Abstract
Circulating cell-free DNA has been recognized as an accurate marker for the diagnosis of prostate cancer, whereas the role of urine cell-free DNA (UCF DNA) has never been evaluated in this setting. It is known that normal apoptotic cells produce highly fragmented DNA while cancer cells release longer DNA. We thus verified the potential role of UCF DNA integrity for early prostate cancer diagnosis. UCF DNA was isolated from 29 prostate cancer patients and 25 healthy volunteers. Sequences longer than 250 bp (c-Myc, BCAS1, and HER2) were quantified by real-time PCR to verify UCF DNA integrity. Receiver operating characteristic (ROC) curve analysis revealed an area under the curve of 0.7959 (95% CI 0.6729–0.9188). At the best cut-off value of 0.04 ng/μL, UCF DNA integrity analysis showed a sensitivity of 0.79 (95% CI 0.62–0.90) and a specificity of 0.84 (95% CI 0.65–0.94). These preliminary findings indicate that UCF DNA integrity could be a promising noninvasive marker for the early diagnosis of prostate cancer and pave the way for further research into this area.
1. Introduction
Early diagnosis plays an important role in the treatability of patients with different tumor types in terms of disease-free and overall survival. Prostate cancer has a high incidence and represents the second cause of death from cancer in men after lung cancer. Early diagnosis is thus essential, especially in view of the slow natural history of the disease and its potential curability in the initial hormone-dependent phase. Non-invasive diagnostic procedures have a higher patient compliance and a lower cost than invasive screening programs. At present, the only noninvasive approach currently used for the diagnosis of prostate cancer is the determination of PSA (prostate-specific antigen) in blood, which has been shown to reduce prostate cancer mortality. However, the use of PSA has recently been questioned because of its low accuracy, especially in terms of specificity. False positive results lead to overtreatment in individuals, with consequently higher healthcare costs and psychological distress [1–5]. Although great efforts have been made to improve the diagnostic accuracy of PSA, the search continues for new molecular markers, proteins, or genetic and epigenetic alterations [6, 7] to be used in this setting.
New accurate and cost-effective diagnostic approaches are needed to enhance or replace standard techniques for prostate cancer diagnosis. Cell-free nucleic acids have proven useful for early cancer diagnosis and positive results have also been published on serum and plasma cell-free DNA and RNA as sources of tumor-specific markers [8, 9]. Circulating cell-free DNA has been shown to play an important diagnostic role in colon [10] and lung cancer [11], and a number of studies have also highlighted its potential usefulness in prostate cancer [12–14]. Urine cell-free (UCF) DNA as a source of tumor biomarkers has not been adequately investigated in prostate cancer and only a few recent studies have discussed its potential importance for early bladder cancer diagnosis [15–17].
It has been shown that DNA from normal apoptotic cells is highly fragmented, whereas DNA from necrotic cancer cells maintains its integrity [18]. Taking this into account and also considering recent results on bladder cancer highlighting the importance of UCF DNA integrity for early diagnosis [15], we investigated the ability of this marker to distinguish between prostate cancer patients and healthy individuals by analyzing UCF-DNA fragments longer than 250 bp in 3 regions is known to be frequently amplified in solid tumors, including prostate cancer: c-Myc (8q24.21), HER2 (17q12.1), and BCAS1 (20q13.2) [19–21].
2. Materials and Methods
2.1. Case Series
This pilot study was composed of 54 individuals, 29 at first diagnosis of prostate cancer and 25 healthy individuals (control group) matched to patients for age. Subjects with previous or concomitant urogenital diseases or cancers were excluded from the study. Healthy individuals underwent transrectal ultrasound (TRUS) to exclude the presence of prostate cancer. Participants were recruited from the Department of Urology of Morgagni, Pierantoni Hospital (Forli) and all provided written informed consent to take part in the study, which was reviewed and approved by the local Ethics Committee. Median age was 65 years for patients and 66 for healthy individuals. All patients underwent radical prostatectomy. The Gleason score and pathological stage were evaluated after surgical removal of the tumor. Twelve patients had a Gleason score of ≤6 and 17 patients had a score of >6. Two patients had pT2a tumors, 14 pT2b, 10 pT3a, and 3 pT3c. The median PSA value was 7.5 (range 3.19–33) (Table 1).
Table 1.
Case series.
| Number | Age (yrs) | Median PSA (range) | Gleason score | Pathological stage | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| <70 | ≥70 | ≤6 | >6 | pT2a | pT2b | pT3a | pT3c | |||
| Healthy individuals | 25 | 15 | 10 | — | — | — | — | — | — | — |
| Prostate cancer patients | 29 | 20 | 9 | 7.5 ( 3.19–33) | 12 | 17 | 2 | 14 | 10 | 3 |
2.2. Urine Collection
First-morning-void urine samples were collected for UCF DNA analysis. For prostate cancer patients, specimens were collected before radical prostatectomy. All patients and controls were instructed to give clean-catch urine samples, which were maintained at 4°C for a maximum of 3 hours. Thirty milliliter aliquots of urine were centrifuged at 850 g for 10 minutes and the supernatants were transferred to cryovials and immediately stored at −80°C until use.
2.3. UCF DNA Analysis
DNA was extracted and purified from 2 mL of supernatant by Qiamp DNA minikit (Qiagen, Milan, Italy) according to the manufacturer's instructions. At the same time, DNA was extracted from a human bladder cancer cell line (MCR) using the same minikit and quantified by spectrophotometry (NanoDrop ND-1000, Celbio, Milan, Italy).
Real-time PCR reactions were carried out by Rotor Gene 6000 detection system (Corbett Research, St. Neots, UK) using IQ SYBR green (Biorad, Milan, Italy). Sequences longer than 250 bp corresponding to 3 oncogenes were analyzed as follows: c-Myc (locus 8q24.21, amplification product 264 bp), BCAS1 (locus 20q.13.2, amplification product 266 bp), and HER2 (locus 17q12.1, amplification product 295 bp). A short 125 bp fragment of STOX1 (locus 10q21.3) was analyzed to check for potential PCR inhibition. Primer sequences were as follows: c-Myc fw TGGAGTAGGGACCGCATATC, rev ACCCAACACCACGTCCTAAC; BCAS1 fw GGGTCAGAGCTTCCTGTGAG, rev CGTTGTCCTGAAACAGAGCA; HER2 fw CCAGGGTGTTCCTCAGTTGT, rev TCAGTATGGCCTCACCCTTC; STOX1 fw GAAAACAGGGCAGCAAGAAG, rev CAGACAGCATGGAGGTGAGA. PCR conditions for the oncogenes were as follows: 95°C for 3 minutes followed by 45 cycles at 94°C for 40 seconds, 56°C for 40 seconds, and 72°C for 1 minute. PCR conditions for the short STOX1 sequence were as follows: 95°C for 90 seconds followed by 45 cycles at 94°C for 40 seconds and 54°C for 1 minute. All real-time PCR reactions were performed in duplicate on 10 ng of each UCF DNA sample. Various amounts of DNA from the MCR cell line (0.01, 0.1, 1, 5, 10, and 20 ng) were also analyzed to construct a standard curve. The UCF DNA value for each sample was obtained by Rotor Gene 6000 detection system software using standard curve interpolation. The analysis was repeated if the difference between duplicate samples was greater than 1 cycle threshold. The final UCF DNA integrity value was obtained by summing the three oncogene values. Real-time experiments were performed independently in duplicate on the same 8 samples to test assay variability. The coefficients of variation (CV) were then calculated for c-Myc, HER2, BCAS, and STOX1. Real-time PCR analyses were performed in accordance with MIQE guidelines (remarks to the MIQE checklist are included as Supplementary Table 1 available online at http://dx.doi.org/10.1155/2013/270457) [22].
2.4. Statistical Analysis
The relationship between UCF DNA integrity values in the two subgroups was analyzed using a nonparametric ranking statistic test. The most discriminating cut-off values between healthy individuals and cancer patients were identified using receiver operating characteristic (ROC) curve analysis. True positive rates (sensitivity) were plotted against false positive rates (1-specificity) for all classification points. Accuracy was measured by the area under the ROC curve (AUC), which represents an average probability of correctly classifying a case chosen at random. Study endpoints were sensitivity (the proportion of cancer patients who were correctly identified by the test or procedures) and specificity (the proportion of healthy individuals who were correctly identified), with their 95% confidence intervals (CIs) [23]. P values <0.05 were considered statistically significant. Statistical analyses were performed using SPSS statistical software (version 12.0, SPSS GmbH Software).
3. Results
Total free DNA showed a median value of 6 ng/μL (range 2–36 ng/μL). There was no statistically significant difference between total urine cell free DNA in cancer patients and healthy individuals (P = 0.1200 Wilcoxon-Mann-Whitney test).
The ROC curve for total free DNA showed an AUC of 0.6262 (Supplementary Figure 1). UCF DNA integrity analysis was feasible and results were evaluable for all 54 individuals. The 125 bp STOX1 sequence was amplified in all samples, thus excluding the presence of PCR inhibitors. Values showed a wide variability in both healthy individuals and cancer patients, with a partial overlapping. However, median values were significantly lower (about 20-fold, P = 0.0004) in healthy than in cancer patients (Table 2).
Table 2.
UCF DNA integrity in healthy individuals and prostate cancer patients.
| UCF DNA integrity (ng/μL) | ||||
|---|---|---|---|---|
| Number | Median values (range) | Mean values (range) | P* | |
| Healthy individuals | 25 | 0.007 (0–0.141) | 0.023 (0–0.141) | 0.0004 |
| Prostate cancer patients | 29 | 0.129 (0–5.379) | 0.533 (0–5.379) | |
*Wilcoxon-Mann-Whitney test.
ROC curve analysis of UCF DNA integrity showed an AUC of 0.7959 (0.6729–0.9188) for healthy individuals and cancer patients (Figure 1). Detailed analysis of sensitivity and specificity highlighted a different accuracy for the various UCF DNA cut-off values, with a sensitivity of 0.79 for 0.03 and 0.04 cutoffs which decreased at the highest cut-off values and a specificity of 0.84 which remained consistent for all cutoffs from 0.03 to 0.06 (Table 3).
Figure 1.
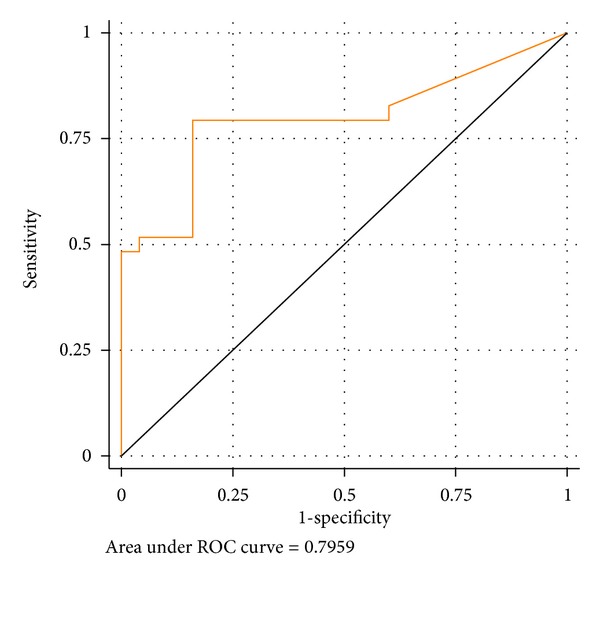
ROC curve of UCF DNA integrity.
Table 3.
Sensitivity and specificity of different UCF DNA integrity cut-off values.
| Cutoff (ng/μL) | Sensitivity | Specificity |
|---|---|---|
| 0.03 | ||
| n | 23/29 | 21/25 |
| Rate (95% CI) | 0.79 (0.62–0.90) | 0.84 (0.65–0.94) |
| 0.04 | ||
| n | 23/29 | 21/25 |
| Rate (95% CI) | 0.79 (0.62–0.90) | 0.84 (0.65–0.94) |
| 0.05 | ||
| n | 19/29 | 21/25 |
| Rate (95% CI) | 0.66 (0.47–0.80) | 0.84 (0.65–0.94) |
| 0.06 | ||
| n | 17/29 | 21/25 |
| Rate (95% CI) | 0.59 (0.41–0.74) | 0.84 (0.65–0.94) |
UCF DNA integrity did not significantly vary between younger (<70 years) and older individuals (data not shown). The analysis of UCF DNA as a function of tumor characteristics did not highlight any significant differences between patients with a Gleason score of ≤6 and those with a score of >6 or between pT2 and pT3 patients (data not shown).
The median PSA value in the patients analyzed was 7.5. Sixteen patients had a PSA value between 4 and 10, considered as a “gray zone,” and 12 of these had a positive UCF DNA result, with a sensitivity of 0.75 (data not shown). We also performed ROC curve analysis for each gene in order to verify the role of single genes in determining test accuracy; AUC values were as follows: 0.7862 for c-MYC (95% CI: 0.6595–0.9129) 0.7779 for HER2 (95% CI: 0.6625–0.8934) and 0.7076 for BCAS (95% CI: 0.5771–0.8381) (Table 4). However, the AUC values observed for the different genes were not statistically different (chi-square test).
Table 4.
Area under ROC curve for each single gene and for UCF DNA integrity.
| AUC (95% CI) | P* | |
|---|---|---|
| c-Myc | 0.7862 (0.6595–0.9129) | |
| BCAS1 | 0.7076 (0.5771–0.8381) | NS** |
| HER2 | 0.7779 (0.6625–0.8934) | |
| UCF DNA integrity | 0.7959 (0.6729–0.9188) |
*Chi-square test.
**NS: not significant.
4. Discussion
In recent years increasing efforts have been made to identify new diagnostic markers and to develop noninvasive diagnostic approaches that can be used additionally or as an alternative to common invasive tests to increase diagnostic accuracy for solid tumors. Important results have been obtained for lung [11] and colon cancer [10]. In a urological setting, studies performed to improve the early noninvasive diagnosis of prostate cancer have highlighted the usefulness of specific DNA alterations (methylation or mutations) in blood to identify cancer patients [12, 24, 25], but the potential of cell-free DNA in urine has been never investigated.
Starting from recent results on the diagnostic relevance of urine cell free DNA for bladder cancer [15–17], we extended the research to prostate cancer, hypothesizing that long DNA in urine may have two different origins: necrotic bladder cancer cells [15] or necrotic prostate cancer cells. We excluded that DNA fragments passing through the glomerular filtration barrier could influence our analysis because these fragments are short, as demonstrated by SU and coworkers [26]. Our results also showed that urine DNA integrity is capable of distinguishing between prostate cancer patients and healthy individuals with an accuracy of about 80%, similar to that observed for bladder cancer. Such findings are seemingly in contrast to those of Ellinger and coworkers who found a positive correlation between prostate cancer and the presence of short DNA fragments in blood [25]. The difference between cell free DNA in urine or blood remains unclear and should be investigated by comparing DNA integrity determined in blood and urine samples from the same patients.
We also analyzed the diagnostic accuracy of DNA integrity of three oncogenes (c-MYC, HER2, and BCAS1) which are known to be involved in the development of bladder cancer. A comparison of ROC curves revealed that c-MYC had the highest AUC, a finding supported by evidence that c-MYC is involved in prostate tumorigenesis [27]. Furthermore, literature data on CGH array and copy number alterations highlight a high frequency of gain at 8q24 region where the c-MYC oncogene maps [19, 28–30], which could explain the higher number of copies of long c-MYC fragments in urine samples from prostate cancer patients than in those from healthy individuals. Lower diagnostic accuracy was observed for HER2 and decreased further for BCAS1, but the AUC values observed for the different genes were not significantly different.
The main limitation of this potentially important diagnostic finding is that it was obtained from a pilot study on a relatively small number of individuals. However, our results are being validated in a large confirmatory study ongoing at our institute. The advantage of the proposed approach is that cell free DNA, as previously shown [15], can be easily detected in a very small amount of urine. Moreover, unlike protein or RNA, it has good stability and is an inexpensive noninvasive method whose results are obtainable in about two working days. In the future it could be used as a test on its own or, thanks to its high specificity, could help to unmask cases of false positive PSA, especially in the subgroup of individuals with grey zone PSA values, thus reducing the number of unnecessary invasive diagnostic tests (e.g., prostate biopsy) carried out.
5. Conclusions
The results obtained in the present work indicate that urine cell-free DNA integrity is a potentially good marker for the early diagnosis of noninvasive prostate cancers, with an overall diagnostic accuracy of about 80%. This preliminary finding paves the way for confirmatory studies on larger case series.
Supplementary Material
Supplementary Table: Remarks to MIQE checklist.
Supplementary Figure: Receiver operating characteristic (ROC) curve analysis of total urine cell free DNA quantified with Nanodrop ND-1000.
Conflict of Interests
The authors have no conflict of interests to declare.
Acknowledgment
The authors thank Grainne Tierney for editing the paper.
References
- 1.Strope SA, Andriole GL. Prostate cancer screening: current status and future perspectives. Nature Reviews Urology. 2010;7(9):487–493. doi: 10.1038/nrurol.2010.120. [DOI] [PubMed] [Google Scholar]
- 2.Schröder FH, Hugosson J, Roobol MJ, et al. Prostate-cancer mortality at 11 years of follow-up. New England Journal of Medicine. 2012;366(11):981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schröder FH, Hugosson J, Tammela TLJ, et al. ERSPC Screening and prostate-cancer mortality in a randomized European study. New England Journal of Medicine. 2009;360(13):1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 4.Heijnsdijk EAM, Der Kinderen A, Wever EM, Draisma G, Roobol MJ, De Koning HJ. Overdetection, overtreatment and costs in prostate-specific antigen screening for prostate cancer. British Journal of Cancer. 2009;101(11):1833–1838. doi: 10.1038/sj.bjc.6605422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brawley OW, Ankerst DP, Thompson IM. Screening for prostate cancer. CA Cancer Journal for Clinicians. 2009;59(4):264–273. doi: 10.3322/caac.20026. [DOI] [PubMed] [Google Scholar]
- 6.Groskopf J, Aubin SMJ, Deras IL, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clinical Chemistry. 2006;52(6):1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 7.Wu T, Giovannucci E, Welge J, Mallick P, Tang WY, Ho SM. Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: a meta-analysis. British Journal of Cancer. 2011;105(1):65–73. doi: 10.1038/bjc.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature Reviews Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 9.Jung K, Fleischhacker M, Rabien A. Cell-free DNA in the blood as a solid tumor biomarker-A critical appraisal of the literature. Clinica Chimica Acta. 2010;411(21-22):1611–1624. doi: 10.1016/j.cca.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 10.Mead R, Duku M, Bhandari P, Cree IA. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. British Journal of Cancer. 2011;105(2):239–245. doi: 10.1038/bjc.2011.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulivi P, Mercatali L, Zoli W, et al. Serum free DNA and COX-2 mRNA expression in peripheral blood for lung cancer detection. Thorax. 2008;63(9):843–844. doi: 10.1136/thx.2008.102178. [DOI] [PubMed] [Google Scholar]
- 12.Ellinger J, Müller DC, Müller SC, et al. Circulating mitochondrial DNA in serum: a universal diagnostic biomarker for patients with urological malignancies. Urologic Oncology. 2012;30(4):509–515. doi: 10.1016/j.urolonc.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Gordian E, Ramachandran K, Reis IM, Manoharan M, Soloway MS, Singal R. Serum free circulating DNA is a useful biomarker to distinguish benign versus malignant prostate disease. Cancer Epidemiology Biomarkers and Prevention. 2010;19(8):1984–1991. doi: 10.1158/1055-9965.EPI-10-0287. [DOI] [PubMed] [Google Scholar]
- 14.Schwarzenbach H, Alix-Panabières C, Müller I, Letang N, Vendrell J-P, Rebillard X. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clinical Cancer Research. 2009;15(3):1032–1038. doi: 10.1158/1078-0432.CCR-08-1910. [DOI] [PubMed] [Google Scholar]
- 15.Casadio V, Calistri D, Tebaldi M, et al. Urine Cell-Free DNA integrity as a marker for early bladder cancer diagnosis: preliminary data. Urologic Oncology. 2012 doi: 10.1016/j.urolonc.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 16.Zancan M, Galdi F, Di Tonno F, et al. Evaluation of cell-free DNA in urine as a marker for bladder cancer diagnosis. International Journal of Biological Markers. 2009;24(3):147–155. doi: 10.1177/172460080902400304. [DOI] [PubMed] [Google Scholar]
- 17.Szarvas T, Kovalszky I, Bedi K, et al. Deletion analysis of tumor and urinary DNA to detect bladder cancer: urine supernatant versus urine sediment. Oncology Reports. 2007;18(2):405–409. [PubMed] [Google Scholar]
- 18.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Research. 2001;61(4):1659–1665. [PubMed] [Google Scholar]
- 19.Ishkanian AS, Mallof CA, Ho J, et al. High-resolution array CGH identifies novel regions of genomic alteration in intermediate-risk prostate cancer. Prostate. 2009;69(10):1091–1100. doi: 10.1002/pros.20959. [DOI] [PubMed] [Google Scholar]
- 20.Oxley JD, Winkler MH, Gillatt DA, Peat DS. Her-2/neu oncogene amplification in clinically localised prostate cancer. Journal of Clinical Pathology. 2002;55(2):118–120. doi: 10.1136/jcp.55.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabach Y, Sakin IK, Buganim Y, et al. Amplification of the 20q chromosomal arm occurs early in tumorigenic transformation and may initiate cancer. PLoS ONE. 2011;6(1) doi: 10.1371/journal.pone.0014632.e14632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 23.Bossuyt PM, Reitsma JB, Bruns DE, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Clinical Chemistry. 2003;49(1):1–6. doi: 10.1373/49.1.1. [DOI] [PubMed] [Google Scholar]
- 24.Altimari A, Grigioni AD, Benedettini E, et al. Diagnostic role of circulating free plasma DNA detection in patients with localized prostate cancer. American Journal of Clinical Pathology. 2008;129(5):756–762. doi: 10.1309/DBPX1MFNDDJBW1FL. [DOI] [PubMed] [Google Scholar]
- 25.Ellinger J, Bastian PJ, Haan KI, et al. Noncancerous PTGS2 DNA fragments of apoptotic origin in sera of prostate cancer patients qualify as diagnostic and prognostic indicators. International Journal of Cancer. 2008;122(1):138–143. doi: 10.1002/ijc.23057. [DOI] [PubMed] [Google Scholar]
- 26.Su YH, Wang M, Brenner DE, et al. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be used in the detection of colorectal cancer. Journal of Molecular Diagnostics. 2004;6(2):101–107. doi: 10.1016/S1525-1578(10)60497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeager M, Chatterjee N, Ciampa J, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nature Genetics. 2009;41(10):1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Liu W, Adams TS, et al. DNA copy number alterations in prostate cancers: a combined analysis of published CGH studies. Prostate. 2007;67(7):692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Xie CC, Zhu Y, et al. Homozygous deletions and recurrent amplications implicate new genes involved in prostate cancer. Neoplasia. 2008;10(8):897–907. doi: 10.1593/neo.08428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Duin M, van Marion R, Vissers K, et al. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes and Cancer. 2005;44(4):438–449. doi: 10.1002/gcc.20259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table: Remarks to MIQE checklist.
Supplementary Figure: Receiver operating characteristic (ROC) curve analysis of total urine cell free DNA quantified with Nanodrop ND-1000.


