Abstract
Enumeration of circulating tumor cells (CTCs) has proved valuable for early detection and prognosis in cancer treatment. This paper describes an automated high-throughput counting method for CTCs based on microfluidics and line-confocal microscopy. Peripheral blood was directly labeled with multiple antibodies, each conjugated with a different fluorophore, pneumatically pumped through a microfluidic channel and interrogated by a line-confocal microscope. Based on the fluorescence signals and labeling schemes, the count of CTCs was automatically reported. Due to the high flow rate, 1 mL of whole blood can be analyzed in less than 30 minutes. We applied this method in analyzing CTCs from 90 stage IV breast-cancer patient samples, and performed a side-by-side comparison with the results of the CellSearch assay, which is the only method approved by U.S. Food and Drug Administration (FDA) at present for enumeration of CTCs. This method has a recovery rate for cultured breast cancer cells of 94% (n=9), with an average of 1.2 counts/mL of background level of detected CTCs from healthy donors. It detected CTCs from breast-cancer patients, ranging from 15 to 3375 counts/7.5 mL. Using this method, we also demonstrate the ability to enumerate CTCs from breast-cancer patients that were positive for Her2 or CD44+/CD24−, which is a putative cancer stem cell marker. This automated method can enumerate CTCs from peripheral blood with high throughput and sensitivity. It could potentially benefit the clinical diagnosis and prognosis of cancer.
Introduction
Metastasis is generally considered the most important clinical indicator of cancer. Over 90% of deaths associated with cancer are directly attributable to metastasis1–2. The release of cancer cells from the primary tumor may happen at early stages of the disease3–4, which is the basis for the later development of metastasis. Circulating Tumor Cells (CTCs), first reported 150 years ago5, are widely considered to be an important factor in this process6–8, and they have been found in patients having many different types of cancer9, such as breast, lung, pancreatic, prostate, liver and colon. Detection and analysis of CTCs can benefit the early diagnosis of cancers10–11, management of clinical treatment of cancer patients12, development of personalized medicine13, and exploring the mechanism of metastasis14, which is thought to be the most poorly understood component of cancer pathogenesis2.
Generally, CTCs are very rare in whole blood; for example, they have been detected at concentrations as low as 1 to 10 CTCs per 1 billion blood cells15–17. As a result, there are many challenges in developing a suitable analytical method. Technologies, such as flow cytometry18–20, have been used to detect CTCs; however, due to their very low concentration, the processing time for one sample can take more than 24 hours18. In another approach, blood samples were lysed before analysis19, which improved throughput but could result in sample contamination and cell stress. A variety of techniques have been developed in recent years, based on immuno-magnetic enrichment9, 21–22, surface capture with antibody immobilized on microfluidic chips23–25, size based filtration26–30, density gradient analysis31, and negative selection32. Nearly all of these methods require an initial enrichment step to improve the sensitivity and throughput, and the enrichment ratio determines to a large extent their analytical performance and potential applications. The throughput of these methods varies considerably, and fluorescence imaging, which is used in most CTC technologies, usually takes a long time and requires manual confirmation of cells.
We recently developed a new method called ensemble-decision aliquot ranking (eDAR) to analyze rare cells from peripheral blood33. The functions of detection, isolation, identification, and downstream analysis are integrated into one microfluidic chip. It is highly sensitive, with a detection limit close to 1 cell/mL, and the throughput is high enough for clinical applications (20 minutes for 1 mL whole blood)33. Due to the open-access design of the chip, it is easy to selectively extract captured cells and perform various downstream analyses. However, the whole process of eDAR still requires some manual manipulation, because cells trapped on the integrated filter need to be imaged and identified manually, and cell harvesting requires micropipetting by a skilled operator. This implies that eDAR might not be highly efficient for some special applications, such as a fast “coarse screening” of CTCs that may not require very detailed information of cellular and molecular profiles.
In this paper, we report the development of a fast and automated screening method based on the multicolor line-confocal detection technique used in eDAR33 for CTCs. The sample processing and experimental procedures – which do not include any enrichment steps – were optimized to maximize throughput. Also, because labeled blood samples are simply flowed through the chip and not immobilized, destroyed, or physically bound to any substrate, this method is highly compatible with other downstream analysis techniques. Indeed, the same sample can be re-analyzed with eDAR or other techniques after this initial “coarse screening” for the presence of CTCs. The overall time of analysis for 1 mL whole blood can be less than 1 hour, including sample preparation, microfluidic detection, and data analysis. The average background level for healthy blood samples was 1.2 counts /mL, and the recovery rate of our method was high, at around 94% (n=9). 90 samples from stage IV breast cancer patients were analyzed using this method and compared side-by-side with the results of the FDA-approval CellSearch method. Based on the results, we believe the reported method represents a more effective approach to fast screening of CTCs for early detection and prognosis in cancer treatment.
Experimental sections
Design and fabrication of microfluidic devices
The microfluidic channel was 200 µm wide, 50 µm tall, and 3 cm long. The polydimethylsiloxane (PDMS) device was prepared by photolithography and replica molding methods described previously34. The features were designed in AutoCAD 2010 (Autodesk, San Rafael, CA), and then written to a transparency mask (FineLine imaging, Colorado springs, CO). To create masters for replica molding, silicon wafers were spin-coated with SU-8 3050 (MicroChem, Newton, MA), forming a 50-µm thick film. The features were developed and then silanized with tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (Sigma-Aldrich, St. Louis, MO) to prevent PDMS from sticking to the wafer. Uncured PDMS was poured onto the surface of the master, and then incubated at 75 °C for 4 hours. The PDMS chip was sealed to a No. 4 cover glass (Thermo Fisher Scientific, Portsmouth, NH) by oxygen plasma bonding.
Biological and clinical materials
Isoton buffer, purchased from Beckman Coulter (Miami, FL), was used in all experiments. Prior to use, the buffer was filtered using a 50 mL Steri-flip filter (Milipore, Billerica, MA). Two breast cancer cell lines, MCF-7 and SKBr-3, were purchased from American Type Culture Collection (ATCC). Both were used to characterize and optimize the continuous flow system. Cells were maintained and cultured in the recommended culture media (McCoy’s 5A or EMEM), which contained 2 mM L-glutamine, 10% fetal bovine serum (FBS) (ATCC), and 50 µg/mL penicillin/streptomycin (ATCC) at 37 °C with 5% CO2 in a humidified environment.
Human whole blood, individually drawn from healthy donors, was provided by Plasma Lab International (Everett, WA) and stored at 4°C upon arrival. Each 20-mL draw was collected into four 5 mL Vacutainer tubes coated with EDTA as anti-coagulant. The first tube of each draw was discarded to avoid potential contamination from skin cells.
Clinical samples were collected from Stage IV metastatic breast cancer patients according to a protocol approved by University of Washington’s institutional review board. Blood was drawn at the Seattle Cancer Care Alliance and multiple tubes were collected in each draw. One tube was collected in a Veridex CellSave tube for enumeration of CTCs by the CellSearch system (Veridex, Raritan, NJ). The second one was collected in a Vacutainer tube containing EDTA for the flow detection analysis. The sample was stored at 4°C after the draw and analyzed within 4 h.
Line-confocal detection scheme
To detect a single CTC in a background of nanoliters of blood, we developed a line-confocal detection scheme34–35 with a probe volume that spanned the width (200 µm) and height (50 µm) of the micro-channel. Our system had two lasers (488 nm and 633 nm), whose outputs were shaped by cylindrical lenses and focused into a 20× objective to form a 200 µm by 5 µm line (Fig. 1a). Fluorescence from this region was collected through a rectangular confocal aperture and a series of dichroics and filters to fiber-coupled avalanche photodiodes (APDs) (Excelitas Technologies, Waltham, MA), operating in the single-photon counting mode34. In our current setup, APD1 detected the yellow fluorescence (560–590 nm) from the monoclonal antibody labeled with phycoerythrin (PE); APD2 was used as a negative control for the green wavelength range, such as fluorescein isothiocyanate (FITC) (500–550 nm) to eliminate false positives from broadly emitting fluorescent contaminants or the antibody conjugated with FITC; and APD3 detected the antibody labeled with Alexa-647 in the red wavelength band (640–690 nm).
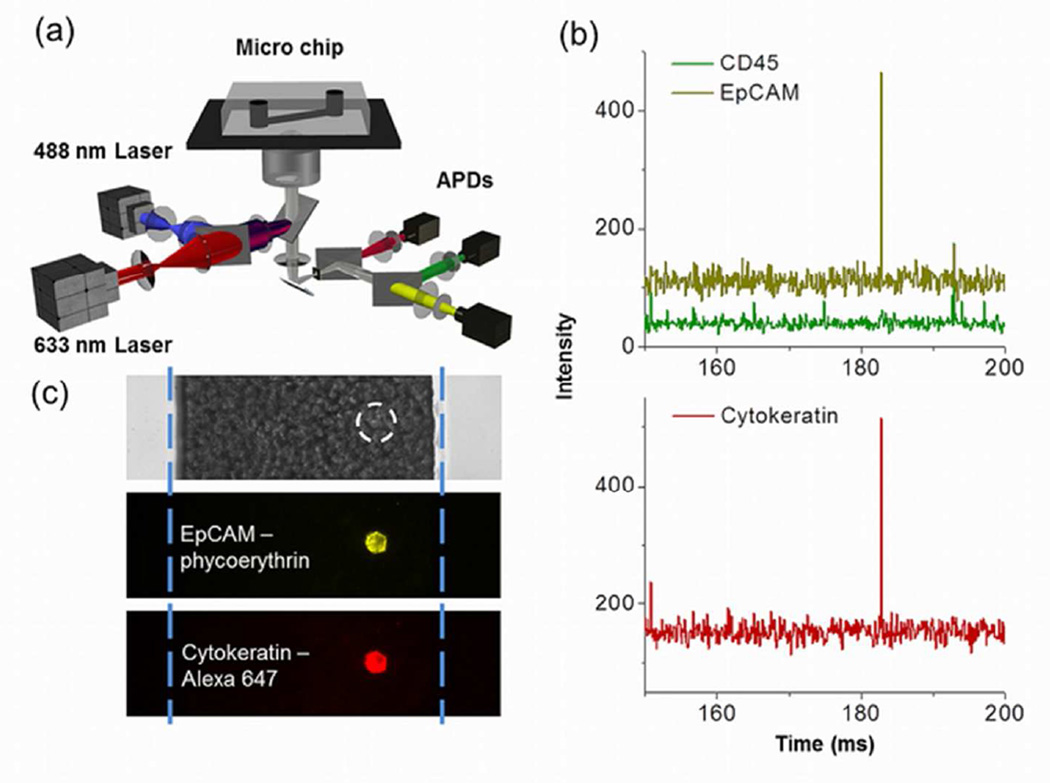
Figure 1.
Schematic and data illustrating the flow-detection platform. a) Depiction of the microfluidics and optics. b) CTC detection and identification scheme using APD signals. A typical CTC event at 183 ms is positive for EpCAM (Yellow signal) and cytokeratin (Red signal), but negative for CD45 (Green signal). c) A cultured MCF-7 cell imaged in the microfluidic channel filled with whole blood. The top panel shows a bright field image of the blood in the microchannel. The white dashed circle shows the location of the MCF-7 cell that is not visible beneath the many blood cells. The blue dashed lines show the location of the microchannel walls. Fluorescence images of the same location show the MCF-7 cell labeled with both anti-EpCAM and anti-Cytokeratin.
Sample preparation and experimental procedures
Antibodies were centrifuged (14000 rpm, 5 min) to remove possible aggregates before any labeling procedure. In each clinical run, 0.5 to 1 mL of blood was added to a 15-mL polypropylene conical centrifuge tube (Becton Dickinson, Franklin Lakes, NJ). 60 µL of PE-anti-epithelial cell adhesion molecule (EpCAM) (BioLegend, San Diego, CA, Catalog # 324206), 80 µL of Alexa-647-anti-Cytokeratin (Cell Signalling technologies, Danvers, MA, Catlog # 4528), and 20 µL of FITC-anti-CD45 (BioLegend, San Diego, CA, Catolog # 304006) were added to the blood in the dark and incubated at room temperature for half an hour. When cytokeratin was used as the second positive marker for breast cancer cells, the blood was fixed before labeling. 84 µL paraformaldehyde (PFA) (20%) were used to fix the blood at room temperature for 15 minutes. Then 44 µL of Surfynol® 465 (Air product, Allentown, PA) were added to help anti-cytokeratin penetrate the cell membrane. When Her2 was used as the second positive marker, there was no fixing step before labeling. 10 µL of Alexa-647-anti-Her2 (BioLegend, San Diego, CA, Catolog # 324412) were added to blood with the same amount of anti-EpCAM and anti-CD45 as described above. The labeling parameters were characterized and optimized in our previous work33.
The labeled blood was diluted to 14 mL and then centrifuged (500 rpm, 10 minutes) to remove the free antibodies (See more details in figure 1S). The final volume was adjusted to be the same as the initial volume for both clinical and control samples. After that, the sample was loaded into the microfluidic chip pneumatically and analyzed using the home-built line confocal system. The flow rate was controlled by the gas pressure regulator and measured by an electrical scale (ACCULAB Measurement Standards, Danvers, MA), which was controlled by a custom LabVIEW (National Instruments, Austin, TX) program. APD traces were collected by a PCI data acquisition card (PCI 6602, National Instruments, Austin, TX) and analyzed by a MATLAB (MathWorks, Natick, MA) script developed in-house. The threshold of signal-to-noise ratio for all the 3 channels was set to 5 to ensure the highest possible sensitivity.
Results and discussion
General description of the system and data analysis
In order to detect CTCs, whole blood was directly labeled with antibodies, loaded into the microfluidic chip pneumatically, and analyzed by the line confocal detection system (Figure 1a). When the labeled blood flowed through the detection area, fluorescence was excited by the combined two-color laser beam (488 nm and 633 nm), and detected by APDs. The collected APD traces were used to determine and enumerate cancer cells in the blood.
Surface markers such as EpCAM are widely used to select CTCs36; however, previous research has shown that simply relying on one biomarker may not be adequate to define the whole population of CTCs. For example, normal-like breast cancer cells do not express EpCAM and thus would be missed by the CellSearch method that relies on EpCAM for CTC isolation37. As a result, a set of criteria is necessary to identify CTCs. The CellSearch assay defines a CTC as a cell that is EpCAM positive, cytokeratin positive, CD45 negative and nuclear stain positive, with a certain nuclear to cytoplasmic ratio and cell morphology character. In our approach, we followed these widely used criteria to enumerate CTCs in all patient samples, except that we did not use a nuclear stain, since we would need another APD detector. Whole blood was labeled with PE-anti-EpCAM, Alexa647-anti-pancytokeratin, and FITC-anti-CD45. Any cell deemed a CTC had positive fluorescence signals representing expression of both EpCAM and cytokeratin, but no signal associated with CD45. As an example, Fig. 1b shows a small portion of APD traces from the analysis of a clinical sample (Sample #16). The event at 183 ms has peaks in both the EpCAM and cytokeratin channels without any significant signal in the CD45 channel, indicating that there was a tumor cell flowing through the detection window at that moment.
Imaging results (Fig. 1c) show a cultured MCF-7 cell labeled with anti-EpCAM and anti-cytokeratin in the microfluidic channel. The bright field image shows that the MCF-7 cell was in the channel surrounded by numerous blood cells and could not be identified in this context. However, using epi-fluorescence imaging, the cell had strong yellow (PE-anti-EpCAM) and red (Alexa647-anti-cytokeratin) emission, which made it distinguishable from the blood cells in the channel.
While the initial signal to noise (S/N) ratio (> 20) of the fluorescence detection was high at the experimental flow rate (50 to 60 µL/min), it was important to optimize the data analysis so that the system could be operated at the highest throughput and would be robust enough to handle the variability found in patient samples undergoing different clinical treatments. For example, to eliminate cross talk between different wavelength bands, spectral un-mixing is used in the MATLAB script to analyze the data. To reduce the noise and smooth the baseline, a sigmoidal burst detection filter was applied in the script for data analysis (Supporting information, Figure 2s).
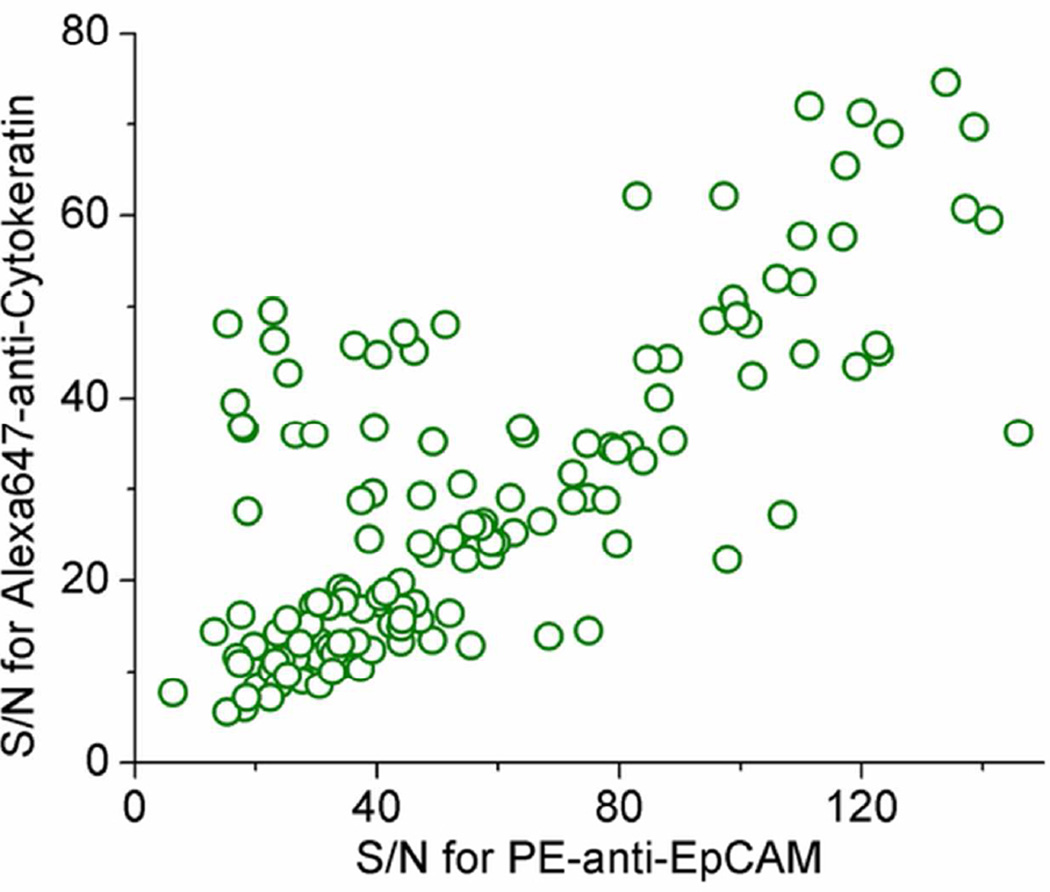
When the flow rate was increased, the mean time for a cancer cell to pass the detection region decreased, thus the S/N was lowered accordingly. To ensure that the S/N value was high enough (>10) to detect CTCs in whole blood with a flow rate between 50 to 80 µL/min, we measured S/N values for both spiked-in cells and clinical samples. Figure 2 shows the distribution of S/N values from a clinical sample (Sample ID 77) tested using EpCAM and cytokeratin as positive markers and CD45 as a negative marker. Over 95% of the data points have a S/N value higher than 20, which is high enough to identify peaks. For this sample, the average and median values of S/N in EpCAM channel are 56 and 44, respectively; the average and median values of S/N in cytokeratin channel are 29 and 24, respectively.
Figure 2.
The distribution of signal-to-noise ratio (S/N) of a breast-cancer sample analyzed by the EpCAM/cytokeratin/CD45 method. All the data points were two-color events (EpCAM+/Cytokeratin+/CD45-), which were considered to be CTCs.
Background level of detected CTCs from healthy donors and recovery values
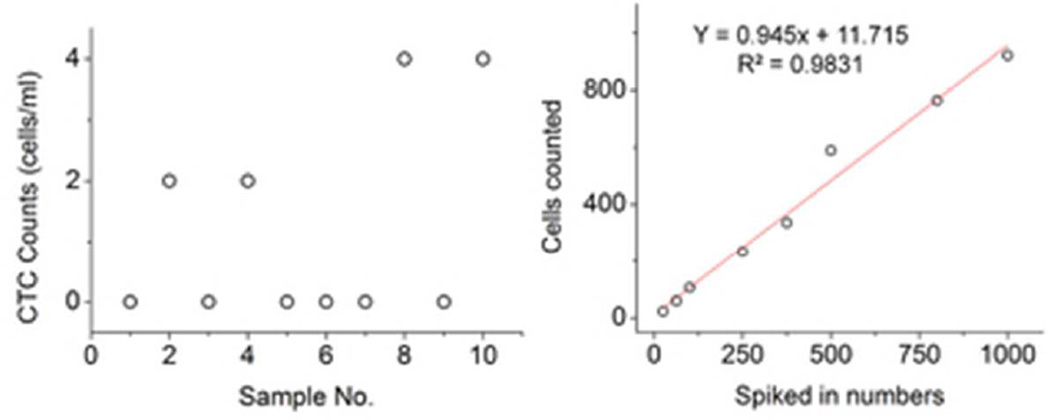
Blood is a highly complex biological fluid, so it is necessary to evaluate the background levels for any method designed to detect CTCs. In this study, 10 samples of whole blood from healthy donors were analyzed under the experimental conditions used for clinical samples. Each sample had an initial volume of 0.5 mL and the results were normalized to 1mL of blood. Similar to many other CTC detection methods, which have reported a non-zero background level20, 38–41, this method has an average background level for healthy donor blood samples of 1.2 counts/mL (Figure 3a). This value is slightly lower than, for example, a recently developed CTC capture chip that has an average background level of 2.2 cells/mL with a range of 0 to 12 cells/mL41.
Figure 3.
False positive and recovery performance. a) On average 1.2 cells were found per mL of healthy donor blood with 60% of the samples reporting zero cells. b) MCF-7 cells with known numbers were spiked into a healthy donor’s blood, which was then labeled with PE-anti-EpCAM, Alexa647-anti-cytokeratin, and FITC-anti-CD45. Enumeration results showed an average 94% recovery.
Background CTC counts may result from several different sources. It could be caused by the process of measurement. For example, similar to flow cytometry, antibodies in solution can form aggregates due to random co-localization, which lead to false positive signals. Antibody aggregation may be more prone to occur when the blood samples are fixed and permeabilized. This issue can be addressed by using more biomarkers with more fluorescent colors, thus lowering the probability of detecting a random event, i.e., antibody aggregates, with the desired combination of colors. Indeed, this type of multi-parameter analysis has been used to significantly reduce false positive rates in flow cytometry42. Alternatively, background levels may be false positive events that resulted from fluorescent dust particles in blood samples that show relatively wide emission spectra compared to labeled cells. However, the possibility of this occurring in our experiment was relatively low, because we require that the green channel (CD45; FITC) be non-fluorescent. Finally, it is possible that the background level was caused by the fact that some circulating epithelial cells are present in healthy individuals15. While this scenario is possible, we believe it is unlikely based on our previous studies33 and as long as the first tube of drawn blood is discarded to avoid contamination of epithelial cells during the venipuncture process. In any case, as long as the background levels are much fewer than in cancer patients, it should not affect the assay.
Because the background was above zero, accurate fast screening of CTCs in actual clinical practice will require a threshold to be determined by statistical analysis of the experimentally measured background, i.e., the sample is considered as CTC-positive only if the number of events is higher than that threshold. This type of threshold is used in conventional flow cytometry43. It is also applied in other published works for the enumeration of CTCs. For example39, Stott, SL et al. showed their method has a mean background level of 3 cells/mL, ranging from 0 to 8 cells/mL, in their analysis of samples from healthy donors. As a result, they set up a cut-off value at 10 cells/mL to prevent false positive detection. If the sample count falls below the threshold in our method, it can be further analyzed by a zero-false-positive method such as eDAR (0 CTCs/mL, n=9)33, because the labeled sample is not damaged during the flow detection test.
To determine the recovery efficiency of this method, 25 to 1000 SKBr-3 cells were spiked into 8 blood samples (1 mL for each sample) from healthy donors and then analyzed using the same preparation procedure used for patient samples. The average recovery was about 94% with a R2 value of 0.9831 (Figure 3b). A similar recovery rate was observed for MCF-7 cells (data not shown). This value is consistent with the recovery value of eDAR (93%)33, and is acceptable for screening CTCs from clinical samples.
Alternative labeling and detection schemes
We have also tested other possible labeling and detection schemes. Because Her2 (Human Epidermal Growth Factor Receptor 2) is a widely used biomarker in breast cancer studies (~ 25% of breast cancer patients are Her2 positive) and is the target of the monoclonal antibody trastuzumab, we designed a scheme to enumerate Her2 positive CTCs from breast cancer patients. In this scheme, the blood sample was labeled with PE-anti-EpCAM, Alexa647-anti-Her2, and FITC-anti-CD45, so a Her2 positive CTC should be positive for EpCAM but negative for CD45 (Supporting information, Figure 3S).
To test the applicability of this scheme, we did an analysis on the distribution of S/N values for another patient sample (Sample ID 103) analyzed by EpCAM/Her2/CD45, which had average S/N ratios of 57 and 35 for EpCAM and Her2 channels, respectively (Supporting information, Figure 4S). False positive studies were also performed using 1 mL of whole blood as the initial volume of each sample (n=5). No CTCs were found in 3 (60%) samples, and 1 CTC per 1 mL of whole blood was found in the other 2 (40%) samples (Figure 5S). The average background value of this scheme is about 0.4 cells/mL. The difference between the false positive values of the two strategies (EpCAM+/Her2+/CD45- and EpCAM+/Cytokeratin+/CD45-) was possibly due to the difference in the sample preparation procedure. Since Her2 is a surface antibody, the blood does not need to be fixed and permeabilized for labeling. This gave us a generally higher signal-to-noise value and a lower false positive rate, which indicates that if we use cell surface markers instead of cytoplasmic markers, we can reduce the false positive rate as well.
To demonstrate the flexibility of our labeling scheme, we also enumerated circulating cells that were CD44+/CD24-/EpCAM+, because CD44+/CD24- is a marker that has been used to identify stem cell like CTCs in previous works44. CD44+ can also be present on some blood cells, however, and our enumeration relied on a single marker, EpCAM+, to distinguish CTCs that were CD44+/CD24- from these blood cells. Therefore, to more definitively confirm these are indeed cancer stem cells, one would need to use additional markers (e.g. CD45- and Cytokeratin+) to minimize the possibility of false positives. At present, our setup is only capable of detecting 3 markers and thus this experiment also shows the advantage of being able to monitor additional markers by adding more detectors and by utilizing fluorophores with narrower emission spectra to facilitate multiplexing.
Results of clinical samples
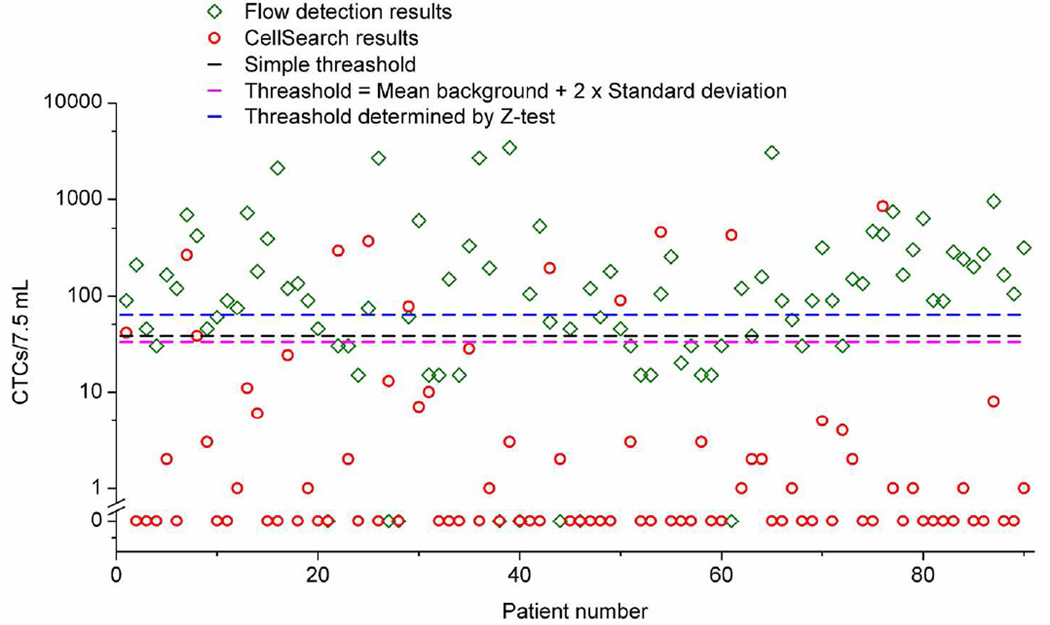
Over a two-year period, we collected 90 blood samples from 24 patients with stage IV metastatic breast cancer and performed a side-by-side study comparing the high-throughput flow detection method with the FDA-approved CellSearch system (Figure 4). CTCs in all the 90 samples reported in this paper were identified as EpCAM+/cytokeratin+/CD45-, similar to the criteria used in CellSearch analysis, which has one more nuclear stain (DAPI) marker, and does not image the EpCAM expression of the captured cells.
Figure 4.
Clinical results from the CTC flow counting system and CellSearch. Our method found a median of 90 CTCs per 7.5 mL of blood compared to a median of zero for the CellSearch system. The black dashed line is a simple threshold (38 counts/7.5 mL) set based on the range of detected CTCs in healthy donors. The magenta dashed line is the threshold (33 counts/7.5 mL) set using the mean background level plus two times of its standard deviation. The blue dashed line is the threshold (63 counts/7.5 mL) determined by Z-test with a 95% confidence level.
CTC positive events were found using the high-throughput flow detection method in 82 samples (91%), ranging from 15 to 3375 counts per 7.5 mL of blood, with an average CTC level of 305 counts per 7.5 mL of blood, and a median value of 90 counts/7.5 mL. The CellSearch system found CTCs in 44% of samples, ranging from 1 to 846 cells per 7.5 mL of blood, with an average CTC level of 36 cells per 7.5 mL of blood, and a median value of 0 cells/7.5 mL. Previous studies proposed that the relevant prognostic cutoff level of CTCs in metastatic breast cancer is 5 cells per 7.5 mL of blood as determined by the CellSearch method. In our study, only 22% of the samples had the level of CTCs higher than that threshold using the CellSearch system.
However, considering our current background level (1.2 counts/mL), it is necessary to set a threshold with a certain confidence level, so we can exclude the data points that might represent false positive signals. As we discussed previously, there are several different approaches we can follow to set this threshold. For example, we might use a simple threshold that is above the range of the detected background level as reported previously39; if we follow this approach, our threshold would be set at 38 counts/7.5 mL. Another possibility is to use a threshold that is the mean background count plus 2 standard deviations, a method reported in the flow cytometry literature43; our threshold would be set at 33 counts/7.5 mL, if we use this method. Finally, a third method is to employ the one dimensional Z-test and assumed that any error in the measurements of the clinical sample was Poisson distributed, and the threshold was for a one-sided test (more details of the statistical consideration can be found in supporting information.). For this method, the threshold would be much higher at 63 counts/7.5 mL for a 95% confidence level. Based on this more stringent threshold, our method found that 60% of the patient samples were positive to CTCs with a 95% confidence level. A detailed list of all the patient samples and CTC counts using CellSearch and our method is provided in supporting information (Table 1S). Figure 4 shows the result and the corresponding thresholds set using the three methods described above.
Of the 90 patient samples, 30 were additionally analyzed using EpCAM and CD44 as positive markers, as well as CD24 as a negative marker to enumerate CTCs with stem cell characteristics. Figure 6S (see supporting information) shows the side by side comparison of the stem cell counts and regular CTC numbers enumerated by the flow detection method, as well as the CTC numbers determined by CellSearch method. EpCAM+/CD44+/CD24- events were found in 90% of the samples with average and median values of 150 cells/7.5mL and 53 cells/7.5 mL, respectively.
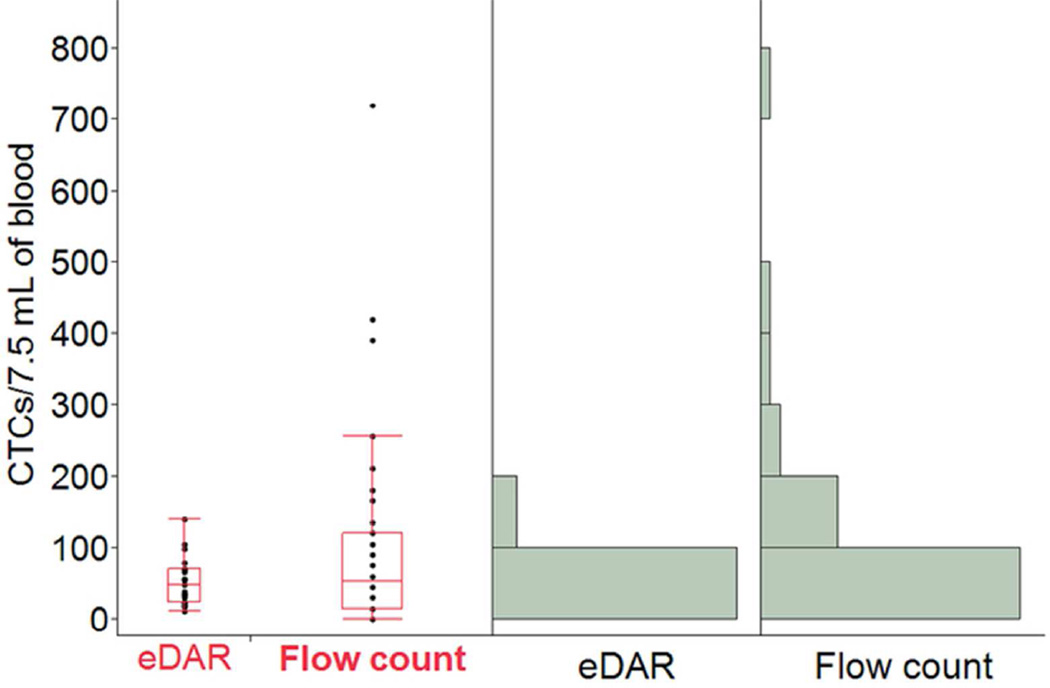
It is also interesting to compare this flow detection method with eDAR. CTC counts collected from the eDAR approach (n=22, from 9 patients) 33 and the flow detection approach (n=40, from the same 9 patients) were analyzed with Analysis of Variance (ANOVA). Figure 5 shows the boxplots and histograms for each dataset. The dataset from flow detection averaged 102 CTCs/7.5 mL, whereas that from eDAR averaged 52 CTCs/7.5 mL. However, given the large variance in the flow detection dataset, eDAR and flow detection are not significantly different (p = 0.14, α = 0.05, ANOVA test). This consistency is due to the similarity of fluidics and fluorescence detection schemes between the two methods.
Figure 5.
Comparison of the CTC enumeration results from the same set of patients using eDAR and flow detection system. The left part is the box plots that show the smallest observation, lower quartile, median, upper quartile, and the largest observation of the two data sets (eDAR vs Flow count), respectively. The right part shows the histograms of the two data sets.
Conclusion
The enumeration of CTCs is an important component of monitoring cancer progression, and the reported CTC flow counting system is an automated, quick, and sensitive method. Additionally, the system is very flexible in the sense that it can be used with a variety of markers and any user-defined fluorescence-based criterion for enumerating cells of interest can be designed. We demonstrated the ability to enumerate spiked-in cancer cells from whole blood with a high recovery efficiency of 94%. A side-by-side clinical study of 90 patient samples showed that our method is sensitive and efficient for CTC detection in patients with stage IV metastatic breast cancer. Flow detection technique is free of enrichment steps, which are required in most CTC analysis techniques, so the sample preparation is minimized and the detection and enumeration analysis is fully automated. Moreover, because it is a non-destructive analysis technique (samples simply flowed through a straight channel), samples can be re-analyzed and verified using other CTC methods. This compatibility would be ideal for a high-throughput screening application, because the positive samples may need to be further tested by other techniques to obtain more cellular and molecular information. In summary, we believe this simple, fast, and robust method is useful for the enumeration of CTCs from patient blood, and it may be potentially used for other rare-cell enumeration studies.
Supplementary Material
Acknowledgement
We wish to thank the patients who enabled this research, as well as Gabriele Shuster for arranging patient samples. We thank Dr. Wyatt Nelson for his help on the manuscript writing, and Dr. Bryant Fujimoto for his help on the statistical analysis. We gratefully acknowledge support from the Life Sciences Discovery Fund and the National Institutes of Health (R21CA147831).
Reference
- 1.Steeg PS. Nat. Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 2.Chaffer CL, Weinberg RA. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 3.Fidler I. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 4.Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, Forni G, Eils R, Fehm T, RiethmUller G, Klein CA. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Ashworth TR. Aust. Med. J. 1869;14:2. [Google Scholar]
- 6.Maheswaran S, Haber DA. Curr. Opin. Genet. Dev. 2010;20:96–99. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mostert B, Sleijfer S, Foekens JA, Gratama JW. Cancer Treat. Rev. 2009;35:463–474. doi: 10.1016/j.ctrv.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Pantel K, Brakenhoff RH, Brandt B. Nat. Rev. Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 9.Allard W, Matera J, Miller M, Repollet M, Connelly M, Rao C, Tibbe A, Uhr J, Terstappen L. Clin. Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa T, Martinez SR, Goto Y, Koyanagi K, Kitago M, Shingai T, Elashoff DA, Ye X, Singer FR, Giuliano AE, Hoon DSB. Clin. Cancer Res. 2007;13:4105–4110. doi: 10.1158/1078-0432.CCR-07-0419. [DOI] [PubMed] [Google Scholar]
- 11.Hartkopf AD, Banys M, Krawczyk N, Wallwiener M, Schneck H, Neubauer H, Fehm T. Geburtshilfe Frauenheilkd. 2011;71:1067–1072. doi: 10.1055/s-0031-1280463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aurilio G, Sciandivasci A, Munzone E, Sandri MT, Zorzino L, Cassatella MC, Verri E, Rocca MC, Nole F. Expert Rev. Anticancer Ther. 2012;12:203–214. doi: 10.1586/era.11.208. [DOI] [PubMed] [Google Scholar]
- 13.Bono JSd, Ashworth A. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 14.Mego M, Mani SA, Cristofanilli M. Nature Reviews Clinical Oncology. 2010;7:693–701. doi: 10.1038/nrclinonc.2010.171. [DOI] [PubMed] [Google Scholar]
- 15.Paterlini-Brechot P, Benali NL. Cancer Lett. 2007;253:180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Dharmasiri U, Witek MA, Adams AA, Soper SA. Annu. Rev. Anal. Chem. 2010;3:409–431. doi: 10.1146/annurev.anchem.111808.073610. [DOI] [PubMed] [Google Scholar]
- 17.Alunni-Fabbroni M, Sandri MT. Methods. 2010;50:289–297. doi: 10.1016/j.ymeth.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 18.Gross HJ, Verwer B, Houck D, Hoffman RA, Recktenwald D. Proc. Natl. Acad. Sci. U. S. A. 1995;92:537–541. doi: 10.1073/pnas.92.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh HB, Marrinucci D, Bethel K, Curry DN, Humphrey M, Krivacic RT, Kroener J, Kroener L, Ladanyi A, Lazarus N, Kuhn P, Bruce RH, Nieva J. Biosens. Bioelectron. 2006;21:1893–1899. doi: 10.1016/j.bios.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 20.Krivacic RT, Ladanyi A, Curry DN, Hsieh HB, Kuhn P, Bergsrud DE, Kepros JF, Barbera T, Ho MY, Chen LB, Lerner RA, Bruce RH. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10501–10504. doi: 10.1073/pnas.0404036101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riethdorf S, Mueller V, Zhang L, Rau T, Loibl S, Komor M, Roller M, Huober J, Fehm T, Schrader I, Hilfrich J, Holms F, Tesch H, Eidtmann H, Untch M, von Minckwitz G, Pantel K. Clin. Cancer Res. 2010;16:2634–2645. doi: 10.1158/1078-0432.CCR-09-2042. [DOI] [PubMed] [Google Scholar]
- 22.Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, Sheth S, Kurian AW, Ford JM, Stockdale FE, Quake SR, Pease RF, Mindrinos MN, Bhanot G, Dairkee SH, Davis RW, Jeffrey SS. PLoS ONE. 2012;7:1. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dharmasiri U, Njoroge SK, Witek MA, Adebiyi MG, Kamande JW, Hupert ML, Barany F, Soper SA. Anal. Chem. 2011;83:2301–2309. doi: 10.1021/ac103172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ST, Liu K, Liu JA, Yu ZTF, Xu XW, Zhao LB, Lee T, Lee EK, Reiss J, Lee YK, Chung LWK, Huang JT, Rettig M, Seligson D, Duraiswamy KN, Shen CKF, Tseng HR. Angew. Chem. Int. Ed. 2011;50:3084–3088. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. J. Chromatogr. A. 2007;1162:154–161. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 27.Kahn HJ, Presta A, Yang LY, Blondal J, Trudeau M, Lickley L, Holloway C, McCready DR, Maclean D, Marks A. Breast Cancer Res. Treat. 2004;86:237–247. doi: 10.1023/B:BREA.0000036897.92513.72. [DOI] [PubMed] [Google Scholar]
- 28.Kuo JS, Zhao YX, Schiro PG, Ng LY, Lim DSW, Shelby JP, Chiu DT. Lab Chip. 2010;10:837–842. doi: 10.1039/b922301k. [DOI] [PubMed] [Google Scholar]
- 29.Pinzani P, Salvadori B, Simi L, Bianchi S, Distante V, Cataliotti L, Pazzagli M, Orlando C. Hum. Pathol. 2006;37:711–718. doi: 10.1016/j.humpath.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 30.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, Fleisher M, Stadler W, Datar RH, Tai Y-C, Cote RJ. Clin. Cancer Res. 2010;16:5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balic M, Dandachi N, Hofmann G, Samonigg H, Loibner H, Obwaller A, van der Kooi A, Tibbe AGJ, Doyle GV, Terstappen L, Bauernhofer T. Cytometry Part B-Clinical Cytometry. 2005;68B:25–30. doi: 10.1002/cyto.b.20065. [DOI] [PubMed] [Google Scholar]
- 32.Balasubramanian P, Yang L, Lang JC, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ. Mol. Pharm. 2009;6:1402–1408. doi: 10.1021/mp9000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiro PG, Zhao M, Kuo JS, Koehler KM, Sabath DE, Chiu DT. Angew. Chem. Int. Ed. 2012;51:4618–4622. doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiro PG, Kuyper CL, Chiu DT. Electrophoresis. 2007;28:2430–2438. doi: 10.1002/elps.200600730. [DOI] [PubMed] [Google Scholar]
- 35.Jeffries GDM, Lorenz RM, Chiu DT. Anal. Chem. 2010;82:9948–9954. doi: 10.1021/ac102173m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patriarca C, Macchi RM, Marschner AK, Mellstedt H. Cancer Treat. Rev. 2012;38:68–75. doi: 10.1016/j.ctrv.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JWM, Gratama JW, Sleijfer S, Foekens JA. J. Natl. Cancer Inst. 2009;101:61–66. doi: 10.1093/jnci/djn419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goda K, Ayazi A, Gossett DR, Sadasivam J, Lonappan CK, Sollier E, Fard AM, Hur SC, Adam J, Murray C, Wang C, Brackbill N, Di Carlo D, Jalali B. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11630–11635. doi: 10.1073/pnas.1204718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L, Ko AH, Korn WM, Schram E, Coward M, Yang X, Metzner T, Lamy R, Honnatti M, Yoshioka C, Kunken J, Petrova Y, Sok D, Nelson D, Kuhn P. Phys. Biol. 2012;9 [Google Scholar]
- 41.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, Moore AL, Tsukrov DI, Kempner ME, Dahl DM, Wu C-L, Iafrate AJ, Smith MR, Tompkins RG, Sequist LV, Toner M, Haber DA, Maheswaran S. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daugherty PS, Iverson BL, Georgiou G. J. Immunol. Methods. 2000;243:211–227. doi: 10.1016/s0022-1759(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 43.Bottcher S, Ritgen M, Pott C, Bruggemann M, Raff T, Stilgenbauer S, Dohner H, Dreger P, Kneba M. Leukemia. 2004;18:1637–1645. doi: 10.1038/sj.leu.2403478. [DOI] [PubMed] [Google Scholar]
- 44.Theodoropoulos PA, Polioudaki H, Agelaki S, Kallergi G, Saridaki Z, Mavroudis D, Georgoulias V. Cancer Lett. 2010;288:99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.