Abstract
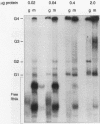
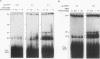
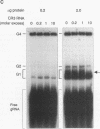
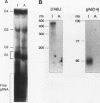
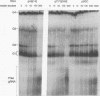
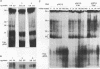
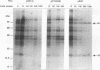
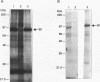
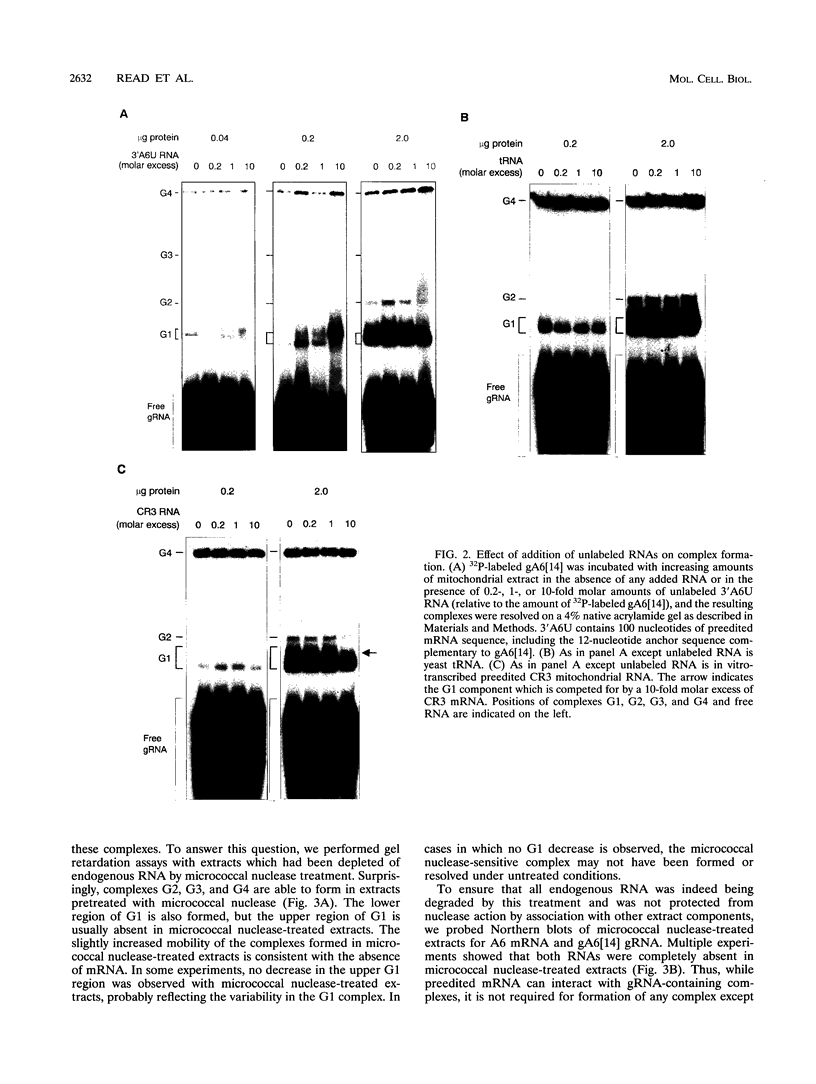
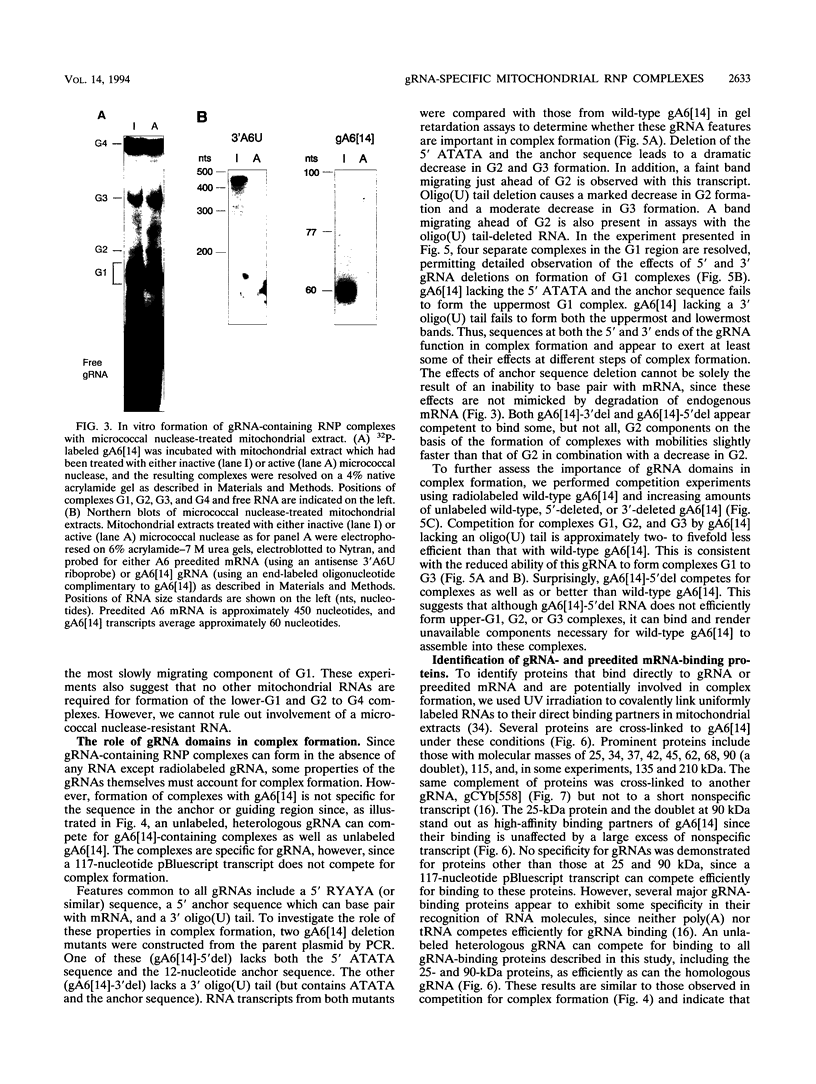
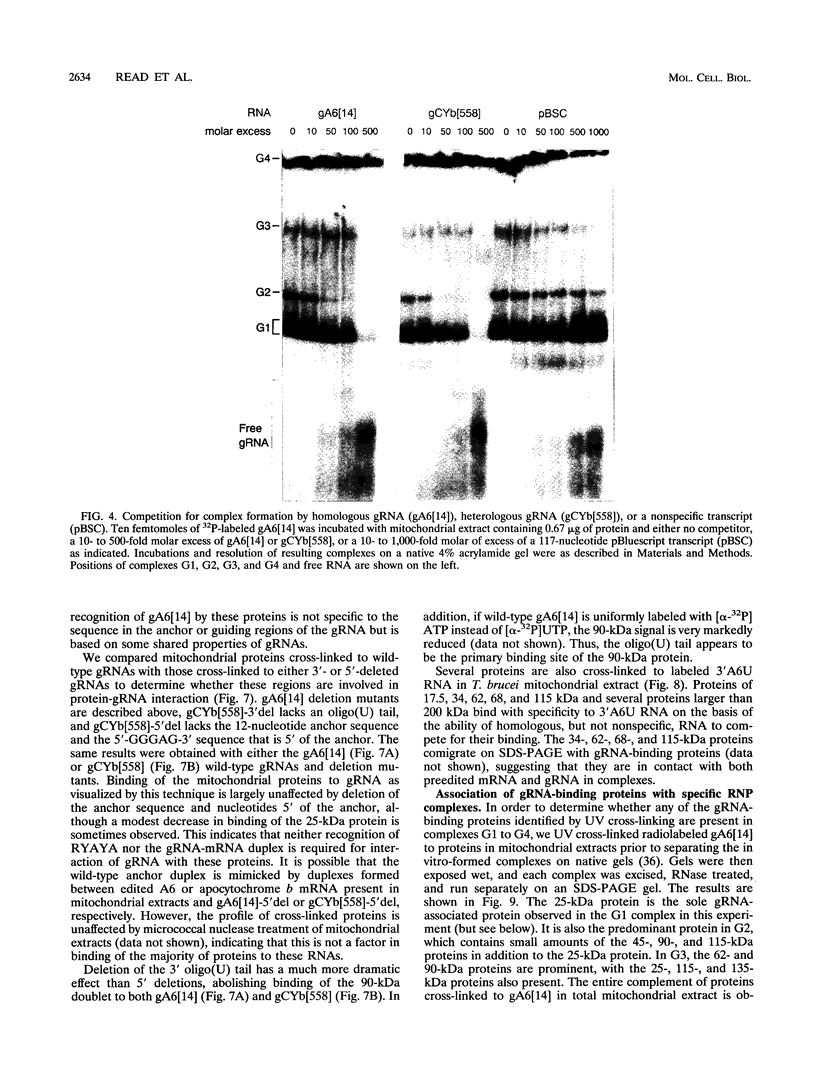
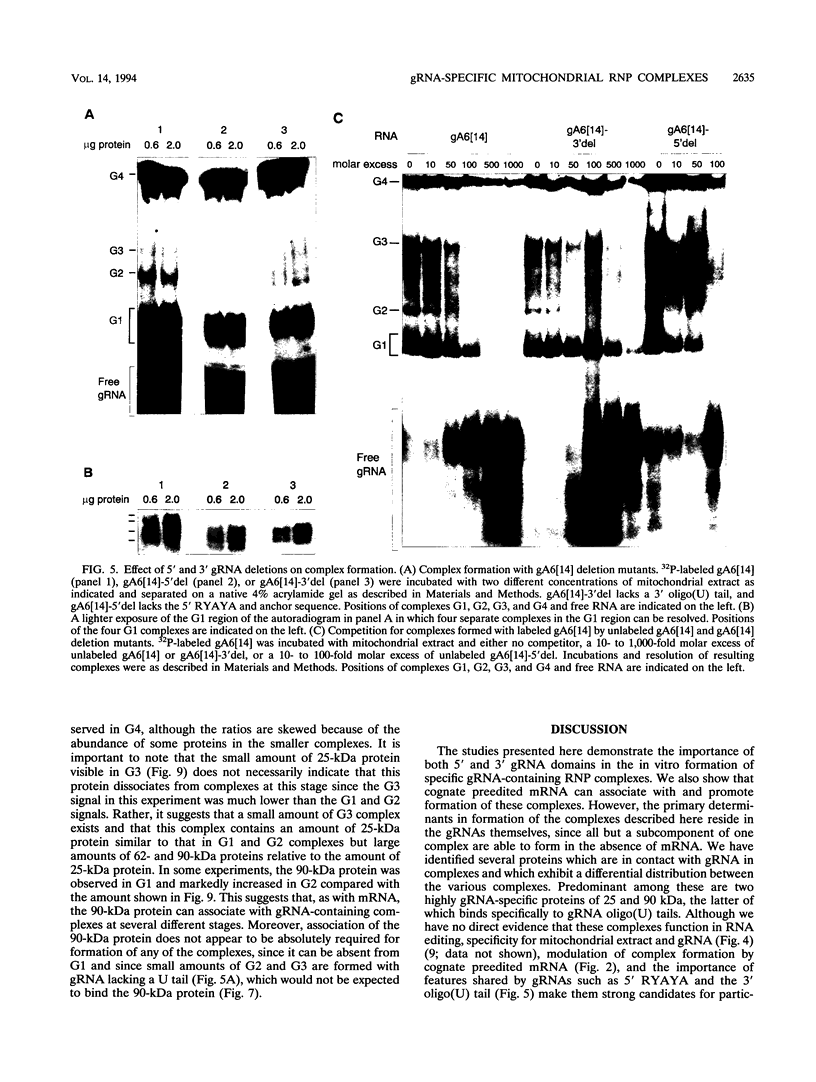
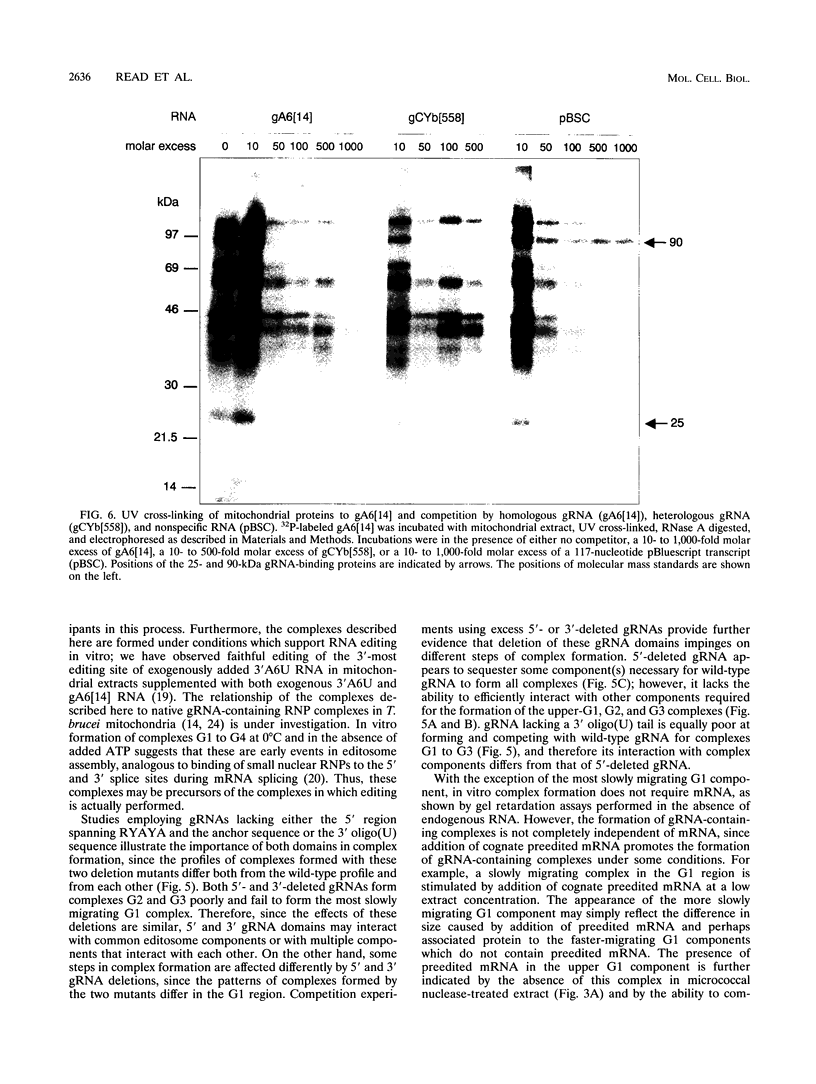
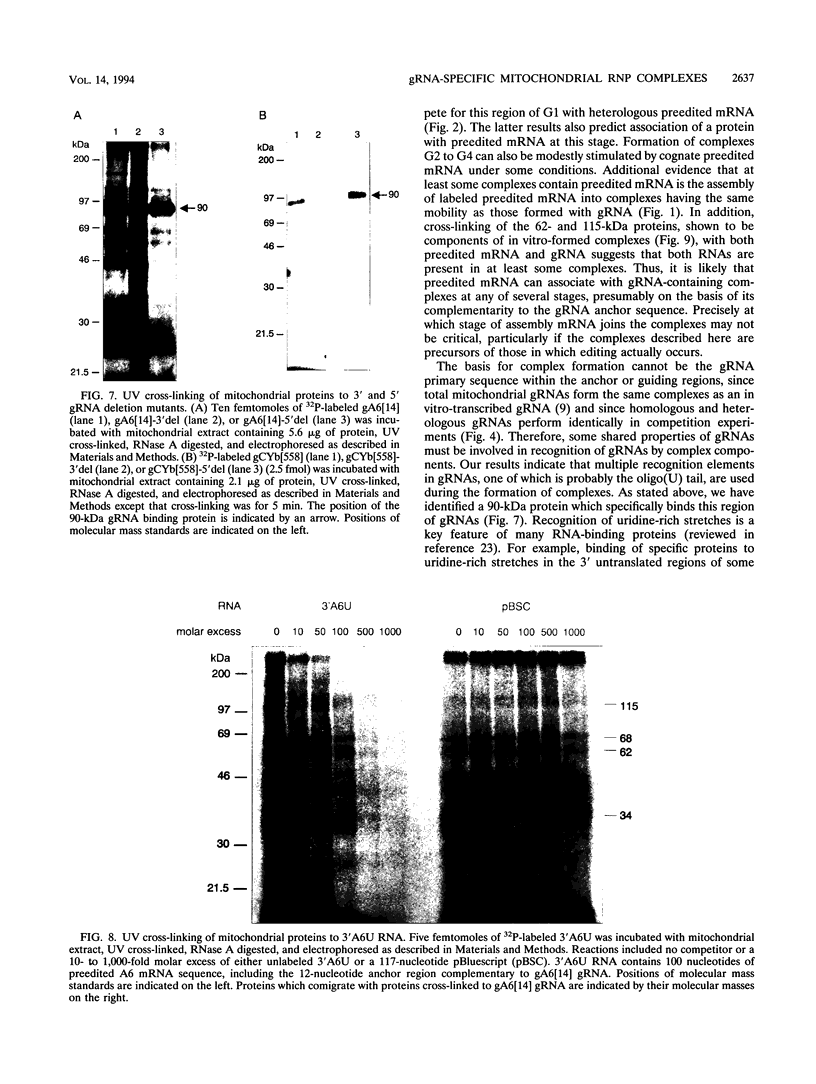
RNA editing in kinetoplastids probably employs a macromolecular complex, the editosome, that is likely to include the guide RNAs (gRNAs) which specify the edited sequence. Specific ribonucleoprotein (RNP) complexes which form in vitro with gRNAs (H. U. Göringer, D. J. Koslowsky, T. H. Morales, and K. D. Stuart, Proc. Natl. Acad. Sci. USA, in press) are potential editosomes or their precursors. We find that several factors are important for in vitro formation of these RNP complexes and identify specific gRNA-binding proteins present in the complexes. Preedited mRNA promotes the in vitro formation of the four major gRNA-containing RNP complexes under some conditions but is required for the formation of only a subcomponent of one complex. The 5' gRNA sequence encompassing the RYAYA and anchor regions and the 3' gRNA oligo(U) tail are both important in complex formation, since their deletion results in a dramatic decrease of some complexes and the absence of others. UV cross-linking experiments identify several proteins which are in contact with gRNA and preedited mRNA in mitochondrial extracts. Proteins of 25 and 90 kDa are highly specific for gRNAs, and the 90-kDa protein binds specifically to gRNA oligo(U) tails. The gRNA-binding proteins exhibit a differential distribution between the four in vitro-formed complexes. These experiments reveal several proteins potentially involved in RNA editing and indicate that multiple recognition elements in gRNAs are used for complex formation.
Full text
PDF


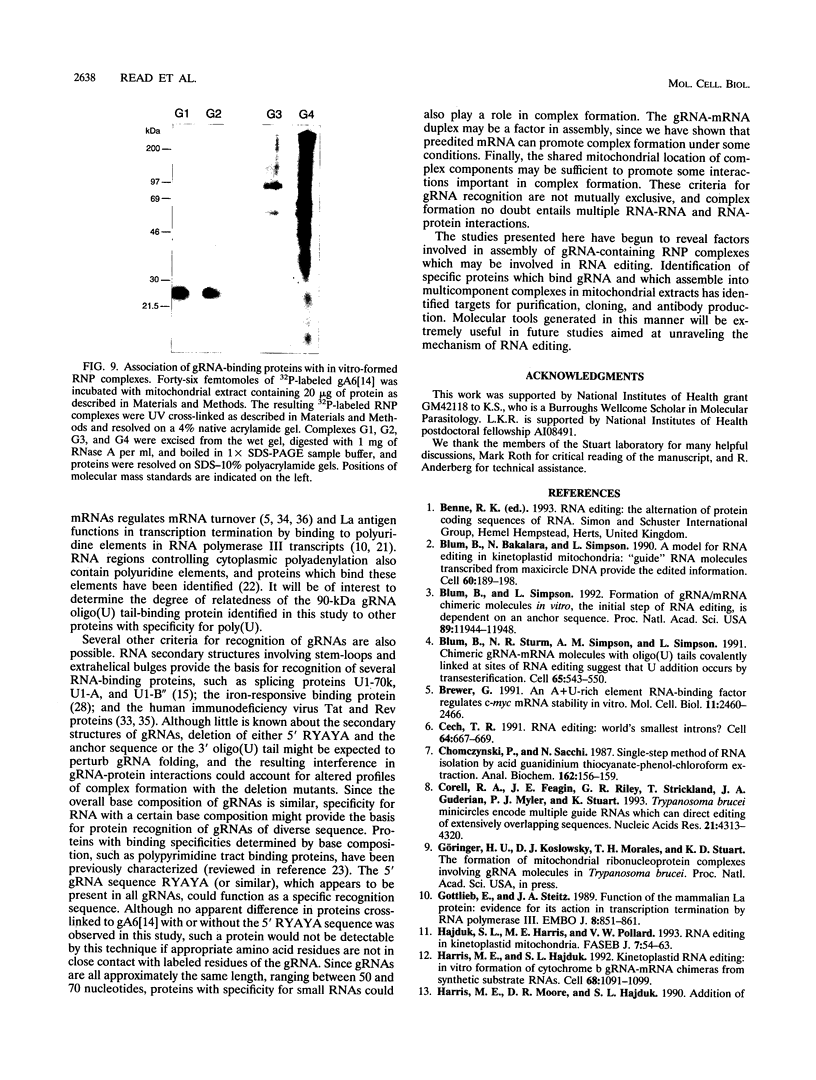

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blum B., Bakalara N., Simpson L. A model for RNA editing in kinetoplastid mitochondria: "guide" RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990 Jan 26;60(2):189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Blum B., Simpson L. Formation of guide RNA/messenger RNA chimeric molecules in vitro, the initial step of RNA editing, is dependent on an anchor sequence. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11944–11948. doi: 10.1073/pnas.89.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum B., Sturm N. R., Simpson A. M., Simpson L. Chimeric gRNA-mRNA molecules with oligo(U) tails covalently linked at sites of RNA editing suggest that U addition occurs by transesterification. Cell. 1991 May 17;65(4):543–550. doi: 10.1016/0092-8674(91)90087-f. [DOI] [PubMed] [Google Scholar]
- Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991 May;11(5):2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. RNA editing: world's smallest introns? Cell. 1991 Feb 22;64(4):667–669. doi: 10.1016/0092-8674(91)90494-j. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Corell R. A., Feagin J. E., Riley G. R., Strickland T., Guderian J. A., Myler P. J., Stuart K. Trypanosoma brucei minicircles encode multiple guide RNAs which can direct editing of extensively overlapping sequences. Nucleic Acids Res. 1993 Sep 11;21(18):4313–4320. doi: 10.1093/nar/21.18.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E., Steitz J. A. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J. 1989 Mar;8(3):851–861. doi: 10.1002/j.1460-2075.1989.tb03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk S. L., Harris M. E., Pollard V. W. RNA editing in kinetoplastid mitochondria. FASEB J. 1993 Jan;7(1):54–63. doi: 10.1096/fasebj.7.1.8422975. [DOI] [PubMed] [Google Scholar]
- Harris M. E., Hajduk S. L. Kinetoplastid RNA editing: in vitro formation of cytochrome b gRNA-mRNA chimeras from synthetic substrate RNAs. Cell. 1992 Mar 20;68(6):1091–1099. doi: 10.1016/0092-8674(92)90080-v. [DOI] [PubMed] [Google Scholar]
- Harris M. E., Moore D. R., Hajduk S. L. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J Biol Chem. 1990 Jul 5;265(19):11368–11376. [PubMed] [Google Scholar]
- Kenan D. J., Query C. C., Keene J. D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991 Jun;16(6):214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Koslowsky D. J., Göringer H. U., Morales T. H., Stuart K. In vitro guide RNA/mRNA chimaera formation in Trypanosoma brucei RNA editing. Nature. 1992 Apr 30;356(6372):807–809. doi: 10.1038/356807a0. [DOI] [PubMed] [Google Scholar]
- Koslowsky D. J., Riley G. R., Feagin J. E., Stuart K. Guide RNAs for transcripts with developmentally regulated RNA editing are present in both life cycle stages of Trypanosoma brucei. Mol Cell Biol. 1992 May;12(5):2043–2049. doi: 10.1128/mcb.12.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Mathews M. B., Francoeur A. M. La antigen recognizes and binds to the 3'-oligouridylate tail of a small RNA. Mol Cell Biol. 1984 Jun;4(6):1134–1140. doi: 10.1128/mcb.4.6.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew L. L., Richter J. D. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990 Nov;9(11):3743–3751. doi: 10.1002/j.1460-2075.1990.tb07587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. R., Kakegawa T., Kaspar R. L., White M. W. Polypyrimidine tracts and their binding proteins: regulatory sites for posttranscriptional modulation of gene expression. Biochemistry. 1993 Mar 30;32(12):2931–2937. doi: 10.1021/bi00063a001. [DOI] [PubMed] [Google Scholar]
- Pollard V. W., Harris M. E., Hajduk S. L. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 1992 Dec;11(12):4429–4438. doi: 10.1002/j.1460-2075.1992.tb05543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V. W., Rohrer S. P., Michelotti E. F., Hancock K., Hajduk S. L. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell. 1990 Nov 16;63(4):783–790. doi: 10.1016/0092-8674(90)90144-4. [DOI] [PubMed] [Google Scholar]
- Read L. K., Corell R. A., Stuart K. Chimeric and truncated RNAs in Trypanosoma brucei suggest transesterifications at non-consecutive sites during RNA editing. Nucleic Acids Res. 1992 May 11;20(9):2341–2347. doi: 10.1093/nar/20.9.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A., Tang C. K., Kaptain S., Burgess W. H., Haile D. J., Samaniego F., McBride O. W., Harford J. B., Klausner R. D. Cloning of the cDNA encoding an RNA regulatory protein--the human iron-responsive element-binding protein. Proc Natl Acad Sci U S A. 1990 Oct;87(20):7958–7962. doi: 10.1073/pnas.87.20.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K. D., Gelvin S. B. Localization of kinetoplast DNA maxicircle transcripts in bloodstream and procyclic form Trypanosoma brucei. Mol Cell Biol. 1982 Jul;2(7):845–852. doi: 10.1128/mcb.2.7.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K., Gobright E., Jenni L., Milhausen M., Thomashow L., Agabian N. The IsTaR 1 serodeme of Trypanosoma brucei: development of a new serodeme. J Parasitol. 1984 Oct;70(5):747–754. [PubMed] [Google Scholar]
- Stuart K. RNA editing in trypanosomatid mitochondria. Annu Rev Microbiol. 1991;45:327–344. doi: 10.1146/annurev.mi.45.100191.001551. [DOI] [PubMed] [Google Scholar]
- Stuart K. The RNA editing process in Trypanosoma brucei. Semin Cell Biol. 1993 Aug;4(4):251–260. doi: 10.1006/scel.1993.1030. [DOI] [PubMed] [Google Scholar]
- Tiley L. S., Malim M. H., Tewary H. K., Stockley P. G., Cullen B. R. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):758–762. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakalopoulou E., Schaack J., Shenk T. A 32-kilodalton protein binds to AU-rich domains in the 3' untranslated regions of rapidly degraded mRNAs. Mol Cell Biol. 1991 Jun;11(6):3355–3364. doi: 10.1128/mcb.11.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- You Y., Chen C. Y., Shyu A. B. U-rich sequence-binding proteins (URBPs) interacting with a 20-nucleotide U-rich sequence in the 3' untranslated region of c-fos mRNA may be involved in the first step of c-fos mRNA degradation. Mol Cell Biol. 1992 Jul;12(7):2931–2940. doi: 10.1128/mcb.12.7.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]