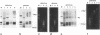Abstract
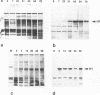
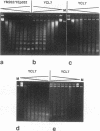
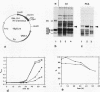
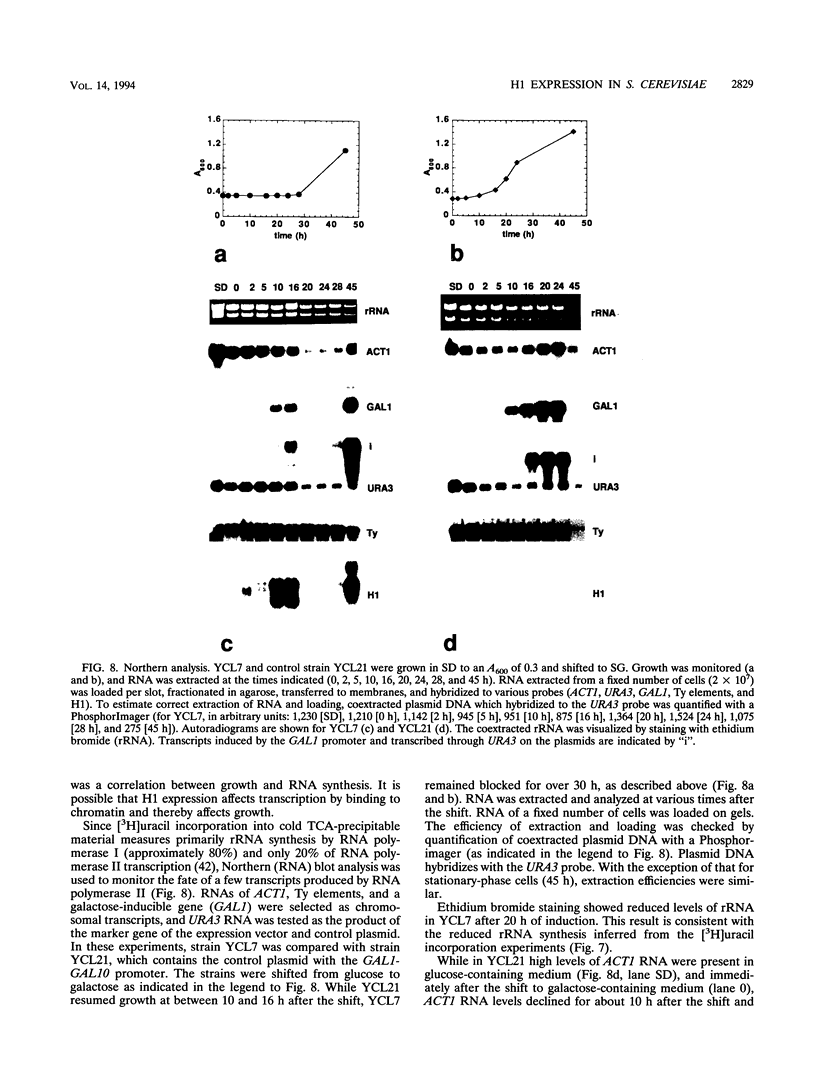
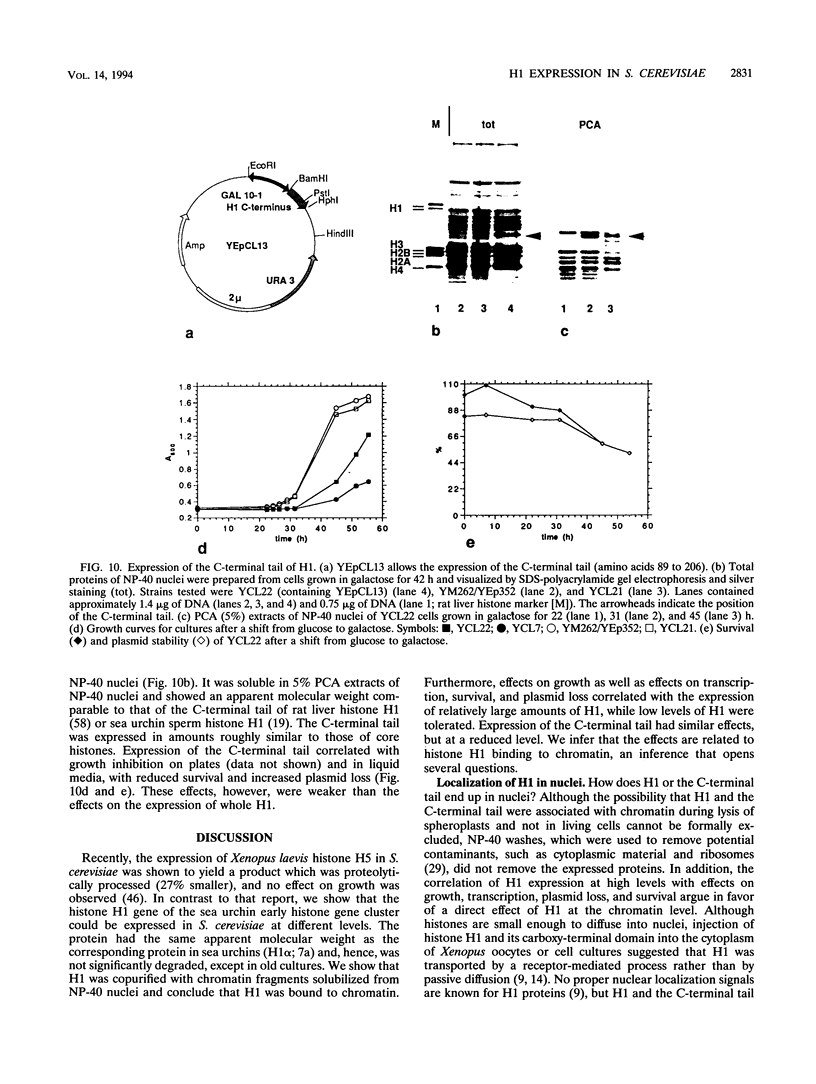
Histone H1 is proposed to serve a structural role in nucleosomes and chromatin fibers, to affect the spacing of nucleosomes, and to act as a general repressor of transcription. To test these hypotheses, a gene coding for a sea urchin histone H1 was expressed from the inducible GAL1 promoter in Saccharomyces cerevisiae by use of a YEp vector for high expression levels (strain YCL7) and a centromere vector for low expression levels (strain YCL1). The H1 protein was identified by its inducibility in galactose, its apparent molecular weight, and its solubility in 5% perchloric acid. When YCL7 was shifted from glucose to galactose for more than 40 h to achieve maximal levels of H1, H1 could be copurified in approximately stoichiometric amounts with core histones of Nonidet P-40-washed nuclei and with soluble chromatin fractionated on sucrose gradients. While S. cerevisiae tolerated the expression of low levels of H1 in YCL1 without an obvious phenotype, the expression of high levels of H1 correlated with greatly reduced survival, inhibition of growth, and increased plasmid loss but no obvious change in the nucleosomal repeat length. After an initial induction, RNA levels for GAL1 and H1 were drastically reduced, suggesting that H1 acts by the repression of galactose-induced genes. Similar effects, but to a lower extent, were observed when the C-terminal tail of H1 was expressed.
Full text
PDF


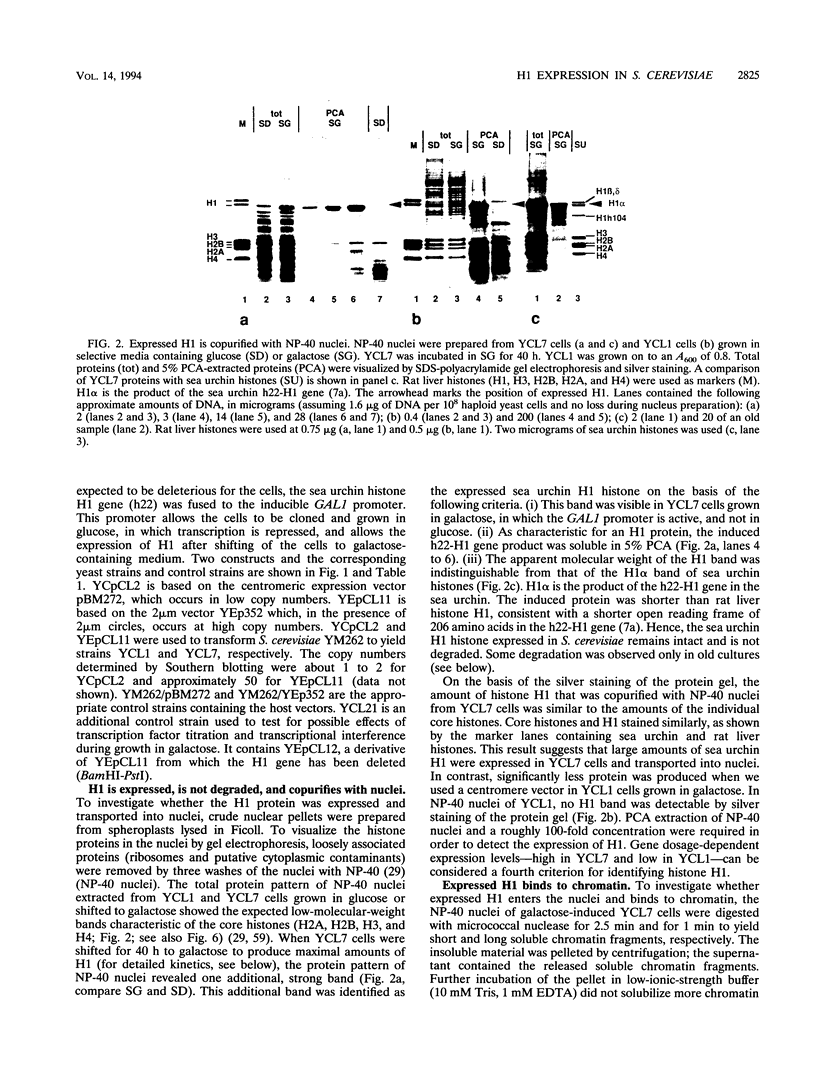
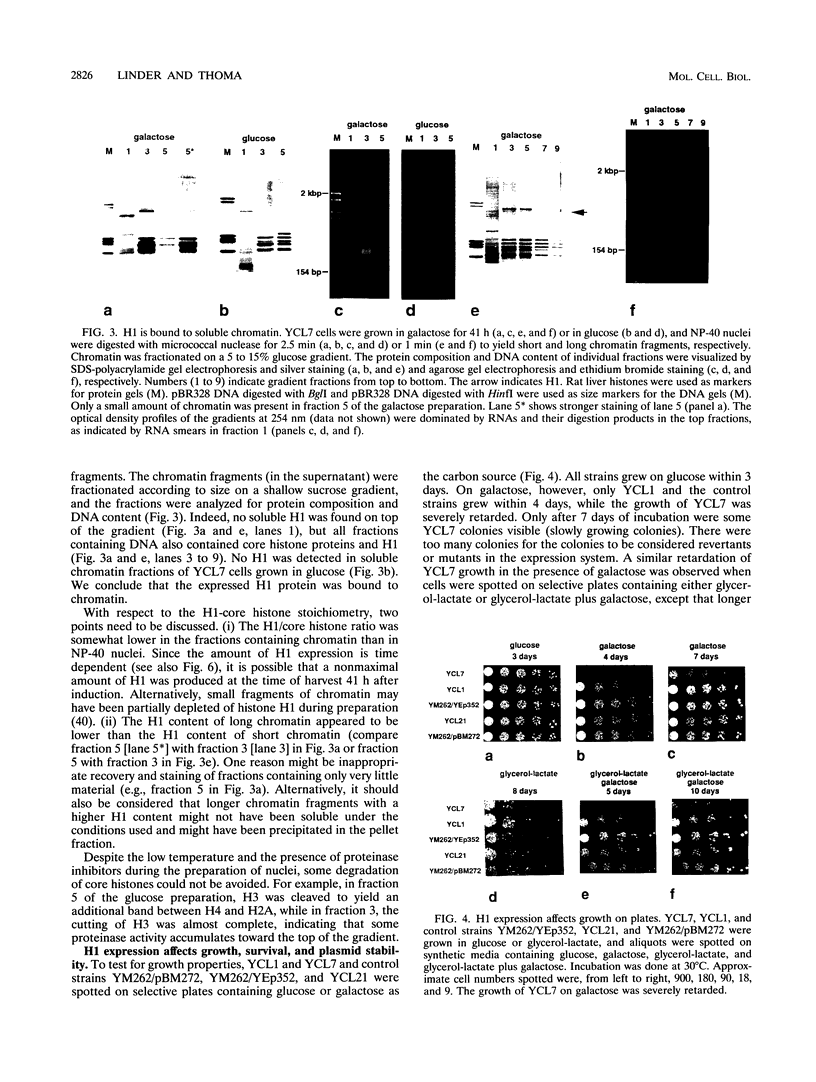
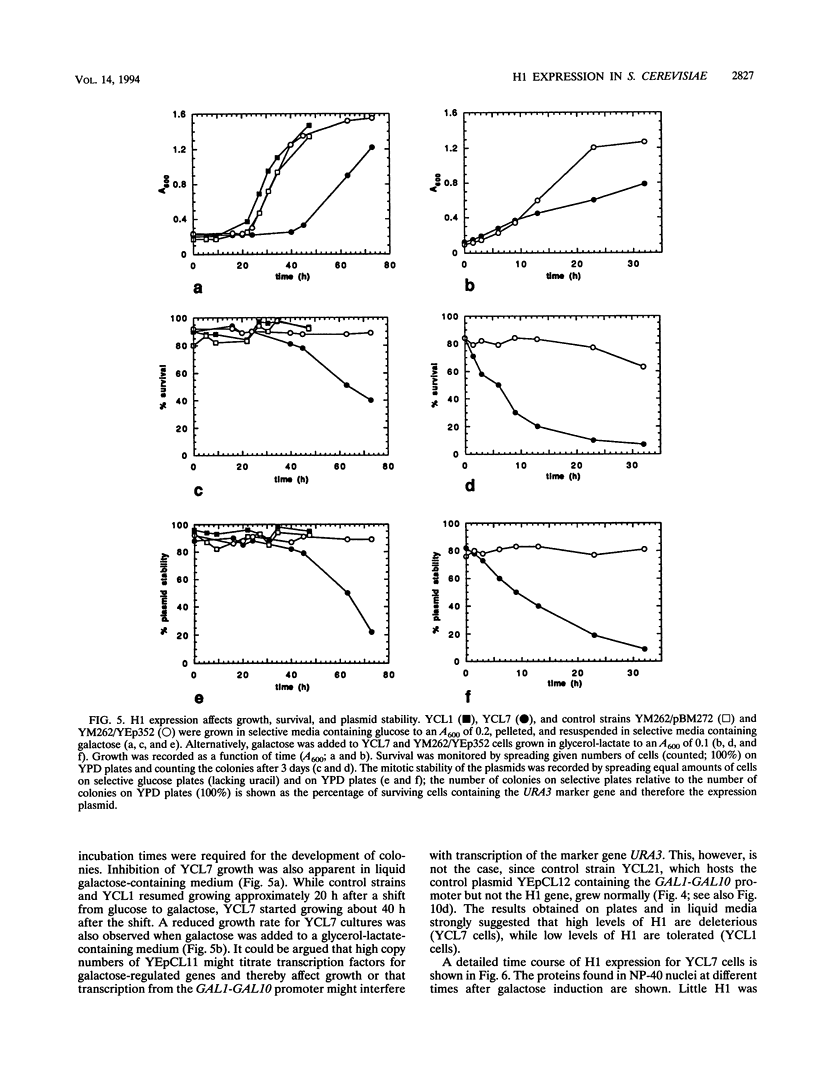
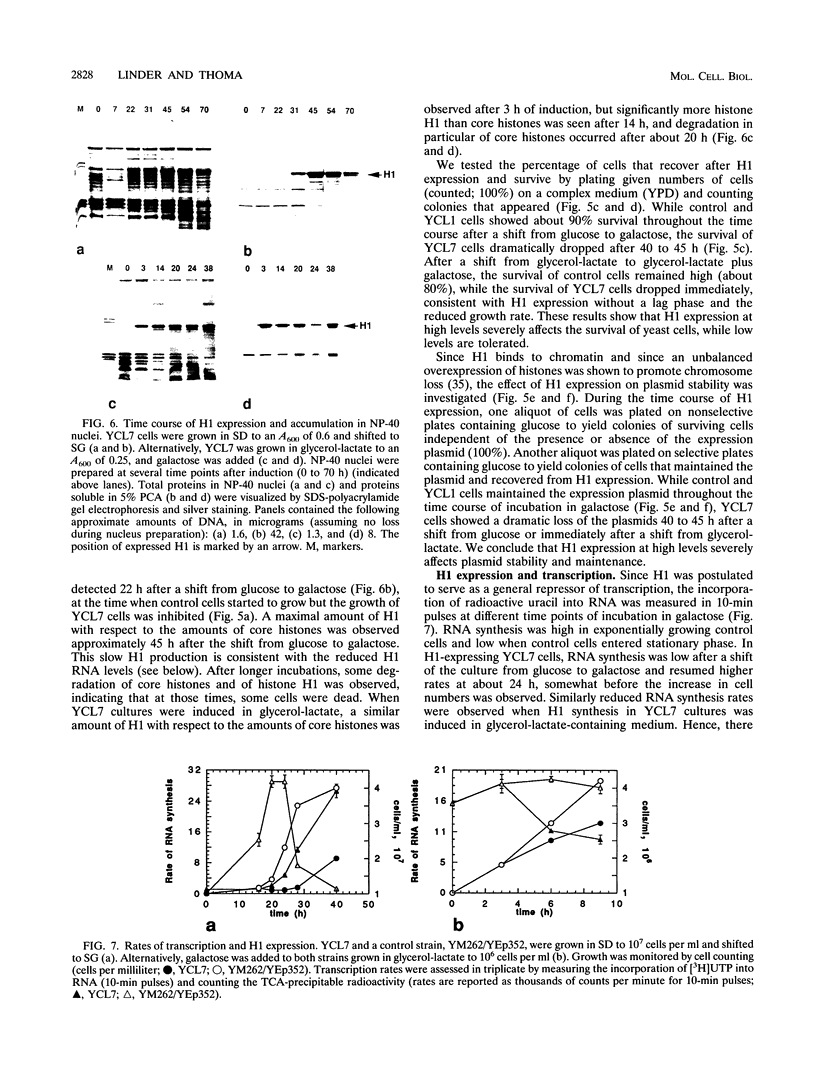





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Arceci R. J., Gross P. R. Histone variants and chromatin structure during sea urchin development. Dev Biol. 1980 Nov;80(1):186–209. doi: 10.1016/0012-1606(80)90508-4. [DOI] [PubMed] [Google Scholar]
- Axelrod J. D., Reagan M. S., Majors J. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes Dev. 1993 May;7(5):857–869. doi: 10.1101/gad.7.5.857. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Butler P. J., Pearson E. C., Thomas J. O. Stability of the higher-order structure of chicken-erythrocyte chromatin in solution. Eur J Biochem. 1981 Oct;119(3):469–476. doi: 10.1111/j.1432-1033.1981.tb05631.x. [DOI] [PubMed] [Google Scholar]
- Bates D. L., Thomas J. O. Histones H1 and H5: one or two molecules per nucleosome? Nucleic Acids Res. 1981 Nov 25;9(22):5883–5894. doi: 10.1093/nar/9.22.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavykin S. G., Usachenko S. I., Lishanskaya A. I., Shick V. V., Belyavsky A. V., Undritsov I. M., Strokov A. A., Zalenskaya I. A., Mirzabekov A. D. Primary organization of nucleosomal core particles is invariable in repressed and active nuclei from animal, plant and yeast cells. Nucleic Acids Res. 1985 May 24;13(10):3439–3459. doi: 10.1093/nar/13.10.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi F., Koller T., Thoma F. The ade6 gene of the fission yeast Schizosaccharomyces pombe has the same chromatin structure in the chromosome and in plasmids. Yeast. 1991 Aug-Sep;7(6):547–558. doi: 10.1002/yea.320070603. [DOI] [PubMed] [Google Scholar]
- Boulikas T., Wiseman J. M., Garrard W. T. Points of contact between histone H1 and the histone octamer. Proc Natl Acad Sci U S A. 1980 Jan;77(1):127–131. doi: 10.1073/pnas.77.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer M., Goldfarb D. S. Facilitated nuclear transport of histone H1 and other small nucleophilic proteins. Cell. 1990 Mar 23;60(6):999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- Brill S. J., DiNardo S., Voelkel-Meiman K., Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. 1987 Mar 26-Apr 1Nature. 326(6111):414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Cavalli G., Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993 Dec;12(12):4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croston G. E., Laybourn P. J., Paranjape S. M., Kadonaga J. T. Mechanism of transcriptional antirepression by GAL4-VP16. Genes Dev. 1992 Dec;6(12A):2270–2281. doi: 10.1101/gad.6.12a.2270. [DOI] [PubMed] [Google Scholar]
- Davie J. R., Saunders C. A., Walsh J. M., Weber S. C. Histone modifications in the yeast S. Cerevisiae. Nucleic Acids Res. 1981 Jul 10;9(13):3205–3216. doi: 10.1093/nar/9.13.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Allan J. Accumulation of the isolated carboxy-terminal domain of histone H1 in the Xenopus oocyte nucleus. EMBO J. 1984 Sep;3(9):1933–1937. doi: 10.1002/j.1460-2075.1984.tb02072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Kornberg R. D. Upstream activation sequence-dependent alteration of chromatin structure and transcription activation of the yeast GAL1-GAL10 genes. Mol Cell Biol. 1989 Apr;9(4):1721–1732. doi: 10.1128/mcb.9.4.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frado L. L., Mura C. V., Stollar B. D., Woodcock C. L. Mapping of histone H5 sites on nucleosomes using immunoelectron microscopy. J Biol Chem. 1983 Oct 10;258(19):11984–11990. [PubMed] [Google Scholar]
- Gerace L., Burke B. Functional organization of the nuclear envelope. Annu Rev Cell Biol. 1988;4:335–374. doi: 10.1146/annurev.cb.04.110188.002003. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Hill C. S., Martin S. R., Thomas J. O. A stable alpha-helical element in the carboxy-terminal domain of free and chromatin-bound histone H1 from sea urchin sperm. EMBO J. 1989 Sep;8(9):2591–2599. doi: 10.1002/j.1460-2075.1989.tb08398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R., Sprague G. F., Jr, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci U S A. 1983 May;80(10):3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Davis R. W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., Bulger M., Kadonaga J. T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993 Sep;7(9):1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- Künzler P., Stein A. Histone H5 can increase the internucleosome spacing in dinucleosomes to nativelike values. Biochemistry. 1983 Apr 12;22(8):1783–1789. doi: 10.1021/bi00277a007. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lennard A. C., Thomas J. O. The arrangement of H5 molecules in extended and condensed chicken erythrocyte chromatin. EMBO J. 1985 Dec 16;4(13A):3455–3462. doi: 10.1002/j.1460-2075.1985.tb04104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Kovacic R. T., Van Holde K. E. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977 Feb 8;16(3):463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Lohr D. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 1984 Nov 26;12(22):8457–8474. doi: 10.1093/nar/12.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Torchia T., Hopper J. The regulatory protein GAL80 is a determinant of the chromatin structure of the yeast GAL1-10 control region. J Biol Chem. 1987 Nov 15;262(32):15589–15597. [PubMed] [Google Scholar]
- Losa R., Omari S., Thoma F. Poly(dA).poly(dT) rich sequences are not sufficient to exclude nucleosome formation in a constitutive yeast promoter. Nucleic Acids Res. 1990 Jun 25;18(12):3495–3502. doi: 10.1093/nar/18.12.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losa R., Thoma F., Koller T. Involvement of the globular domain of histone H1 in the higher order structures of chromatin. J Mol Biol. 1984 Jun 5;175(4):529–551. doi: 10.1016/0022-2836(84)90183-9. [DOI] [PubMed] [Google Scholar]
- Mann R. K., Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 1992 Sep;11(9):3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D., Hartwell L. H. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986 Jan 17;44(1):43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- Moreland R. B., Langevin G. L., Singer R. H., Garcea R. L., Hereford L. M. Amino acid sequences that determine the nuclear localization of yeast histone 2B. Mol Cell Biol. 1987 Nov;7(11):4048–4057. doi: 10.1128/mcb.7.11.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEELIN J. M., CALLAHAN P. X., LAMB D. C., MURRAY K. THE HISTONES OF CHICKEN ERYTHROCYTE NUCLEI. Can J Biochem. 1964 Dec;42:1743–1752. doi: 10.1139/o64-185. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Thoma F., Simpson R. T. Core particle, fiber, and transcriptionally active chromatin structure. Annu Rev Cell Biol. 1986;2:117–147. doi: 10.1146/annurev.cb.02.110186.001001. [DOI] [PubMed] [Google Scholar]
- Renz M., Nehls P., Hozier J. Involvement of histone H1 in the organization of the chromosome fiber. Proc Natl Acad Sci U S A. 1977 May;74(5):1879–1883. doi: 10.1073/pnas.74.5.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago T. C., Purvis I. J., Bettany A. J., Brown A. J. The relationship between mRNA stability and length in Saccharomyces cerevisiae. Nucleic Acids Res. 1986 Nov 11;14(21):8347–8360. doi: 10.1093/nar/14.21.8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W., Kunz G., Daetwyler H., Telford J., Smith H. O., Birnstiel M. L. Genes and spacers of cloned sea urchin histone DNA analyzed by sequencing. Cell. 1978 Jul;14(3):655–671. doi: 10.1016/0092-8674(78)90249-0. [DOI] [PubMed] [Google Scholar]
- Shaw B. R., Cognetti G., Sholes W. M., Richards R. G. Shift in nucleosome populations during embryogenesis: microheterogeneity in nucleosomes during development of the sea urchin embryo. Biochemistry. 1981 Aug 18;20(17):4971–4978. doi: 10.1021/bi00520a025. [DOI] [PubMed] [Google Scholar]
- Shwed P. S., Neelin J. M., Seligy V. L. Expression of Xenopus laevis histone H5 gene in yeast. Biochim Biophys Acta. 1992 Jun 15;1131(2):152–160. doi: 10.1016/0167-4781(92)90070-g. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Snyder M., Sapolsky R. J., Davis R. W. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1988 May;8(5):2184–2194. doi: 10.1128/mcb.8.5.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Sun J. M., Ali Z., Lurz R., Ruiz-Carrillo A. Replacement of histone H1 by H5 in vivo does not change the nucleosome repeat length of chromatin but increases its stability. EMBO J. 1990 May;9(5):1651–1658. doi: 10.1002/j.1460-2075.1990.tb08285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. M., Wiaderkiewicz R., Ruiz-Carrillo A. Histone H5 in the control of DNA synthesis and cell proliferation. Science. 1989 Jul 7;245(4913):68–71. doi: 10.1126/science.2740916. [DOI] [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977 Sep;12(1):101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T. Unravelled nucleosomes, nucleosome beads and higher order structures of chromatin: influence of non-histone components and histone H1. J Mol Biol. 1981 Jul 15;149(4):709–733. doi: 10.1016/0022-2836(81)90354-5. [DOI] [PubMed] [Google Scholar]
- Thoma F., Losa R., Koller T. Involvement of the domains of histones H1 and H5 in the structural organization of soluble chromatin. J Mol Biol. 1983 Jul 5;167(3):619–640. doi: 10.1016/s0022-2836(83)80102-8. [DOI] [PubMed] [Google Scholar]
- Thoma F. Protein-DNA interactions and nuclease-sensitive regions determine nucleosome positions on yeast plasmid chromatin. J Mol Biol. 1986 Jul 20;190(2):177–190. doi: 10.1016/0022-2836(86)90291-3. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Furber V. Yeast chromatin structure. FEBS Lett. 1976 Jul 15;66(2):274–280. doi: 10.1016/0014-5793(76)80521-2. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Tremethick D. J., Frommer M. Partial purification, from Xenopus laevis oocytes, of an ATP-dependent activity required for nucleosome spacing in vitro. J Biol Chem. 1992 Jul 25;267(21):15041–15048. [PubMed] [Google Scholar]
- Workman J. L., Kingston R. E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992 Dec 11;258(5089):1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]