Abstract
Readout of information from the genome depends on intricate regulation of how DNA is packaged by proteins. The great endeavour to reveal how this packaging operates pan-genomically is now under way.
A new era is opening for biologists involved in understanding cellular systems. It is exemplified by papers by Mikkelsen et al. (page 553 of this issue)1 and Barski et al. (published in Cell)2 — they describe the kind of unprecedented insights that are emerging from investigations of how a single mammalian genome can be regulated to produce different cell types.
The technical and biological advances described in these studies extend the remarkable accomplishments of elucidating the structure3, then the sequence4,5, of the human genome; and they reflect a growing, ‘post-genomic’, appreciation of the complexities of genome structure and function (Fig. 1). The intriguing — and daunting — challenge now is to understand the process of how and when specific DNA regions are controlled to produce the cellular diversity that underpins the development and maintenance of a single organism.
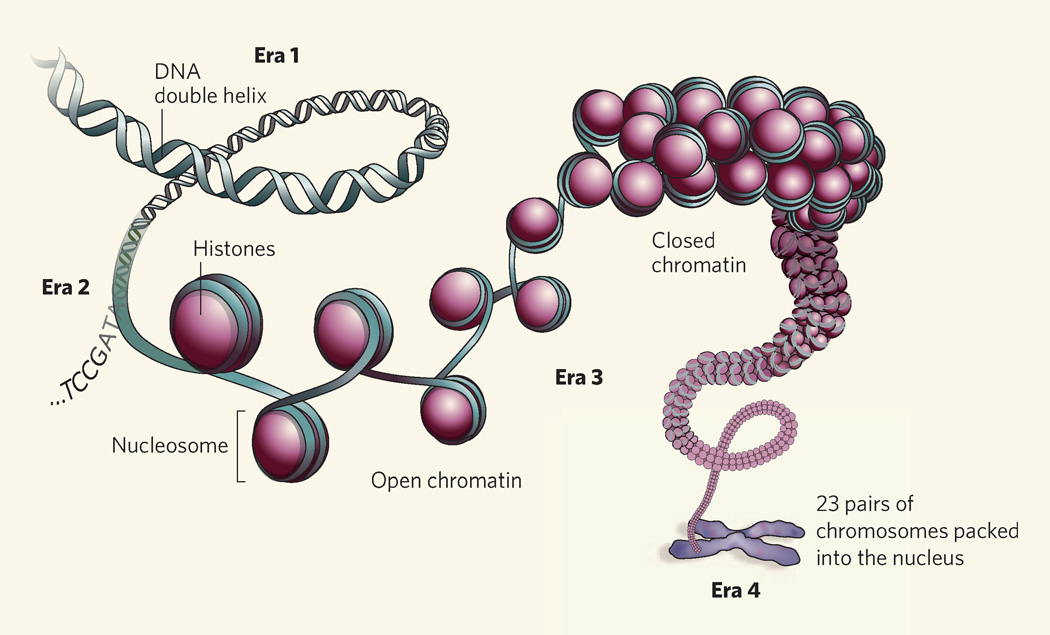
Figure 1. Genomic architecture, and eras of investigation.
Eras 1 and 2 were launched by the elucidation of the structure of DNA, and then of the sequence of the human genome. DNA is packaged with histone proteins to form chromatin, the central unit of which is the nucleosome. As examples of work in the unfolding era 3 — the era of the epigenome — the papers of Mikkelsen et al.1 and Barski et al.2 provide genome-wide linear maps relating histone modifications (‘marks’) to the active or inactive sites where various types of regulatory RNA, as well as messenger RNA, are produced. Nucleosomes are further organized to create open and closed regions of chromatin, which in turn create three-dimensional structures that encompass different levels of gene organization in the chromosomes packed into a cell nucleus. A likely era 4 will involve generation of maps of this ‘genome topography’ to reveal how a single genome can produce such a diversity of cell types.
Central to this challenge is the task of enumerating the dizzying number of proteins interacting with the genome, and the functions they subserve. These proteins, called histones, form a combination with DNA that is termed chromatin. It is chromatin that provides the software packaging for the readout of the DNA hard drive. If alterations in genome heritable states occur through a change in the hard drive (that is, through a change in the primary sequence of DNA), a genetic alteration or mutation has occurred. This contrasts with an epigenetic change, which is an alteration in the heritable states of DNA function produced by altering the chromatin software. Epigenetic changes lie at the heart of how organisms generate different types of tissue under different circumstances — in embryonic development, in regulating cell renewal in adults, and in the cellular responses of the organism to environmental factors and stress. Moreover, disease states such as cancer are associated with a combination of both genetic and epigenetic abnormalities.
The central unit of chromatin is the nucleosome, which is constructed from short regions of DNA wound around an octet of histone proteins. This unit can modulate the readout from DNA in at least three ways.
First, nucleosomes can be physically rearranged on DNA by complexes known as chromatin-remodelling proteins6 — generally, the greater the distance between nucleosomes, and so the ‘openness’ of chromatin, the higher the likelihood that such regions of DNA will be transcribed into RNA. Second, many nucleosomes can be compacted into higher-order aggregates to form ‘closed’ chromatin, or hetero chromatin6, thereby preventing transcription. The balance between the open and closed parts of the genome facilitates proper gene-expression patterns in given cell types, and also prevents unwanted gene transcription.
Third, there is a complex interplay between enzymes that can modify particular amino acids in the histone component of the nucleosomes, and those that reverse the modifications. The modifications, or histone ‘marks’, interact with proteins that bind to and interpret them. The marks were initially seen as a ‘histone code’, the idea being that a restricted number of them would specify the ‘on’ or ‘off ’ state of RNA production from DNA7. This concept was a most useful starting point. But it is increasingly recognized that the constituents of chromatin, and nucleosome structure, position and modification, are highly complex. It is a balance between these factors that marks an individual gene, or groups of genes, for various levels and states of expression8. That is, there is no simple on–off code.
All of which brings us to the papers by Mikkelsen et al.1 and Barski et al.2. Both represent examples of genome ‘tiling’ approaches — the aim being to catalogue, across the entire human genome, the locations not only of key histone modifications but also of proteins that respond to and mediate them. Mikkelsen et al. begin the process of mapping how these parameters change as cells negotiate their conversion from immature to adult states, whereas Barski et al. examine a more mature cell state. The two groups used an ingenious new technology, Solexa 1G sequencing, which allows millions of short DNA ‘sequence tags’ to be assigned to individual histone marks, thus mapping the marks to their precise location in the genome.
The results are remarkably comprehensive linear maps of the principal chromatin constituents across the human genome. The maps highlight the complexity of DNA packaging, and reveal that combinations of histone modifications and positions, rather than single histone marks, correlate most accurately with multiple levels of the genes’ transcriptional states. Histone characteristics can define the immediate start sites of genes, which are often regulatory in nature. But they can also define discrete but distant regions that influence gene expression, as well as regions that may encompass an entire gene to prompt its active or repressed transcription.
The papers also provide insights about genomic regions — within genes, or between genes — that are unexpectedly marked for expression activity. These data relate to the recent revelations that much more of our genome than previously thought is engaged in expressing RNA from DNA. The result is production not only of ‘classical’ messenger RNAs (which produce the proteins defined by the initial analyses of the genome sequence), but also of a huge number of regulatory RNAs (which modulate genome readout by producing multi ple forms of the same protein or without producing proteins at all9).
So, are we done with mapping the genome? Hardly. These genome-packaging data1,2 provide a first linear view that can only hint at the three-dimensional aspects of how the genome is organized in the cell nucleus to regulate DNA. We already know, broadly at least, that nucleosome-mediated chromatin domains create three-dimensional structures that surround individual gene-regulatory regions, whole genes, groups of genes and genes encompassed in chromosome territories10 — producing, altogether, what can be seen as a genome topography. Perhaps a complete view of the genome will require a further era of investigation (Fig. 1), in which we generate maps of the genomic topography that characterizes each of the many cell types of which we are constituted. Who knows? We are just at the beginning of exploring how a single genome can spawn multiple epigenomes.
References
- 1.Mikkelsen TS, et al. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barski A, et al. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Watson JD, Crick FHC. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Venter JC, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Carey M, Workman JL. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein BE, Meissner A, Lander ES. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 9.Kapranov P, et al. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 10.Albiez H, et al. Chromosome Res. 2006;14:707–733. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]