Abstract
A great portion of tyrosine kinases are involved in cell development and their structural alteration are intimately involved in associated pathologies of development and oncology. These kinases are one of major groups of targets under investigation for molecular therapeutics. To carry out biochemical and structural biological studies on these kinases, economical production of their purified forms is highly desirable. However over-expressing tyrosine kinases as recombinant forms in bacterial systems and their purification is a significant challenge. Abelson kinase (Abl) has previously been expressed on a large scale to facilitate X-ray crystallography and NMR structure studies mainly in baculovirus infected insect cells. Even though success has been achieved in expression of soluble tyrosine kinases in E. coli with chaperones to improve correct folding, low expression level of kinases are intrinsic in such systems because of diversion of resources to produce chaperones. Here we present a straightforward method to express and purify isolated Abl kinase domain, and SH3-SH2-kinase multi-domain structures. The expressed Abl protein retains its correct folding and biological function. The yield of soluble protein is in a several mg/L range in minimal media. Furthermore we demonstrate that segmental isotopic labelling using expressed protein ligation can be achieved using bacterial expressed Abl kinase domain constructs, which is especially useful in NMR structure/activity studies.
Introduction
Nonreceptor tyrosine kinase c-Abl is ubiquitously expressed and highly conserved in metazoan evolution1, 2. In its N-terminal half of the molecule c-Abl bears a 42% sequence identity to the Src tyrosine kinase family and shares a similar domain organization. It is well known that Bcr-Abl fusion protein has enhanced tyrosine kinase activity and consequently is the major cause of human chronic myeloid leukaemia (CML)3. CML is a hematopoietic stem cell disease whose underlying cause is generally thought to be a reciprocal translocation between the long arms of chromosomes 9 and 22. The chromosomal translocation results in the expression of Bcr-Abl, an aberrant fusion tyrosine kinase with elevated activity 4. The identification of the kinase hyperactivity as the underlying cause of the leukemia enabled the rational development of targeted therapy with tyrosine kinase inhibitors. Imatinib, an inhibitor with selectivity for Bcr-Abl was the first available targeted treatment for patients with newly diagnosed CML5, 6. Imatinib is highly effective in newly diagnosed CML patients but many patients have their Abl kinase ultimately evolve to drug-resistant forms7–11. Studies have revealed that the small molecule Imatinib is susceptible to resistance in patients because of amino acid substitutions in the target Abl kinase domain that sterically hinder drug occupancy of the active site11. The majority of relapsed patients frequently harbour point mutations within Bcr-Abl kinase domain12, where imatinib binds to the ATP binding site of the kinase domain with high affinity and specificity13. Search for novel therapeutic approaches may eventually develop inhibitors which can effectively suppress evolved molecular resistance, and significant effort has been devoted to this7, 14, 15.
To design inhibitors with specificity toward Abl tyrosine kinase activity, or to other kinases, one needs to fully understand the subtle differences in conformation and dynamics that distinguish it from the other structurally closely related kinases, or to discover and utilize its unique regulatory mechanism, to provide selectivity and potency16, 17. As for Bcr-Abl, there is a need to better comprehend the effects of resistant mutations on drug binding and on the regulatory functions of those neighbouring domains on the enzymatic activity of kinase domain to search for alternative therapeutic target sites from the ATP-binding pocket, possibly distant from the kinase domain itself18. This goal can best be achieved by combination of in vivo studies and in vitro biochemical, biophysical, and structural investigation. Whereas biochemical studies might require only minute amounts of protein, many biophysical and structural studies demand proteins on the milligram scale. The availability of sufficient amounts of homogenous protein of exceptional stability and purity is often the bottleneck for these studies. Currently the most often used method for large scale production of Abl tyrosine kinase is insect cell culture19–22. Even though insect cell culture gives high protein yields, its lengthy time and high cost can be concerns. Obtaining isotope enriched samples in such culture is cost prohibitive for most labs pursuing solution structural studies of Abl. Many attempts have been carried out but only limited protocols for bacterial expression of Abl either in its viral form v-Abl23, or in cellular form c-Abl24, 25 have been published. Problems faced by these methods are that either the bacterial expressed recombinant Abl has very low solubility, or lacks evidence of its quality adequate for NMR studies. Even though through chaperone over-expression of the soluble portion of Abl kinase domain is increased in bacterial cell lysate, the total expression level of it is decreased due to large portion of limited nutrition resource being consumed by producing large quantity of chaperones with large molecular size. Such intrinsic conflicting features present a significant challenge for high yields of c-Able kinase.
Here we present a straightforward method for expressing and purifying wild type, biologically active c-Abl kinase domain in E. coli. Either isolated c-Abl kinase domain or SH3-SH2-kinase triple domain construct was expressed as a C-terminal fusion of Maltose-binding protein MBP. Multi-milligrams of purified protein can be generated from 1 l of E. coli culture in minimum medium M9. We believe that this is one of the first works demonstrating that an active tyrosine kinase domain has been expressed in bacterial cells with both high quantity and quality suitable for NMR studies. The purified Abl kinase domain is very soluble and stable in buffered solution. Such an economic approach permits us to carry out NMR studies on Abl. Moreover the domains were constructed as intein fusions which allow us to carry out protein ligation. Such reaction permits us to produce domain/segment specifically isotope labelled samples for NMR studies.
Results
c-Abl kinase expressed in E. coli
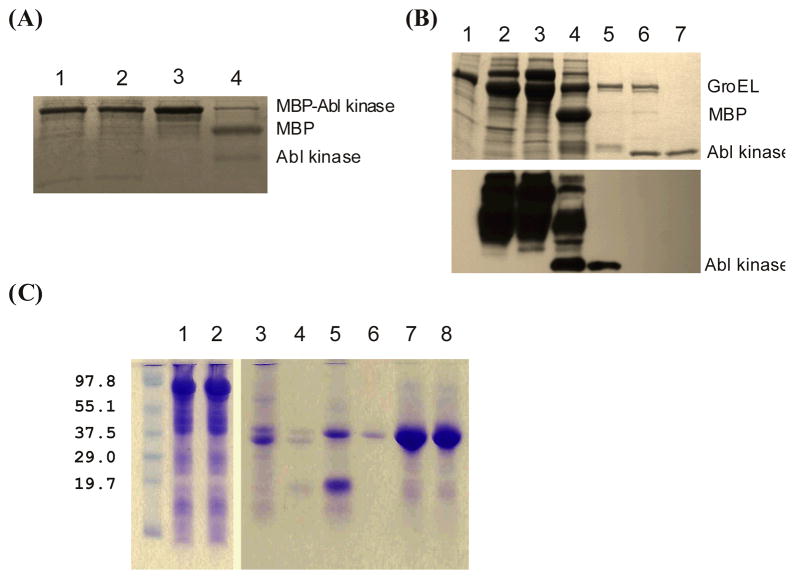
To express c-Abl kinase domain in bacteria we have tried several expression systems including GST fusion, MBP fusion, and Intein fusions. In general the expression level of Abl kinase was significant, judged by SDS gel analysis. However in most cases, the recombinant Abl kinase was mainly found to be insoluble. Only when Abl kinase was expressed as an MBP fusion, did its solubility dramatically increase. More than 90% of such recombinant fusion remained in soluble cell lysate after 30 m centrifugation at 15,000g (Figure 1A). To improve the affinity binding efficiency His6-tag/Co2+ was chosen over MBP/amylose. Isolated Abl kinase domain on itself tends to aggregation. Adding its inhibitors such as PD166326 or Imatinib depending on its phosphorylation status can improve the situation dramatically. Overproducing the bacterial protein chaperone GroEL/ES machinery 26 during Abl Kinase expression did not further increase its solubility. Similarly SH3-SH2-kinase was expressed as an MBP fusion, and high yield was obtained for purified soluble and stable recombinant protein (Figure 1C).
Figure 1.
Abl kinase domain and Abl SH3-SH2-Kinase single chain multi-domain expression in bacteria and their purification analysed by SDS-PAGE and Western blot. (A) SDS-PAGE of Abl kinase: whole-cell lysate (lane 1); soluble proteins after centrifuge (lane 2); proteins bound on Co2+ resin (lane3); retaining proteins on Co2+ resin after thrombin cleavage (lane 4). (B) Tyrosine phosphorylation status: whole-cell lysate of BL21 with GroEL expression (lane 1); whole-cell lysate of BL21 with GroEL and Abl kinase domain co-expression (lane 2); proteins bound on Co2+ resin (lane 3); proteins on Co2+ resin after thrombin cleavage (lane 4); proteins eluted from the affinity resin (lane 5); protein from Lane 5 after CIP treatment (lane 6); dephosphorylated Abl kinase domain after purification (lane 7). The upper panel is Coomassie blue stained SDS-PAGE, the lower panel is an immunoblot with anti-phosphotyrosine antibody. (C) SDS-PAGE of Abl SH3-SH2-kinase: cell lysate before and after centrifuge (land 1 and 2); proteins remained on and eluted from the TALON resin after thrombin cleavage (land 3 and 4); after ionic exchange purification (lane 5); after both ionic exchange and size exclusion purification (lane 6); NMR sample before and after measurement (lane 7 and 8).
Tyrosine phosphorylation activity detected
The kinase released from TALON® resin was subjected to Q column for further purification. It was eluted from the ion exchange column through a large NaCl concentration range. The mass spectrum indicated that the purified recombinant protein was possibly phosphorylated, since the measured mass was higher than the calculated one and showed a broadened peak. To identify the phosphorylation site(s), Western blots were carried out. Only anti-pTyr antibody showed a strong positive response (Figure 1B). A faint signal of phosphorylated serine/threonine was detected on Abl kinase domain (data not shown with other antibodies). The results were the same for Abl kinase expressed in bacterial cells with or without GroEL/ES over-expression. Without Abl kinase there was no detective phosphorylated tyrosine in E. coli cells after protein expression induction (Figure 1B, lane 1). The bacterial cells harboured pREP4groESL was used as a control. Upon Abl kinase domain expression, however, not only the kinase itself was phosphorylated, many bacterial proteins were also heavily phosphorylated (lane 2 and 3). When CIP was applied the kinase was dephosphorylate (Figure 1B, lane 6 and 7). The gel band of the kinase became focused and migrated faster compared to one without CIP treatment (Figure 1B, lane 5 and 6). After CIP treatment the kinase domain eluted from Q column as a single peak.
Phosphorylation sites in bacterial expression
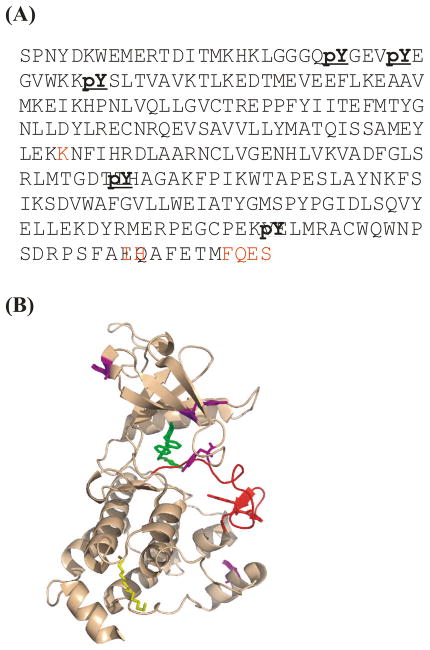
The mass spectrum indicated the phosphorylation of recombinant Abl kinase was not homogeneous, which prompted us to seek its location. Partial trypsin and subtilisin digestion of Abl kinase prior to phosphatase treatment generated overlapping peptide fragments. Due to the different preferences for cutting sites, the two digestions generate complementary sets of peptide fragments depending on the degree of completion 27. LC-MS/MS 28 analyses of the digest peptides of two proteases recovered fragments corresponding to 97.8% of the sequence in the construct, including all tyrosine and threonine as shown in Figure 2(A). Briefly, the mixed peptide fragments were separated by standard liquid chromatographic methods and individual masses determined. The entire set of MS data was compared to the genomic database 29. Five out of 16 tyrosine residues were found in phosphorylated form, which are Y272, Y276, Y283, Y412, and Y468. The key residue for Abl kinase activation, Y412 in the activation loop, through auto-phosphorylation, was among them. Four other phosphorylated tyrosines were located near the protein surface (Figure 2(B)). One of the two threonine residues flanking Y412 was also found phosphorylated at very low level. Information on the possible biological role of such phospho-threonine in Abl kinase activity is beyond the scope of this paper.
Figure 2.
Locating phosphorylation sites in Abl kinase via trypsin/subtilisin digestion and mass analysis. (A) Mass spectrometry identified phospho-tyrosine residues in primary sequence, marked in bold, in bacterial-expressed Abl kinase domain. Sequences undetected by mass analyses are marked in red. These seven are not subject to possible phosphorylation. (B) Locations of phosphorylated residues in its 3-D structure (PDB code 1OPJ). The active loop is colored red. All other phosphorylated sites are labelled in magenta.
Inhibitors and ligand binding
Our purification of Abl kinase domain was carried out with PD166326 addition in the cell lysate. If Imatinib was used instead very little purified kinase could be obtained under otherwise same conditions. It suggested that the bacterial expressed recombinant protein bound to PD166326, but not Imatinib, before CIP treatment. However after complete de-phosphorylation the purified Abl kinase domain could either bind to PD166326 or to Imatinib as our NMR data indicated. This is in total agreement with the fact that activated and inactivated Abl kinase adopt different conformation30. While imatinib only binds inactive conformer, PD166326 binds to both inactive and active ones.
Two dimensional NMR studies
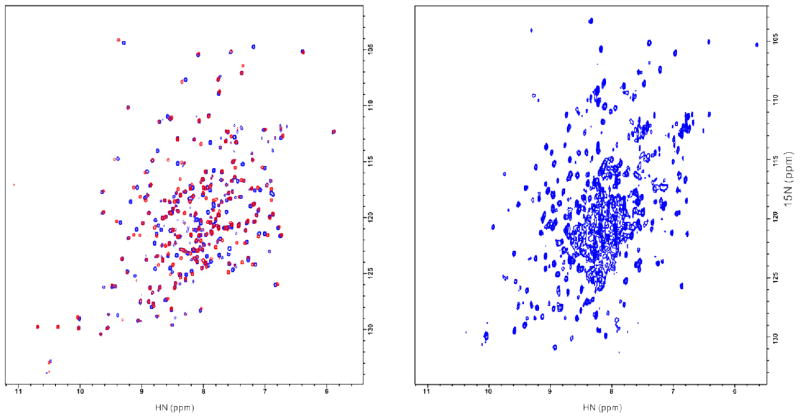
The two dimensional NMR spectra of Abl kinase in complex with PD166326 and with imatinib were taken on 15N uniformly labelled samples. The 15N HSQC maps of both complexes show a well dispersed chemical shift pattern, which indicates that this bacterial expressed Abl kinase domain is folded (Figure 3A). Compared to the HSQC/TROSY spectra for a c-Abl kinase expressed in baculovirus-infected insect cells with either amino-acid-type selective or uniform labelling21, 22, 31, our spectra have similar corresponding chemical shifts. This data provides concrete support that our Abl kinase domain expressed in bacterial cells has the same folding as those expressed in high level eukaryotic cells.
Figure 3.
(A) [15N-1H]-HSQC spectra for Abl kinase domain expressed in bacteria. Signals for Abl kinase in complex with PD166326 is in blue, and with imatinib in red. (B) [15N-1H]-TROSY spectrum of bacterial expressed Abl SH3-SH2-kinase in complex with PD166326 and 2BP.
There are some large chemical shift changes between the spectrum of Abl kinase in complex with PD166326 and one with Imatinib. This is in total agreement with the fact that when Abl kinase binds to PD166326 or Imatinib it adopts different conformation.
Abl kinase domain with its regulatory domains SH3 and SH2 in the native single chain form was also successfully expressed in E. coli. The TROSY spectrum of 15N, 2H-uniformly labelled Abl SH3-SH2-kinase in complex with PD166326 and 2PB, a peptide as SH2 domain ligand, is shown in Figure 3B. The data indicates that this bacterial expressed multi-domain protein also adopts a well folded form. Similarly replacing PD166326 by Imatinib in the complex also induces chemical change (data not shown). 2BP was added to reduce the degree of protein self-association.
Protein ligation
Abl SH3-SH2 dual domain was expressed with an intein fused to its C-terminus. Such construction allows SH3-SH2 to form a C-terminal thioester on addition of a suitable thiol.
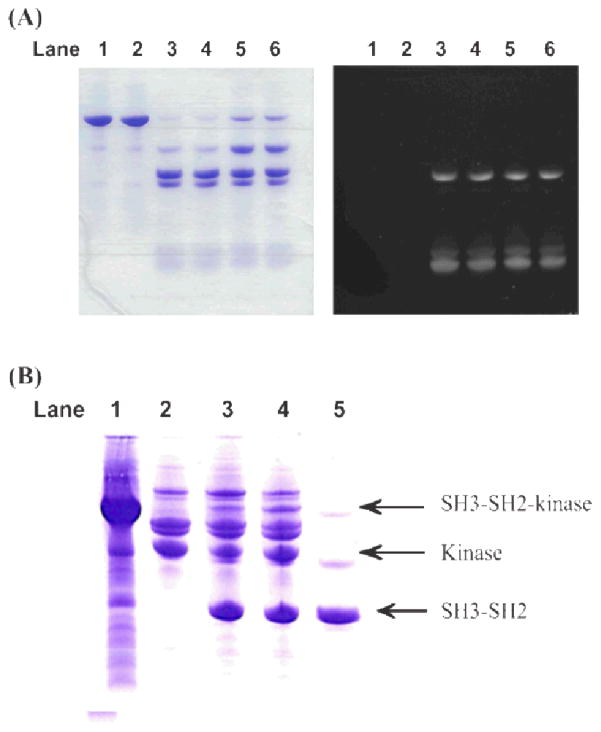
To test the activity of SH3-SH2-intein-CBD in thio-ester formation and subsequent protein ligation at the C-terminus of SH3-SH2, a synthetic fluorescent peptide Flu-P1 containing an N-terminal cysteine and a fluorescein-labelled lysine, NH2-CDPEK(Fl)DS-CONHa, was used as reaction reporter. This synthetic peptide contains an N-terminal cysteine and a fluorescein-labelled lysine in meddle. The N-terminal free cysteine is a requirement for such ligation. The fluorophore allowed one to monitor the reaction and to visualize any ligation product formation. MASNA (sodium 2-mercaptoethanesulfonate) was used to release SH3-SH2 from the intein and to form chemically active thioester derivative. The test ligations were under two pH conditions, 7.0 and 8.0. More than 2/3 of the SH3-SH2 reactant was incorporated in the ligation product after 20 hours (Figure 4(A)). SDS gel indicated that the SH3-SH2 was fully active in protein ligation. The visible difference between pH 7.0 and 8.0 was that the latter weakened the binding of CBD to Chitin Beads.
Figure 4.
SDS-PAGE gel analysis of Abl SH3-SH2 and kinase domain ligation. (A) SH3-SH2 ligated with a test peptide Flu-P1. Lane 1 and 2 are SH3-SH2-intein-CBD bound on Chitin Beads; Lane 3 and 4 are the liquid phase of ligation mixture at pH 7.0 after 20 hour reaction; Lane 5 and 6 are the same as 3 and 4, except the reaction was carried out at pH 8.0. Left panel is the gel visualized by Coomassie blue stain, and right panel is the same gel under UV light. (B) SH3-SH2 ligated to kinase. Lane 1 is SH3-SH2-intein-CBD bound to Chitin Beads before mixed with cys-kinase; lane 2 is partially purified cys-kinase; lane 3 is the ligation mixture of 100 μl (~100 μM) of cys-kinase added to 200 μl SH3-SH2-intein-CBD (~200 μM) bound Chitin Beads, and lane 4 is the ligation mixture of 150 μl of cys-kinase added to 200 μl SH3-SH2-intein-CBD bound Chitin Beads, after 20 hours reaction; lane 5 is SH3-SH2-intein-CBD bound Chitin Beads suspended in 150 μl ligation buffer without cys-kinase as a control.
With active SH3-SH2 in hand, generating single chain Abl SH3-SH2-Kinase from separately expressed and purified SH3-SH2 and kinase was attempted. When Abl kinase was constructed as an intein fusion in pMAL/SspDnaAblCkinase, a free N-terminal cysteine was introduced to it after temperature-sensitive cleavage. Upon mixing bead-bound SH3-SH2-intein-CBD and Cys-kinase, with 300 mM MASNA in the system, ligation product SH3-SH2-kinase was clearly visible on SDS-PAGE gel (Figure (B)).
Discussion and conclusions
Even though expression level was never a major issue for many engineered protein expression systems in E. coli which we have tested, only MBP fusion construct showed significant improvement in recombinant Abl kinase solubility. Its inhibitor PD166326 further stabilized it and made purification and large quantity of this kinase domain for NMR study feasible. When using this construct, over-expression of bacterial chaperone GroEL/ES is not required to enhance the solubility of expressed Abl kinase. This in turn allows the nutrition resources in a bacterial cell to be used mainly to produce Abl kinase after induction, and therefore reaches a higher recombinant protein expression level.
While there is no readily detectable tyrosine phosphotransferase activity in E. coli BL21 cells, many proteins were heavily phosphorylated at their tyrosine sites after expression of the recombinant Abl kinase was induced in our study (see Figure 1(B) Lane 1 and 2). Mass analysis found out that the auto-phosphorylation site in this bacterially expressed recombinant Abl kinase active loop, Y412, was indeed phosphorylated. This is direct evidence that the Abl kinase domain we expressed auto-phosphorylated and activated itself, which in turn further randomly phosphorylated itself at some surface accessible tyrosine sites as well as other proteins in the bacterial system. The data solidly supports that this bacterial produced kinase domain is enzymatically active.
All phosphate groups on this recombinant kinase can be removed by CIP treatment (Figure 1(B) Lane 6 and 7). On the other hand both Abl kinase specific inhibitors, PD166326 and imatinib, did inhibit such auto-phosphorylation after Abl kinase forms a complex with either of them. The tyrosine kinase activity of Abl in the bacterial host cells did interfere with cell growth. Successful recombinant Abl kinase expression here depends on the balance of a careful maintenance of healthy cell growth and a moderate Abl kinase induction rate. While others have shown that co-expression with phosphatases can produce some quantities of Abl kinase18, 24, 25, 32, 33 no NMR spectra are available to assess the fold-ness of the product overall.
In our study there was indirect evidence that the phosphorylated form of Abl kinase in cell lysate did not bind to imatinib. Adding PD166326 to fresh cell lysate let us purify Abl kinase and obtain final homogenous product with yield around several milligrams from one litre bacterial culture starting material. However, keeping all other conditions the same if PD166326 was replaced by imatinib, only negligible amount of purified kinase could be obtained. On the other hand, if large amount of CIP was added to the cell lysate and enough incubation time was allowed before adding imatinib and moving onto purification, similar yield to the PD166326 case could also be achieved. Since PD166326 is a type I inhibitor which binds to Abl kinase in both active and inactive forms, while imatinib only binds to its inactive form34, the stabilization differences are understandable.
Furthermore our NMR data shows the inactive form (dephosphorylated) of bacterial expressed recombinant Abl kinase forms complex with both PD166326 and imatinib in different conformations (Figure 3A). Compared to the HSQC/TROSY spectra for a c-Abl kinase expressed in a baculovirus-infected insect cells with either 15N-Phe, 15N-Tyr, 15N-Val, and 15N-Gly amino-acid-type selective labelling or uniform labelling21, 22, 31, our spectrum has similar corresponding chemical shifts.
From both functional and folding points of view our data provides indicate that the Abl kinase domain expressed in bacterial cells simply as MBP fusion has the same folding as those expressed in high level eukaryotic cells. The yield is in the range of several milligrams per litre of bacterial culture. Such yield is high enough, in an economic sense, for NMR type structural studies, and for some other in vitro functional studies/screening which require large amount of samples. The bacterial expressed Abl SH3-SH2-kinase also adopts a fully folded form as its NMR TROSY spectrum indicates. Without SH2 ligand the concentration suitable for NMR study causes self-association of this multi-domain protein. Adding a SH2 peptide ligand, 2BP, improves the stability.
To demonstrate the possibility of generating segmentally labelled SH3-SH2-kinase, the protein was engineered and expressed in two separated fragment and “sewed” together through protein ligation under native conditions. SH3-SH2 was expressed as N-terminal intein fusion to form active thioester derivative. The kinase domain was expressed as a C-terminal intein fusion with a cysteine at its N-terminus. Again MBP was introduced to produce correctly folded and soluble kinase. Indeed two separately expressed segments can be ligated together in a designed order through peptide linkage. This approach opens doors for us to study the allosteric change through function regulation, inter-domain interaction, ligand/substrate binding, et al, via NMR technique.
Experimental Procedures
PD166326 was purchased from Cayman Chemical (Michigan), and Imatinib was a generous gift from Novartis.
Expression constructs
The gene segment coding c-Abl 1b kinase domain between residues S248 and S519 35 was selected for isolated kinase construct. Its PCR product was sub cloned in pMAL-c2X vector (New England Biolabs) between restriction sites BamHI and SalI. Similarly a longer construct with the PCR product of gene segment coding c-Abl L84 to G533 was inserted in this vector. A cysteine to alanine mutation, C119A, was introduced using QuikChange® Site-Directed Mutagenesis kit (Stratagene). A His6-tag has been constructed in front of the malE gene which codes a Maltose-binding protein MBP 36 to facilitate efficient affinitive purification.
To facilitate protein ligation reaction, the expression plasmids of c-Abl SH3-SH2 harbouring C119A mutation and the kinase domain were constructed as follows. The DNA segment coding residues L84-G246, with C119A mutation, was cloned into pTXB1 (New England Biolabs) between restriction NdeI and SpeI sites to produce SH3-SH2 as N-terminal fusion of Mxe intein/chitin binding domain (CBD). This expression vector is named as pTXB1/AblSH32G_(C2A). For the kinase portion, first the DNA coding residues S248-G533 was inserted into pTWIN1 (New England Biolabs) between restriction sites SapI and EcoRI. A stop codon and a SalI restriction site were introduced right in front of the EcoRI site at the 3’prime end of the insert via primer design and PCR. The new plasmid, pTWIN/Ablkinase was used as template to amplify a DNA sequence coding in-frame fusion of mini Ssp DnaB intein and c-abl kinase. This piece was then cloned in pMAL mentioned above between restriction sites BamHI and SalI to generate expression plasmin pMAL/SspDnaAblCkinase.
Protein expression and purification
Plasmids containing abl kinase, pMAL/Ablkinase, was transformed into E. coli BL21 (Stratagene) competent cells, or BL21 harbouring a GroEL/GroES expression vector pREP4groESL. Plasmid containing SH3-SH2-kinase, pMAL/SH32(C2A)kin and plasmid containing kinase domain for ligation, pMAL/SspDnaAblCkinase were simply transformed into E. coli BL21 for recombinant protein expression. For pTBX1/AblSH32G_(C2A), BL21-codonplus(DE3)RIPL (Stratagene) were used. Bacterial competent cells were transformed by plasmids carrying selected inserts and plated on LB agar. Appropriate ampicillin or combination of antibiotics, ampicillin and kanamycin, was applied for resistance selection. The plate was incubated at 37 °C overnight. Next morning the colonies from the plate were suspended in expression media with antibiotics (50 mg/L for ampicillin, and 30 mg/L for kanamycin), LB for unlabelled protein or M9 in either H2O or D2O with 15NH4Cl for NMR samples. Cultures were grown at 37 °C with 300 rpm shaking. When cell density reached 0.9 OD600nm, IPTG was added to 0.25 mM to induce recombinant protein expression. The induction was carried out at 37 °C for 0.5 h, and transferred to 18 °C for another 16 h for in-LB growth, to 20 °C for 20 h for in-H2O M9 growth, and to 22 °C for 40 h for in-D2O M9 growth. The IPTG induction concentration is 0.5 mM for pTBX1/AblSH32G_(C2A), and its induction was either at 30 °C for 2 h followed by 28 °C for 12 h in D2O or at 37 °C for 5 h in H2O. Cells were harvested by centrifuge at 8,000 g for 20 min. Cell pellets were either stored at -80 °C for future use, or processed immediately to purification.
All MBP fusions were purified via affinity binding of their N-terminal His6-tag with TALON® resin (Clontech). After re-suspension in lysis buffer A (25 mM MOPS pH 7.0, 500 mM NaCl, 1 mM MgCl2, 15% glycerol), cells harbouring recombinant Abl kinase were incubated with lysozyme (0.1 mg/ml) on ice for one hour. They were further lysed by passing through a high pressure homogenizer EF-C3 (Avestin, Inc.). One EDTA-free COMPLETE protease inhibitor tablet (Roche) is used per 50 ml cell lysate. 6-(2,6-dichlorophenyl)-2-[3-(hydroxymethyl)anilino]-8-methylpyrido[2,3-d]pyrimidin-7-one (PD166326)37, 38 (Cayman Chemical, Ann Arbor) at 20 μM was added to form Abl kinase/inhibitor complex. Clear lysate was collected after centrifuging at 15,000 g for 30 min. The clear cell lysate was mixed with TALON® resin pre-equilibrated with lysis buffer. Continued gentle agitation of the mixture on a rotator at 4 °C for > 4 h allowed the His6-tagged fusion protein to bind the resin. Unbound proteins and other contaminants were drained away. The protein-bound resin was thoroughly washed with a high salt buffer (20 mM MOPS pH 7.2, 500 mM NaCl, 1 mM MgCl2, 20% glycerol) and then washed and equilibrated in cleavage buffer A (20 mM MOPS pH 7.6, 150 mM NaCl, 10 mM MgCl2, 20% glycerol). For Abl SH3-SH2-kinase purification, the only difference is that 20 mM NaH2PO4 was included in the lysis buffer and the high salt buffer, and 5 mM of it in the cleavage buffer. Both Abl kinase from pMAL/Ablkinase and SH3-SH2-Kinase from pMAL/SH32(C2A)kinase were released from resin bound protein fusion by thrombin cleavage. Briefly, one volume of resin was suspended in 3 volumes of cleavage buffer with 2.5 mM CaCl2 and 25 U/ml of thrombin (Enzyme Research Laboratories) following standard procedures39. The reaction was carried out at 20 °C for 18 h with gentle agitation to keep the resin suspended. Protein released from MBP fusion by thrombin was eluted from the resin, and 1 mM AEBSF (Allex) was added to stop thrombin activity. Abl kinase was dephosphorylated by calf intestinal alkaline phosphatase (CIP, 20 units/ml) (New England BioLabs) at 20 °C. Abl kinase expressed from pMAL/SspDnaAblCkinase was released from the fusion in the cleavage buffer, with pH adjusted to 7.0, after keeping at room temperature for 24 hours.
SH3-SH2-intein-CBD was purified through chitin affinitive binding. Cells were lysed in lysis buffer B (50 mM Tris pH 8.0, 150 mM NaCl, 0.1 mM EDTA, 0.1% Triton X-100) through a EF-C3 homogenizer. The recombinant fusion protein in the clear cell lysate was bound to Chitin Beads (New England BioLabs). After thorough washing, the fusion protein was left on beads to proceed to the ligation reaction. SH3-SH3 may be released from beads in cleavage buffer B (25 mM MOPS pH 7.2, 150 mM NaCl, 100 mM DTT, 0.1 mM EDTA) for other studies as well.
Subsequent anion exchange chromatography was applied to separate Abl kinase from thrombin and other co-eluted protein contaminants. The dephosphorylated Abl kinase was loaded on a HiTrap™ Q HP column (GE Healthcare), and was eluted with a 55 min linear gradient from 0 to 50 mM NaCl in 20 mM MOPS pH 7.2. Elution fractions containing Abl kinase were collected and concentrated for other analyses. SH3-SH2-kinase was first subjected to HiTrap™ Q HP column linear gradient purification, and then to size exclusive Sephacryl™ S-100 column (GE Healthcare). A Bis-tris/NaH2PO4 buffer (20 mM Bis-tris, 20 mM NaH2PO4, pH 6.8, 100 mM NaCl, 1 mM EDTA) was used as the mobile phase. SH3-SH2 was purified through HiTrap™ SP HP column (GE Healthcare) in 20 mM MOPS pH 7.0 with a linear gradient of NaCl from 0 to 50% in 52 min.
Mass analysis and tyrosine phosphorylation identification
Abl kinase samples at different purification stages were subjected to 12% Tris-HCl SDS-PAGE gels (Bio-Rad), then transferred to PVDF membranes (Bio-Rad) using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). The membranes were blocked by non-fat dry milk. Phosphotyrosine was identified by rabbit anti-phosphotyrosine polyclonal antibody (Zymed Laboratories). The corresponding peroxidase-conjugated second antibodies, goat anti-rabbit IgG (Zymed Laboratories) was applied subsequently. After a thorough wash with 0.05% Tween 20 and rinse with fresh water, the blotting was revealed by ECL Western blotting detection reagents (GE Healthcare) and the images were captured on X-ray films.
The molecular mass of Abl kinase domain was measured by MALDI TOF using sinapinic acid as matrix40.
To identify phosphorylation sites, Abl kinase eluted from TALON® resin was first alkylated, and then separately subjected to trypsin and subtilisin digests. These were analyzed by LC-MS/MS. Phosphorylated peptide identification and the phosphorylated residue location was carried out by Mascot search engine (Matrix Science, Boston, MA).
NMR samples and measurement
[U-15N]Abl kinase in complex with PD166326 was concentrated to about 300 μM in buffer containing 20 mM Bis-Tris, pH 6.5 (25 °C), 100 mM NaCl, 1 mM EDTA, and 2 mM DTT. To form Abl kinase/imatinib complex, 4:1 imatinib to kinase ratio was added to the diluted solution with Abl kinase/PD166326, followed by concentration to replace bound PD166326. The procedure was repeated to complete the replacement.
[U-15N,2H] Abl SH3-SH2-kinase, with C119A mutation, was in complex with PD166326 in 1:1 ratio. A 180 μM solution was prepared in 90% H2O and 10% D2O, 20 mM MOPS, 300 mM NaCl, 1 mM EDTA (pH 7.0). Abl SH2 domain ligand, 2BP with sequence GSG(pY)ENPED41, was also added in 1:2 protein to peptide ratio.
All NMR spectra were acquired at 30 °C on an 800 MHz Bruker Avance-II spectrometer. 1H was referenced relative to the frequency of the 2H lock resonance of water. 15N chemical shifts were referenced indirectly using the 1H/15N frequency ratios of the zero point: 0.101329118. 1H[15N]-HSQC spectrum was acquired for isolated Abl kinase domain, and 1H[15N]-TROSY spectrum for SH3-SH2-kinase multidomain sample. Topspin software was used to process acquired data and generate final spectra.
Expressed protein ligation
To test the reactivity of its potential C-terminal thioester intermediate, 50 μl SH3-SH2-intein-CBD bound Chitin Beads were flushed with 3x bead volume of a testing buffer (25 mM HEPES, 300 mM MESNA, 300 mM NaCl, 20 mM MgCl2, 0.1 mM EDTA, 10% glycerol) with pH at either 7.0 or 8.0. The beads was re-suspended in 50 μl ligation buffer, and 20 μl Flu-P1 peptide (New England Biolabs) was added so that in the liquid phase the composition is MESNA is ~ 200 mM, peptide 570 μM, and SH3-SH2 ~ 200 μM (assuming it was totally released from the beads). The reaction system was kept at room temperature with gentle agitation for 20 h. Reaction mixture was checked on 20% PAGE gel and visualized by UV light and Coomassie blue stain.
Expressed protein ligation was carried out between Abl SH3-SH2 and kinase domains. SH3-SH2-intein-CBD-bound Chitin Beads were flushed with 3 bead volume of Ligation buffer (25 mM MOPS pH 7.0, 300 mM MASNA, 120 mM NaCl, 20 mM MgCl2, 0.1 mM EDTA, 10% glycerol). Partially purified kinase domain expressed from pMAL/SspDnaAblCkinase was exchanged to Ligation buffer, and added to the beads. The reaction was carried out at room temperature with agitation for one day before PAGE gel analysis.
Acknowledgments
We are grateful to Dr. K. Dutta and Dr. M. Goger at the New York Structural Biology Center for their technical advice and support. This study was supported by NIH GM-047021-16.
Notes and references
- 1.Koleske AJ. Landes Bioscience. Springer Science+Business Media; Georgetown, Tex. New York: 2006. Abl family kinases in development and disease. [Google Scholar]
- 2.Woodring PJ, Litwack ED, O’Leary DD, Lucero GR, Wang JY, Hunter T. Journal of Cell Biology. 2002;156:879–892. doi: 10.1083/jcb.200110014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Druker BJ. Blood. 2008;112:4808–4817. doi: 10.1182/blood-2008-07-077958. [DOI] [PubMed] [Google Scholar]
- 4.Pinilla-Ibarz J, Flinn I. Critical reviews in oncology/hematology. 2012 doi: 10.1016/j.critrevonc.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Nature medicine. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 6.Deininger MW, Goldman JM, Lydon N, Melo JV. Blood. 1997;90:3691–3698. [PubMed] [Google Scholar]
- 7.O’Hare T, Deininger MW, Eide CA, Clackson T, Druker BJ. Clin Cancer Res. 2011;17:212–221. doi: 10.1158/1078-0432.CCR-09-3314. [DOI] [PubMed] [Google Scholar]
- 8.Sherbenou DW, Hantschel O, Kaupe I, Willis S, Bumm T, Turaga LP, Lange T, Dao KH, Press RD, Druker BJ, Superti-Furga G, Deininger MW. Blood. 2010;116:3278–3285. doi: 10.1182/blood-2008-10-183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young MA, Shah NP, Chao LH, Seeliger M, Milanov ZV, Biggs WH, 3rd, Treiber DK, Patel HK, Zarrinkar PP, Lockhart DJ, Sawyers CL, Kuriyan J. Cancer Res. 2006;66:1007–1014. doi: 10.1158/0008-5472.CAN-05-2788. [DOI] [PubMed] [Google Scholar]
- 10.Azam M, Daley GQ. Molecular diagnosis & therapy. 2006;10:67–76. doi: 10.1007/BF03256446. [DOI] [PubMed] [Google Scholar]
- 11.Azam M, Nardi V, Shakespeare WC, Metcalf CA, 3rd, Bohacek RS, Wang Y, Sundaramoorthi R, Sliz P, Veach DR, Bornmann WG, Clarkson B, Dalgarno DC, Sawyer TK, Daley GQ. Proc Natl Acad Sci U S A. 2006;103:9244–9249. doi: 10.1073/pnas.0600001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 13.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 14.Eide CA, Adrian LT, Tyner JW, Mac Partlin M, Anderson DJ, Wise SC, Smith BD, Petillo PA, Flynn DL, Deininger MW, O’Hare T, Druker BJ. Cancer Res. 2011;71:3189–3195. doi: 10.1158/0008-5472.CAN-10-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. Nat Biotech. 2011 advance online publication. [Google Scholar]
- 17.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Nat Biotech. 2011 doi: 10.1038/nbt.2017. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, Guo GR, Ding Q, Okram B, Choi Y, Wojciechowski A, Deng X, Liu G, Fendrich G, Strauss A, Vajpai N, Grzesiek S, Tuntland T, Liu Y, Bursulaya B, Azam M, Manley PW, Engen JR, Daley GQ, Warmuth M, Gray NS. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer BJ, Baltimore D. Mol Cell Biol. 1994;14:2883–2894. doi: 10.1128/mcb.14.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 21.Strauss A, Bitsch F, Fendrich G, Graff P, Knecht R, Meyhack B, Jahnke W. Journal of biomolecular NMR. 2005;31:343–349. doi: 10.1007/s10858-005-2451-3. [DOI] [PubMed] [Google Scholar]
- 22.Vajpai N, Strauss A, Fendrich G, Cowan-Jacob SW, Manley PW, Grzesiek S, Jahnke W. The Journal of biological chemistry. 2008;283:18292–18302. doi: 10.1074/jbc.M801337200. [DOI] [PubMed] [Google Scholar]
- 23.Lydon NB, Adams B, Poschet JF, Gutzwiller A, Matter A. Oncogene Res. 1990;5:161–173. [PubMed] [Google Scholar]
- 24.Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haacke A, Fendrich G, Ramage P, Geiser M. Protein Expres Purif. 2009;64:185–193. doi: 10.1016/j.pep.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Amrein KE, Takacs B, Stieger M, Molnos J, Flint NA, Burn P. Proc Natl Acad Sci U S A. 1995;92:1048–1052. doi: 10.1073/pnas.92.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James P, Quadroni M, Carafoli E, Gonnet G. Protein Sci. 1994;3:1347–1350. doi: 10.1002/pro.5560030822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Journal of proteome research. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 29.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Okram B, Nagle A, Adrian FJ, Lee C, Ren P, Wang X, Sim T, Xie Y, Xia G, Spraggon G, Warmuth M, Liu Y, Gray NS. Chem Biol. 2006;13:779–786. doi: 10.1016/j.chembiol.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Vajpai N, Strauss A, Fendrich G, Cowan-Jacob SW, Manley PW, Jahnke W, Grzesiek S. Biomol NMR Assign. 2008;2:41–42. doi: 10.1007/s12104-008-9079-7. [DOI] [PubMed] [Google Scholar]
- 32.Fabbro D, Manley PW, Jahnke W, Liebetanz J, Szyttenholm A, Fendrich G, Strauss A, Zhang J, Gray NS, Adrian F, Warmuth M, Pelle X, Grotzfeld R, Berst F, Marzinzik A, Cowan-Jacob SW, Furet P, Mestan J. Biochim Biophys Acta. 2010;1804:454–462. doi: 10.1016/j.bbapap.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Marimuthu A, Tsai J, Kumar A, Krupka HI, Zhang C, Powell B, Suzuki Y, Nguyen H, Tabrizizad M, Luu C, West BL. Proc Natl Acad Sci U S A. 2006;103:3563–3568. doi: 10.1073/pnas.0600048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McInnes C, Mezna M, Kontopidis G. Chemistry & Biology. 2006;13:693–694. doi: 10.1016/j.chembiol.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Oppi C, Shore SK, Reddy EP. Proc Natl Acad Sci U S A. 1987;84:8200–8204. doi: 10.1073/pnas.84.23.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexandrov A, Dutta K, Pascal SM. BioTechniques. 2001;30:1194–1198. doi: 10.2144/01306bm01. [DOI] [PubMed] [Google Scholar]
- 37.Kraker A, Hartl BG, Amar AN, Barvian MR, Showalter HDH, Moore CW. Biochem Pharmacol. 2000;60:885–898. doi: 10.1016/s0006-2952(00)00405-6. [DOI] [PubMed] [Google Scholar]
- 38.Wolff NC, Veach DR, Tong WP, Bornmann WG, Clarkson B, Ilaria RL., Jr Blood. 2005;105:3995–4003. doi: 10.1182/blood-2004-09-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu D, Xu R, Dutta K, Cowburn D. FEBS Lett. 2008;582:1163–1167. doi: 10.1016/j.febslet.2008.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu CF, Lu Y, Ma J, Mohammadi M, Neubert TA. Mol Cell Proteomics. 2005;4:809–818. doi: 10.1074/mcp.T400019-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Cantley L. J Cell Biochem. 1993:225–225. [Google Scholar]