released 03/03/05) for yeast samples or the rat International Protein Index (IPI, v3.05) database for the brain samples.
Quantitative Analysis using Census
After filtering the results from SEQUEST using DTASelect2, ion chromatograms were generated using an updated version of a program previously written in our lab 22. This software, called Census (manuscript in preparation) is composed of two parts: a quantitative analysis module that generates chromatograms from spectra and calculates peptide ion abundances, and a qualitative module that aids in tandem MS spectra visualization. Census has been developed in Java and is, therefore, operating system independent. It can be deployed on a personnel Windows desktop using a graphical user interface or onto a high performance server and run from the console making it accessible to multiple users. Census is available from the authors for individual use and evaluation through an Institutional Software Transfer Agreement (see http://fields.scripps.edu/Census for details).
Chromatogram Generation
First, the elemental compositions and corresponding isotopic distributions for both the unlabeled and labeled peptides were calculated and this information was then used to determine the appropriate m/z range from which to extract ion intensities, which included all isotopes with greater than 5% of the calculated isotope cluster base peak abundance. MS1 files were used to generate chromatograms from the m/z range surrounding both the unlabeled and labeled precursor peptides. Census can take advantage of high resolution, and high accuracy data (i.e., LTQ-Orbitrap) by accurately predicting both unlabeled and labeled m/z values based on a user-defined mass accuracy tolerance. Using this strategy, noisy peaks or co-eluting peptides can be easily excluded. The mass tolerance in high resolution quantitative mode can be defined in the Census configuration file.
The final step associated with the extraction process is generation of a preprocessed XML file containing the chromatograms, peak information, fragment ion data (for MS/MS quantification experiments) and other miscellaneous data for use in post-extraction analysis. After generating this XML file, the user can investigate the quantification results using the GUI post analysis pages. Upon opening a processed XML file, Census displays the list of quantified proteins in a tabular format. By double clicking on an individual protein in the table, peptide information will be displayed with chromatogram and score information.
Calculation of Peptide and Protein Ratios
Census calculates peptide ion intensity ratios for each pair of extracted ion chromatograms. The heart of the program is a linear least squares correlation that is used to calculate the ratio (i.e., slope of the line) and closeness of fit (i.e., correlation coefficient (r)) between the data points of the unlabeled and labeled ion chromatograms. Measured peptide ratios are then sorted by protein locus and outliers (see Outliers section) are thrown out. Protein ratios are then calculated from the mean of remaining peptide ratios. In the post-extraction analysis, users can visually examine chromatograms generated from peptide pairs, evaluate them based on multiple scores, and apply filters to generate final reports.
Filters and Outliers
Census allows users to filter peptide ratio measurements based on a correlation threshold because the correlation coefficient represents the quality of the correlation between the unlabeled and labeled chromatograms, and can be used to filter out poor quality measurements. In addition, Census provides an automated method for detecting and removing statistical outliers. In brief, standard deviations are calculated for all proteins using their respective peptide ratio measurements. The Grubbs test is then used with a user-defined p-value to remove outlier peptides. The outlier algorithm is used only when there are more than three peptides found in the same proteins, because the algorithm becomes unreliable for a small number of measurements.
Results and Discussion
Modern advances in mass spectrometry technology have resulted in high performance instrumentation capable of extremely high resolution and mass accuracy at a fraction of the cost compared to just a few years ago. In addition to the inherent benefits for peptide identification provided by these instruments, there are also notable benefits to be had for quantitative analysis. One such instrument that has recently been commercialized is the LTQ-Orbitrap hybrid mass spectrometer. As an initial evaluation, we examined a collection of unlabeled and metabolically 15N labeled yeast standards that were mixed in known ratios. These samples provided the basis for a well-controlled set of experiments aimed at probing the quantitative analysis capabilities of the LTQ-Orbitrap. To test the performance of the Orbitrap on challenging real world samples, we also analyzed several developmental stages of rat cerebellum development.
Evaluation of LTQ-Orbitrap Performance Using Yeast Standard Mixtures
Quantitative figures of merit
To evaluate the performance of the LTQ-Orbitrap for relative quantification, we analyzed a collection of unlabeled yeast protein mixtures that were mixed in known ratios (i.e., 1:1, 5:1, 10:1, 50:1 and 100:1) with metabolically 15N-labeled yeast. The 1:1, 5:1, and 10:1 mixtures were analyzed three times each as replicate analyses, while the 50:1, and 100:1 were analyzed only once. Each sample was analyzed using a 6-step MudPIT protocol with salt steps consisting of 10, 20, 30, 50, 70, and 100% 500mM ammonium acetate. After searching the acquired MS/MS spectra with SEQUEST, identifications were filtered with DTASelect2 at a false positive rate of 5%. The resulting peptide identifications were analyzed by Census where chromatograms were generated from both LTQ and Orbitrap MS scans (see methods) for comparison purposes.
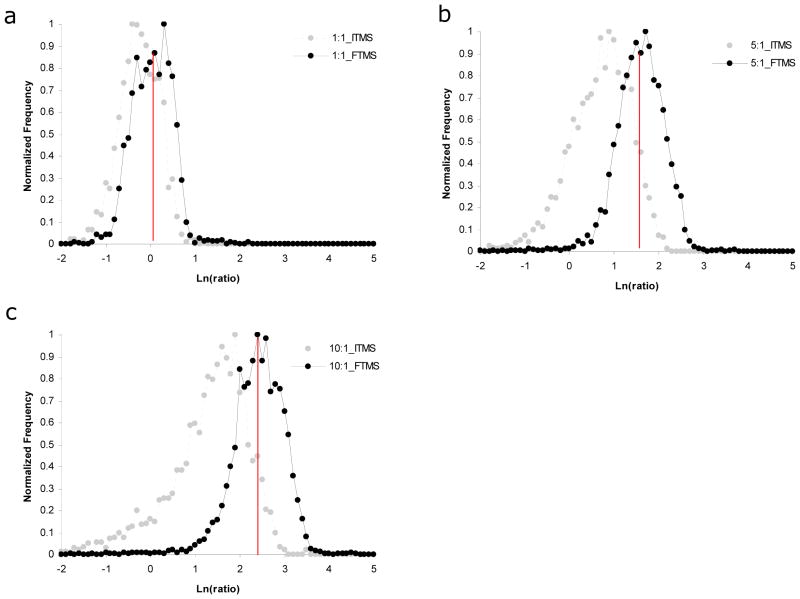
We first examined the accuracy of peptide ion abundance ratios since this is one of the most important criteria for quantitative analysis. In order to assess the entire collection of measurements and not focus on a few select measurements, we plotted the natural log of the measured ion abundance ratios as a series of histograms (Fig 2). Measured ratios were generally accurate for the 1:1 standard regardless of which MS detector was employed. However, peptide ion abundance ratios determined from LTQ MS scans for the other standards (i.e., 5:1, and 10:1) were systematically underestimated resulting in wide distributions with a significant skew. For example, average peptide ion abundance ratios determined from LTQ scans were 0.8, 2.31, and 4.76 for the 1:1, 5:1, and 10:1 standards respectively. Similar results have been published previously 4, 23. In contrast, peptide ion abundance ratios determined from Orbitrap MS scans were generally accurate within these same standard mixtures (i.e., average ratios were 1.07, 5.30, and 12.27 for the 1:1, 5:1, and 10:1 standards respectively). Contrary to other published reports, we had little success quantifying ion abundance differences of 50 fold and higher on the LTQ. However, results were better on the Orbitrap as we were able to quantify (within a factor of two) ~200 peptides within both the 50:1 and 100:1 standard mixtures (data not shown).
Fig. 2.
Accuracy of peptide ion abundance ratios for a collection of yeast standard mixtures that were mixed in known ratios with metabolically 15N-labeled yeast. Histograms were generated showing the distribution of measured ion abundance ratios for (a) 1:1, (b) 5:1, and (c) 10:1 standards. Measurements derived from LTQ MS scans are shown in grey and those derived from Orbitrap MS scans are shown in black.
From these data, it is clear that the Orbitrap provides a larger useful quantitative range for high throughput proteomic analyses compared to an LTQ. There are several potential reasons for the increased performance including higher signal to noise in Orbitrap MS scans. The Orbitrap analyzer is much further from the source of chemical noise then the LTQ and chemical background must pass through two high pressure trapping devices to be detected in the Orbitrap. Additionally, the ion image current detection strategy employed by the Orbitrap inefficiently detects the heterogeneous clusters of solvent ions common to chemical noise 24. Furthermore, the Orbitrap has a higher tolerance for space charge compared to the LTQ, thus it can be operated at much higher ion capacities, which in turn, provides an increased dynamic range. This is especially important for full MS scans where co-eluting peptides or other high abundant ions can effectively take the place of ions of interest.
Dynamic Range
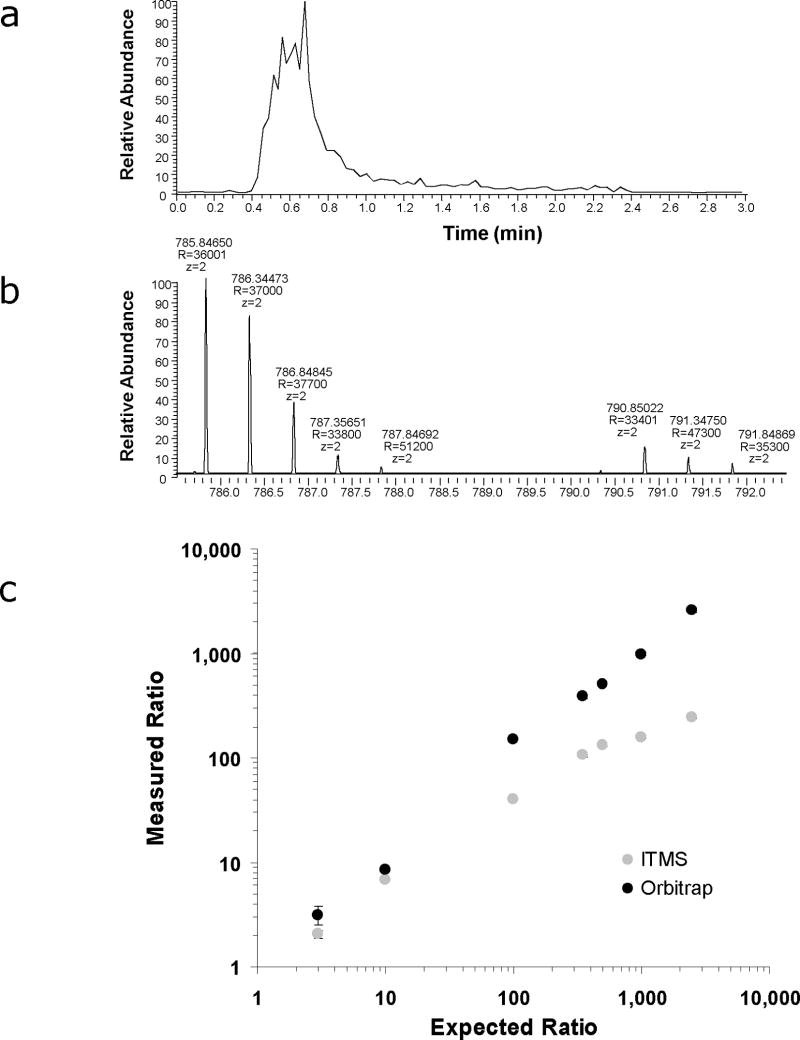
The intra-spectral dynamic range is also an important criterion for evaluating instrumentation to be used in relative quantification experiments because it ultimately defines the largest differences that can be accurately measured. To determine the intra-spectral dynamic range of the Orbitrap, we analyzed a collection of glu-fibrinopeptide B standards that were mixed in known ratios (i.e., 3:1, 10:1, 100:1, 350:1, 500:1, 1000:1, 2500:1) with an isotopically enriched version of the same peptide that contained 13C15N arginine. Each sample was analyzed in triplicate by flow injection and the resulting chromatograms for both the “light” and “heavy” peptides were evaluated using Microsoft Excel by summing the intensities of all signals within the isotope distributions over the time range from 0.3–1.2 minutes (Fig 3A and 3B). The measured ratios were then plotted as a function of the expected ratios (Fig 3C). From this analysis, it is apparent that the intra-spectral dynamic range of the Orbitrap is at least 2,500 and possibly greater although significant underestimation of the measured ratio was observed for a 10,000:1 mixture (data not shown). As expected, the LTQ’s intra-spectral dynamic range was less than that of the Orbitrap and measured ion abundance ratios tended to be underestimated in mixtures with >10 fold difference in abundances of the “light” and “heavy” peptides. These differences in dynamic range can most likely be attributed to the fact that the Orbitrap is much more tolerant of space charge than the LTQ and therefore can be operated at a high level of performance while trapping ~20X more ions.
Fig. 3.
Comparsion of intra-spectral dynamic range between MS scans acquired in LTQ and LTQ-Orbitrap mass spectrometers. Flow injection analysis was used to evaluate the relative abundances of glu-fibrinopeptide B and an isotopically labeled version of the same peptide. (a) Ion chromatograms for both the “light” and “heavy” versions of glu-fibrinopeptide B were constructed from the sum of (b) all ions within their respective isotope distributions. A correlation plot (c) between the measured and expected ion abundance ratios was then generated to compare the dynamic range between the two instruments.
Impact of high mass accuracy on accuracy and precision
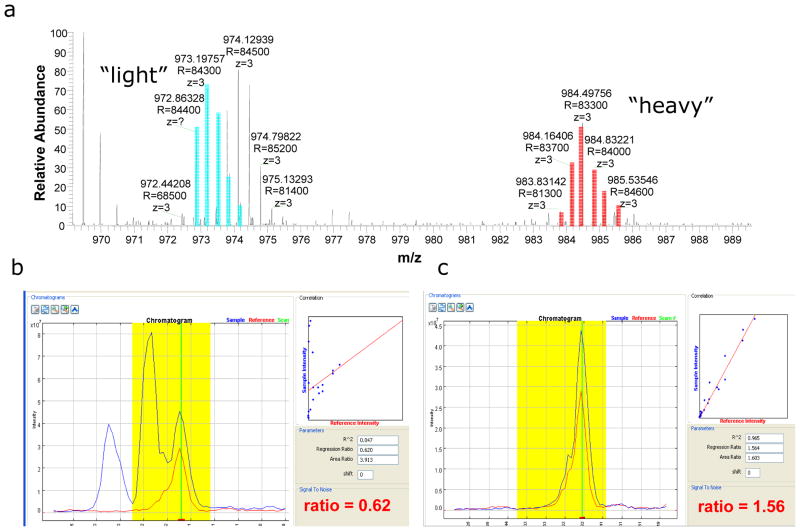
In the course of our evaluation and from comparison with previous experiments, it was evident that the high resolution and high mass accuracy capabilities of the LTQ-Orbitrap resulted in improved signal to noise chromatograms and ultimately a more straightforward and accurate quantification. To illustrate, Figure 4 shows an MS scan as well as a comparison of two pairs of chromatograms generated from Census for one of the peptides identified in our database search from the 1:1 yeast mixture. The first pair of chromatograms (Fig. 4B) was generated using a mass accuracy tolerance (i.e., maximum difference between theoretical and experimental m/z) of 250 ppm. The correlation coefficient is low and there appear to be three peaks in the chromatogram which are most likely derived from an overlapping peptide distribution with the “light” peptide (Fig. 4A). The resulting estimation of the ion abundance ratio is, therefore, inaccurate. The second pair of chromatograms (Fig. 4C) was generated using a mass accuracy tolerance of 5 ppm. In contrast to the previous example, the correlation coefficient is high and the chromatograms are simple and track each other quite well. In addition, the measured ion abundance ratio is ~1.5 which is more in accordance with the ion abundances in Fig. 5a.
Fig.4.
Illustration of the impact the high resolution and high mass accuracy capabilities of the LTQ-Orbitrap have on the quantification process. During our database search, a peptide with sequence KTGVIVGEDVHNLFTYAK was identified and subsequently quantified using full MS scans (a) acquired in the LTQ-Orbitrap. Ions derived from the “light” version of the peptide are highlighted in blue while those derived from the “heavy” version of the peptide are highlighted in red. When a mass accuracy tolerance of (b) 250ppm was employed, the resulting chromatograms contained multiple peaks and were inaccurate. In contrast, when a mass accuracy tolerance of (c) 50ppm was employed, chromatograms were simplistic and quantification was straightforward and accurate.
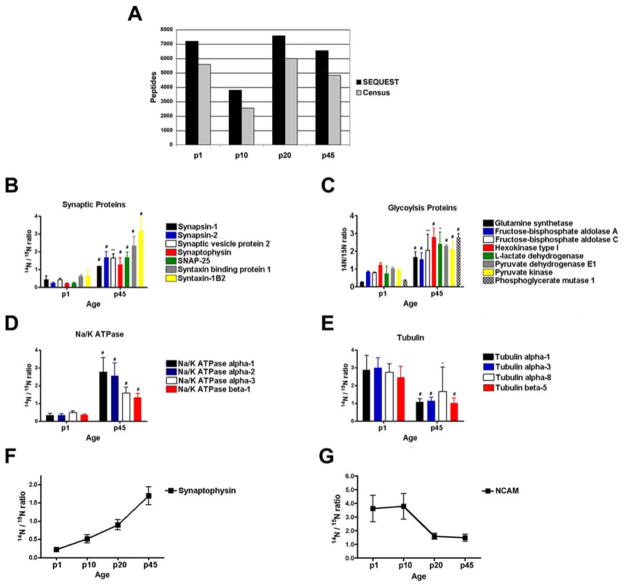
Fig.5.
Quantification of protein expression in a complex tissue using the LTQ-Orbitrap. One 12-step MudPIT was performed on synaptosomes isolated from rat brain at four different developmental periods (p1, p10, p20, and p45). Using Census (a), we were able to quantify an average of 74% of the peptides identified by SEQUEST. Consistent with the literature, we able to identify statistically significant changes between p1 and p45 brains in the 14N/15N ratios of synaptic proteins (b), ATP generating proteins (c), ATP consuming proteins (d), and proteins necessary for neurite outgrowth (e). In addition, we were able to identify statistically significant linear trends of proteins expression during the four developmental periods. The 14N/15N ratio of synaptophysin (slope = 0.17, p < .0001) increases with age, while NCAM (slope = −0.43, p < .0001) decreases with age
In addition to improvements in accuracy, we also noticed a substantial reduction in the errors associated with multiple peptide measurements derived from the same protein. For example Table 1 shows a representative protein that was identified and quantified from the 1:1 standard. Using a mass accuracy tolerance of 50 ppm, the percent relative standard deviation for this collection of 25 peptides was ~9.5%. In contrast, employing a mass accuracy tolerance of 5000 ppm resulted in a percent relative standard deviation of ~28.7%. In addition, the average percent relative standard deviations derived from all of the protein ratios determined in the sample followed the same trend.
Table 1.
Effect of different mass accuracy tolerances on measured ion abundance ratios for 25 peptides derived from the same protein.
| YKL060C FBA1 | ||||
|---|---|---|---|---|
| Peptides | Ratio (50ppm) | r2 | Ratio (5,000ppm) | r2 |
| R.KTGVIVGEDVHNLFTYAK.E | 0.988 | 0.962 | 0.463 | 0.956 |
|
| ||||
| K.TGVIVGEDVHNLFTYAK.E | 0.921 | 0.968 | 0.675 | 0.943 |
|
| ||||
| K.SPIILQTSNGGAAYFAGK.G | 1.154 | 0.99 | 0.598 | 0.874 |
|
| ||||
| K.GISNEGQNASIK.G | 0.884 | 0.946 | 1.29 | 0.875 |
|
| ||||
| K.GAIAAAHYIR.S | 0.848 | 0.968 | 0.8 | 0.962 |
|
| ||||
| R.SIAPAYGIPVVLH.S | 1.111 | 0.944 | 1.365 | 0.913 |
|
| ||||
| R.SIAPAYGIPVVLHSDH.C | 0.985 | 0.942 | 0.937 | 0.95 |
|
| ||||
| R.SIAPAYGIPVVLHSDHCA.K | 0.962 | 0.929 | 0.474 | 0.885 |
|
| ||||
| K.KLLPWFDGMLEADEAYFK.E | 1.041 | 0.989 | 0.867 | 0.986 |
|
| ||||
| K.LLPWFDGMLEADEAYFK.E | 0.889 | 0.973 | 1.147 | 0.987 |
|
| ||||
| K.EHGEPLFSSH.M | 0.877 | 0.972 | 0.611 | 0.838 |
|
| ||||
| K.EHGEPLFSSHMLDLSEETDEENISTCVK.Y | 1.068 | 0.966 | 0.787 | 0.917 |
|
| ||||
| H.GLYAGDIALRPEILAEHQK.Y | 0.869 | 0.973 | 0.611 | 0.831 |
|
| ||||
| L.YAGDIALRPEILAEHQK.Y | 1.051 | 0.985 | 0.826 | 0.835 |
|
| ||||
| R.EQVGCKEEKPLFLVFHGG.S | 0.994 | 0.968 | 1.114 | 0.887 |
|
| ||||
| R.EQVGCKEEKPLFLVFHGGS.G | 1.002 | 0.972 | 0.476 | 0.813 |
|
| ||||
| R.EQVGCKEEKPLFLVFHGGSG.S | 1.257 | 0.952 | 1.092 | 0.979 |
|
| ||||
| R.EQVGCKEEKPLFLVFHGGSGSTVQEFHTGIDNGVK.V | 0.943 | 0.986 | 0.853 | 0.981 |
|
| ||||
| K.EEKPLFLVFHGGSGSTVQEFHTGIDNGVVK.V | 1.031 | 0.985 | 1.29 | 0.961 |
| V.KVNLDTDCQYAYLTGIRDYVLNK.K | 0.901 | 0.947 | 0.793 | 0.963 |
| K.VNLDTDCQYAYLTGIR.D | 0.935 | 0.972 | 0.779 | 0.848 |
| K.VNLDTDCQYAYLTGIRDYVLNKK.D | 0.947 | 0.971 | 0.642 | 0.935 |
| K.KDYIMSPVGNPEGPEKPNK.K | 0.948 | 0.973 | 1.16 | 0.97 |
| K.DYIMSPVGNPEGPEKPNK.K | 0.974 | 0.973 | 1.082 | 0.982 |
| K.DYIMSPVGNPEGPEKPNKK.F | 0.927 | 0.972 | 1.383 | 0.978 |
|
| ||||
| Average | 0.980 | 0.885 | ||
| % r.s.d. | 0.095 | 0.287 | ||
| Average% r.s.d. for whole dataset | 0.152 | 0.368 | ||
Quantification efficiency
A rarely discussed, but extremely important criterion for assessing the efficacy of mass spectral quantification strategies is the percentage of identified peptides that can be successfully quantified. Obviously, this assessment is heavily dependent on what constitutes “successful” quantification, and thus, should be employed using strict guidelines. For our evaluation, we employed a minimum determination factor (i.e., r2) of 0.5 to address noisy chromatograms and a maximum deviation from the expected ratio of 2 fold to filter inaccurate measurements. Table 2 shows a comparison of quantification efficiencies between the LTQ-Orbitrap and LTQ obtained from the analysis of our yeast dataset. For every sample, the quantification efficiency was higher when Orbitrap MS scans were employed than when LTQ MS scans were used. The quantification efficiencies of protein standards mixed in ratios equal to or larger than five showed the most significant differences where the percentage of peptides accurately quantified by LTQ MS scans fell to below 11%. In contrast, the percentage of peptides accurately quantified by Orbitrap MS scans was consistently ~4–5 times higher.
Table 2.
Comparison of quantification efficiencies between LTQ and LTQ-Orbitrap for 11 different 6-step MudPIT experiments.
| Sample | # Spectra Identified | LTQ # Spectra Quantified * | LTQ Quantification Efficiency | Orbitrap # Spectra Quantified* | Orbitrap Quantification Efficiency |
|---|---|---|---|---|---|
| 1:1 | 528 | 266 | 0.50 | 361 | 0.68 |
| 1:1 | 1267 | 595 | 0.47 | 741 | 0.58 |
| 1:1 | 1637 | 832 | 0.51 | 1336 | 0.82 |
| 5:1 | 3099 | 328 | 0.11 | 1662 | 0.54 |
| 5:1 | 1121 | 127 | 0.11 | 591 | 0.53 |
| 5:1 | 2885 | 464 | 0.16 | 1475 | 0.51 |
| 10:1 | 4019 | 378 | 0.09 | 1856 | 0.46 |
| 10:1 | 3900 | 350 | 0.09 | 1683 | 0.43 |
| 10:1 | 2674 | 180 | 0.07 | 839 | 0.31 |
| 50:1 | 2283 | 1 | 0.00 | 210 | 0.09 |
| 100:1 | 2767 | 4 | 0.00 | 224 | 0.08 |
criteria used for quantification include an r2 value > 0.5 and accuracy within 2 fold of the expected ratio. In addition, statistical outliers were removed using grubbs test with an outlier threshold p-value of 0.1.
Quantitative Analysis of Rat Brain Synapses During Development
To test the quantitative analysis capabilities of the LTQ-Orbitrap MS, we chose to examine protein expression in rat brain development, since the brain is arguably the most complex mammalian tissue consisting of many distinct cell types. In addition, the brain has the highest lipid content than any other tissue (excluding adipose) making it particularly challenging for mass spectrometry analysis 25. For this analysis, we analyzed a fraction that was enriched for synapses termed the “synaptosome” fraction. One 12-step MudPIT experiment was performed on each of the synaptosome fractions (i.e., p1, p10, p20, and p45) using the LTQ-Orbitrap. After searching the acquired MS/MS spectra with SEQUEST and filtering with DTASelect2 requiring a 5% false positive rate, we identified 1594 unique proteins from the combined analyses. Census then was employed to determine the 14N/15N ratios for these peptides and corresponding proteins (Fig 5A).
As with the previously described yeast lysates, we were able to successfully quantify a much larger percentage of the identified peptides than we have been able to in the past. The average quantification efficiency of the 4 independent runs was 74%. Similar analyses using an LTQ for quantification resulted in quantification efficiencies <50%. We next performed a two-tail Student’s t test on the 384 proteins that were quantified in both (i.e., the intersection of) the p1 and p45 samples because those time points would be expected to have the greatest difference. Statistical analysis revealed that the expression of 201 proteins were different (p value < 0.05) between the p1 and p45 rat brains. We examined our significant differences for changes described in the literature during brain development. All the quantified resident synaptic proteins (i.e. only localized to the synapse) had larger 14N/15N ratios in the p45 rat brain compared to p1 (Fig 5B), which is consistent with the well documented burst in synaptogenesis between p1 and p45 26. The adult brain requires more energy to support this increase in synaptic activity. Correspondingly, many reports describe an increase in the expression of proteins that generate ATP and proteins that utilize ATP in an adult brain compared to a newborn brain27, 28. Our analysis identified an increase in the expression of proteins directly involved in the generation of ATP (glycolysis) and Na/K ATPase proteins, which are the main consumers of ATP in the brain (Fig 5C and 5D). We also identified proteins whose expression was higher in the p1 brain compared to the p45 brain. For example, neural cell adhesion molecule (NCAM), which is essential for neurite outgrowth, was increased in the p1 brain consistent with greater neurite outgrowth in early postnatal brains compared to adult brains 29. Tubulin is a major constituent of nerve processes, and neurite outgrowth is dependent on the formation of microtubules30. Increased expression of tubulin has been demonstrated in many species, including rat, to correlate with the extent of neurite outgrowth 31–33. Our data demonstrates that the expression of many different tubulin proteins were increased at p1 compared to p45 (Fig 5E).
Next, we sought to determine if we could detect more subtle biological changes by comparing protein expression in all four developmental timepoints. Out of the 362 proteins common in all 4 developmental timepoints, we were able to quantify 324 proteins using Census. To determine if there was a significant difference (p < .05) in the protein expression between the 4 samples, we first performed one-way ANOVA analysis. If a significant difference was detected, we performed an additional statistical test to determine if there was a significant linear trend (p < 0.05) between the samples. From this analysis, we identified 82 proteins that were up-regulated (i.e., positive slope) and 67 proteins that were down-regulated (i.e., negative slope). For example, protein expression of synaptophysin was identified as being up-regulated while the protein expression of NCAM was down-regulated during rat brain development (Figures 6F and G).
Besides describing the power of the LTQ-Orbitrap mass spectrometer to generate accurate and precise quantitative data using stable labeled isotope peptides, we also describe an important tool to study neurodevelopment. The biological changes that occur during development to form the complex synaptic networks in the adult brain are still not well understood. Many neurological diseases, including autism, fragile-X syndrome, Rett syndrome, and schizophrenia, have been hypothesized to manifest from perturbations during neurodevelopment34–37. However, such perturbations during neurodevelopment cannot be readily identified until there is comprehensive description of this process. Thus, the ability of the LTQ-Orbitrap mass spectrometer to accurately quantify protein expression during neurodevelopment provides the potential to develop therapies for many neurological diseases.
Conclusions
The quantitative analysis of complex peptide mixtures using the LTQ-Orbitrap results in a dramatic increase in data quality compared to the LTQ alone. The improved performance of the LTQ-Orbitrap in these quantitative analyses can be attributed to its high mass accuracy, resolution, and dynamic range as well as its larger ion capacity. In order to fully take advantage of these features for quantitation, we have also developed the Census algorithm which is capable of calculating peptide and protein abundances from high resolution LTQ-Orbitrap data in a highly automated fashion. The combination of the LTQ-Orbitrap and Census provides a powerful platform for quantitative proteomic studies. We demonstrate the effectiveness of this platform by measuring protein expression in rat synapse-enriched protein samples collected at different time points in rat brain development. In total, we identified 82 and 67 proteins that were either up-regulated or down-regulated, respectively, during development. A detailed understanding of how changes in protein expression promote normal brain development will be critical in deciphering how the abnormal brain development that is typical in a number of different pathological processes is linked to aberrant protein expression patterns.
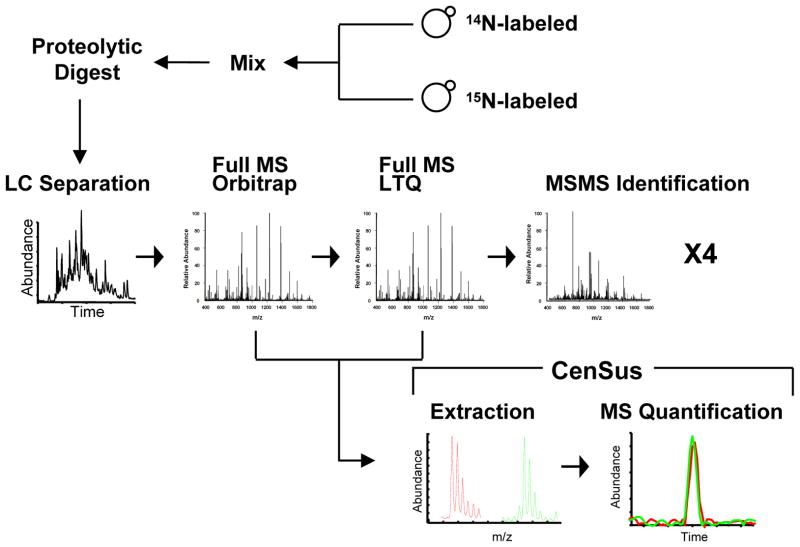
Fig.1.
Strategy for quantitative proteomic analysis of 14N and 15N-labeled yeast protein lysates using LTQ-Orbitrap and Census.
Acknowledgments
The authors would like to thank other members of the Yates laboratory for their comments and discussions. J.D.V. is supported by a National Research Service Award (NIH) fellowship. J.A.W. is supported by a postdoctoral fellowship from the American Cancer Society. J.R.Y. is supported by NIH Grant No. P41RR11823-10 and 5R01 MH067880-02.
References
- 1.MacCoss MJ, Matthews DE. Analytical chemistry. 2005;77:294A–302A. doi: 10.1021/ac053431e. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, Mann M. Nature chemical biology. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 3.Ong S-e, et al. Molecular and Cellular Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 4.Venable JD, Dong M, Wohlschlegel J, III, JRY Nature Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 5.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. Journal of mass spectrometry. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 6.Hardman M, Makarov AA. Analytical chemistry. 2003;75:1699–1705. doi: 10.1021/ac0258047. [DOI] [PubMed] [Google Scholar]
- 7.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Molecular & cellular proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Yates JR, Cociorva D, Liao L, Zabrouskov V. Analytical chemistry. 2006;78:493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 9.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Proc Natl Acad Sci. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu C, MacCoss M, III, JRY Anal Chem. 2004 In press. [Google Scholar]
- 11.MacCoss MJ, Wu CC, Matthews DE, Yates JR., 3rd Anal Chem. 2005;77:7646–7653. doi: 10.1021/ac0508393. [DOI] [PubMed] [Google Scholar]
- 12.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. The Journal of cell biology. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu CC, MacCoss MJ, Howell KE, Yates JR., 3rd Nature biotechnology. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 14.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., III Nature Biotechnology. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 15.McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR. International Journal of Mass Spectrometry. 2002;219:245–251. [Google Scholar]
- 16.MacCoss MJ, Wu CC, Yates JR., III Analytical Chemistry. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 17.McCormack AL, Schieltz DM, Goode B, Yang S, Barnes G, Drubin D, Yates JR., III Analytical Chemistry. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 18.Krijgsveld J. Nat Biotechnology. 2003 advanced online publication. [Google Scholar]
- 19.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, III, JRY Rapid Commun Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 20.Eng JK, McCormack AL, Yates JR., III Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 21.Sadygov RG, Eng J, Durr E, Saraf A, McDonald H, MacCoss MJ, Yates JR., III Journal of Proteome Research. 2002;1:211–215. doi: 10.1021/pr015514r. [DOI] [PubMed] [Google Scholar]
- 22.MacCoss MJ, Wu CC, III, JRY Analytical Chemistry. 2003;75:6912–6921. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- 23.Pan C, Kora G, McDonald WH, Tabb DL, VerBerkmoes NC, Hurst GB, Pelletier DA, Samatova NF, Hettich RL. Analytical Chemistry. 2006 doi: 10.1021/ac060654b. Web 09/13/2006. [DOI] [PubMed] [Google Scholar]
- 24.Yates JR, Cociorva D, Liao L, Zabrouskov V. Anal Chem. 2006;78:493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 25.Yehuda S, Rabinovitz S, Mostofsky DI. J Neurosci Res. 1999;56:565–570. doi: 10.1002/(SICI)1097-4547(19990615)56:6<565::AID-JNR2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 26.Aghajanian GK, Bloom FE. Brain Res. 1967;6:716–727. doi: 10.1016/0006-8993(67)90128-x. [DOI] [PubMed] [Google Scholar]
- 27.Pysh JJ. Brain Res. 1970;18:325–342. doi: 10.1016/0006-8993(70)90332-x. [DOI] [PubMed] [Google Scholar]
- 28.Erecinska M, Cherian S, Silver IA. Prog Neurobiol. 2004;73:397–445. doi: 10.1016/j.pneurobio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Ronn LC, Hartz BP, Bock E. Exp Gerontol. 1998;33:853–864. doi: 10.1016/s0531-5565(98)00040-0. [DOI] [PubMed] [Google Scholar]
- 30.Daniels MP. J Cell Biol. 1972;53:164–176. doi: 10.1083/jcb.53.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bond JF, Farmer SR. Mol Cell Biol. 1983;3:1333–1342. doi: 10.1128/mcb.3.8.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Havercroft JC, Cleveland DW. J Cell Biol. 1984;99:1927–1935. doi: 10.1083/jcb.99.6.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis SA, Lee MG, Cowan NJ. J Cell Biol. 1985;101:852–861. doi: 10.1083/jcb.101.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. J Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickett J, London E. J Neuropathol Exp Neurol. 2005;64:925–935. doi: 10.1097/01.jnen.0000186921.42592.6c. [DOI] [PubMed] [Google Scholar]
- 36.Segawa M, Nomura Y. Curr Opin Neurol. 2005;18:97–104. doi: 10.1097/01.wco.0000162848.99154.9a. [DOI] [PubMed] [Google Scholar]
- 37.Arnold SE, Talbot K, Hahn CG. Prog Brain Res. 2005;147:319–345. doi: 10.1016/S0079-6123(04)47023-X. [DOI] [PubMed] [Google Scholar]