Abstract
We report the synthesis and characterization of a series of ionizable lysine-based lipids (ILL), novel lipids containing a lysine head group linked to a long-chain dialkylamine through an amide linkage at the lysine α-amine. These ILLs contain two ionizable amines and a carboxylate, and exhibit pH-dependent lipid ionization that varies with lipid structure. The synthetic scheme employed allows for the simple, orthogonal manipulation of lipids. This provides a method for the development of a compositionally diverse library with varying ionizable headgroups, tail structures, and linker regions. A focused library of four ILLs was synthesized to determine the impact of hydrophobic fluidity, lipid net charge, and lipid pKa on the biophysical and siRNA transfection characteristics of this new class of lipids. We found that manipulation of lipid structure impacts the protonation behavior, electrostatic driven membrane disruption, and ability to promote siRNA mediated knockdown in vitro. ILL-siRNA liposomal formulations were tested in a murine Factor VII model; however, no significant siRNA-mediated knockdown was observed. These results indicate that ILL may be useful in vitro transfection reagents, but further optimization of this new class of lipids is required to develop an effective in vivo siRNA delivery system.
Keywords: Lipid synthesis, lipid nanoparticles, siRNA delivery, zwitterionic lipids, bioconjugate chemistry
INTRODUCTION
Lipid-based nanoparticles have long been used to deliver biologically active molecules including drugs, DNA, and more recently siRNA in vivo.1,2 Liposomes and lipoplexes alter the pharmacokinetics, biodistribution, and uptake pathways of encapsulated or associated molecules.3 Depending on the therapeutic application and drug properties, lipids with specific biophysical characteristics are required to develop an effective delivery system.4 Due to their anionic charge and large size, siRNA are not able to passively cross cellular membranes to reach their site of action in the cytoplasm.5,6 Therefore, delivery systems that actively promote cellular internalization and endosomal escape are essential.7,8 Because of these properties, lipid-based siRNA delivery systems are generally cationic or ionizable to allow for high efficiency siRNA encapsulation and to promote intracellular membrane disruption and endosomal escape.7,9–16 Recent work on the rational design of amino lipids using structure-activity relationships indicates that fusogenic lipids with pKas in the pH=6.0–7.0 range are promising candidates for siRNA delivery in vivo.7,12 Following internalization, the low-pH environment of the endocytic pathway is believed to ionize these lipids, promoting electrostatic interactions between carrier lipids and naturally occurring anionic membrane lipids. These interactions drive the formation of unstable, non-bilayer structures that lead to membrane disruption and contents delivery to the cytoplasm.17–19 Formulation of nanoparticles using these ionizable lipids at low-pH allows for efficient siRNA encapsulation, and incorporation of helper lipids including phospholipids, sterols, and PEG-lipids alters the size, surface charge, and stability of these formulations, providing some control over the in vivo pharmacokinetic properties of these systems.20,21
Despite recent advances in the field, ionizable and cationic systems are still limited by their immunostimulatory effects and poor pharmacokinetics.14 In contrast to cationic lipids, zwitterionic lipids such as phosphatidylcholine (PC) or phosphatidylethanolamine (PE) show low in vivo cytotoxicity and immunogenicity.22 However, their neutral surface charge and pH-independent biophysical properties limit their ability to efficiently encapsulate and deliver charged, high molecular weight drugs. Previously, we reported a new class of zwitterionic lipids (ZL) with head groups containing a cationic amine and an anionic carboxylate.23 These lipids are non-immunogenic and non-toxic, and are capable of encapsulating siRNA at low-pH. However, ionization of these head groups occurs at a pH below that found in the endosomes, limiting the utility of ZL as siRNA delivery systems.23 Building on this work, we describe the design, synthesis, and characterization of ionizable lysine-based lipids (ILL) containing a lysine head group linked to a long-chain dialkylamine through an amide linkage at the lysine α-amine (Fig. 1). The resulting lipid structure contains a primary and tertiary amine, as well as a single carboxylate, and the overall net charge of the lipid depends upon the ionization state of each protonatable moiety (Fig. 2). The general ILL structure maintains a zwitterionic head group at the bilayer interface, potentially reducing immunogenicity compared to other cationic systems. However, the addition of a second ionizable amine in the lipid core is expected to increase ILL pKa into a physiologically relevant range to promote membrane destabilization, endosomal escape, and siRNA delivery to the cytoplasm. We hypothesized that varying the structure of ILL would impact their ionization behavior, membrane interactions, and siRNA transfection both in vitro and in vivo. A focused library of four ILLs with distinct structures was synthesized to determine the impact of hydrophobic fluidity, lipid net charge, and lipid pKa on the biophysical and transfection characteristics of this new class of lipids.
Figure 1.
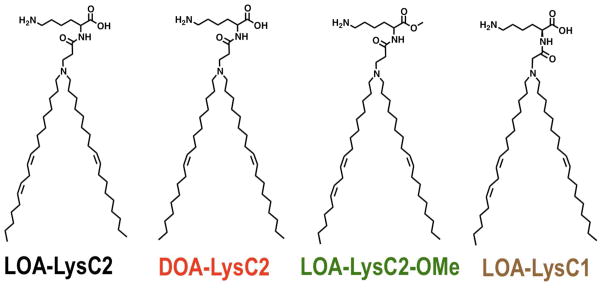
Ionizable lysine-based lipid (ILL) structures
Figure 2.
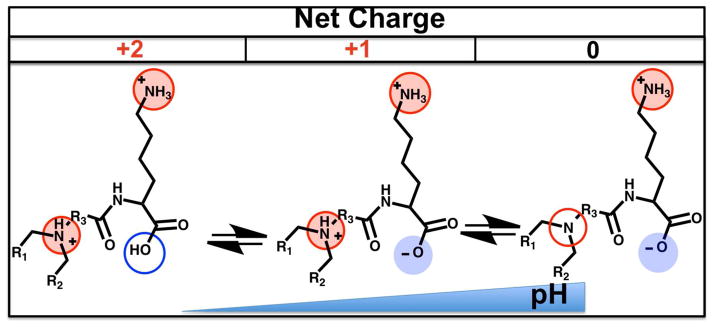
Theoretical pH-dependent ionization of ILL pH
EXPERIMENTAL PROCEDURE
Materials and Chemicals
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-(phosphor-1′-rac-glycerol) (DOPG), 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), and cholesterol (Chol) were purchased from Avanti Polar Lipids (Alabaster, AL). 1-(monomethoxypolyethyleneglycol)-2,3-dimyrystoylglycerol (PEG-DMG) was purchased from NOF-Corporation (Tokyo, Japan). DLinDMA was provided by Pfizer (Cambridge, MA). Boc-Lys-OMe HCl was purchased from Combi-Blocks Inc. (San Diego, CA). Linoleic acid was purchased from TCI-America Inc. (Portland, OR). Quant-iT Ribogreen RNA reagent was purchased from Invitrogen (Carlsbad, CA). The Steady-Glo luciferase assay kit was purchased from Promega (Madison, WI). The Aniara Biophen Factor VII assay kit was purchased from Fisher Scientific (Houston, TX). Lipofectamine 2000 was purchased from Life Technologies (Carlsbad, CA). All other reagents were purchased from Sigma-Aldrich (Milwaukee, WI). Solvents were purchased from VWR (Radnor, PA), and used without further purification. siRNA for in vitro luciferase knockdown experiments was provided by Pfizer (Cambridge, MA). siRNA for in vivo Factor VII knockdown experiments was synthesized by Dharmacon (Lafayette, CO). siRNA sequences are provided in the Supporting Information.
Lipid Synthesis
A library of four ionizable lysine-based lipids (ILL) were synthesized by conjugating lysine to a dialkyl-β-alanine or dialkylglycine through an amide linkage between the α-amine of lysine and the carboxylic acid of the dialkylated amino acid. A detailed scheme for all synthesized lipids can be found in the Supporting Information. TLC analyses were performed on 0.25 mm silica gel F254 plates (Fisher Scientific, Houston, TX) using the described solvent systems. High-performance flash chromatography (HPFC) was performed on a Grace (Deerfield, IL) Reveleris HPFC system with pre-packed Reveleris silica gel columns (70 Å, 40 μm). 1H NMR spectra were acquired using a Bruker (Fremont, CA) Avance 300 MHz instrument. Chemical shifts are expressed as parts per million, and tetramethylsilane was used as an internal standard. MALDI-TOF spectra were acquired using Bruker (Fremont, CA) Microflex workstation.
Liposomal Formulation for Biophysical Assays
Chloroform solutions of lipids were dried in 10 × 25 mm glass test tubes by rotary evaporation under reduced pressure followed by high vacuum for 18 hours. The lipid film was rehydrated with 500 μL of 10 mM Tris buffer containing 150 mM NaCl, pH=7.4 (5 mM final concentration). Each preparation was sonicated 30 min at room temperature to form monodisperse liposomes with diameters in the 80–120 nm range. Particle size was measured using a Malvern (Westborough, MA) Zetasizer NanoZS.
TNS Fluorescence Assay
Ionization behavior of ILL containing liposomes was assayed using a protocol adapted by Zhang et. al.24 A 5 mM solution of ILL liposomes (40:40:15:5, ILL:Chol:POPC:PEG-DMG) was diluted to 2.67 μM in a 10 mM buffer containing 150 mM NaCl, pH=3.5–9.5. Fluorescence was measured (Ex/Em = 320/430 nm) using a Spex Fluorolog Fluorimeter (Horiba Scientific, Edison, NJ) to obtain background, then measured again following the introduction of 0.167 μM TNS to give the background subtracted fluorescent signal (F). All experiments were run in triplicate, and normalized to the background subtracted fluorescent signals of DOTAP and DOPC liposomes to correct for changes in TNS fluorescence as a function of pH. The net charge of the ionizable lipid at a given pH is defined as:
where F is the fluorescence of the sample at the specified pH, FDOPC is the fluorescence of DOPC liposomes (40:40:15:5, DOPC:Chol:POPC:PEG-DMG) at the specified pH, and FDOTAP is the fluorescence of DOTAP liposomes (40:40:15:5, DOTAP:Chol:POPC:PEG-DMG) at the specified pH.
Membrane Lysis Assay
Membrane lysis activity was assayed using a protocol adapted from Zhang et. al.24 A 5 mM solution of membrane mimicking liposomes (45:20:20:15, DOPC:DOPE:DOPG:Chol) encapsulating 10 mM TRIS, 12.5 mM ANTS, 45 mM DPX, 20 mM NaCl was diluted to 3.33 μM in a 10 mM isosmotic buffer containing 150 mM NaCl, pH=5.5, 6.5, or 7.4. A 5 mM solution of ionizable liposomes (40:60, ionizable lipid:POPC) was then diluted in to a concentration of 16.67 μM. Baseline ANTS fluorescence (Ex/Em = 360/530 nm) was measured at t = 0 (Fmin) using a Spex Fluorolog Fluorimeter (Horiba Scientific, Edison, NJ). Following a 30 min incubation at 37 °C, ANTS fluorescence was measured again (F). Maximum ANTS fluorescence was measured following total membrane lysis using 0.5% C12E10 (Fmax). All measurements were done in triplicate. % Lysis is defined as:
In Vitro siRNA Transfection Assay
In vitro transfection was assayed using a protocol adapted from Akinc et. al.25 HeLa cells stably expressing firefly luciferase were seeded (8,000 cells/well) on an opaque black 96-well plate (Greiner Cellstar), and allowed to attach overnight in 90% MEM media with 10% FBS, 100 mcg/ml streptomycin and 100 units/ml penicillin. Lipid/siRNA complexes were made by combining lipid formulations containing 40:40:20 ILL:Chol:DOPE (1 mg/ml total lipid, 25 mM NaOAc buffer, pH=5.0) and siRNA (12.5 μg/ml siRNA, 25 mM NaOAC buffer, pH=5.0) at N:P = 10 for 30 min at room temperature. Cells were transfected with lipoplexes containing anti-luciferase or non-specific siRNA at the given concentrations by adding the lipoplex solution to the cell culture media and incubating for 24 hr. Luciferase expression was measured using the Steady-Glo assay kit (Promega, Madison, WI). Luminescence was measured using an Infinite m1000 Pro microplate reader (Tecan, San Jose, CA). Control experiments were performed with Lipofectamine 2000 (LF2000) using the protocol provided by the vendor (Invitrogen, Carlsbad, CA). Luminescence was normalized to the non-specific siRNA control, and treated wells were compared against untreated wells to evaluate knockdown efficiency. All measurements were done in triplicate.
In Vivo ILL-siRNA Liposomal Formulation
ILL-siRNA formulations were prepared using a batch mixing process as previously described.23 Formulations contained ILL, Chol, DSPC, and PEG-DMG (40:40:10:10). 2 mg total lipid was dissolved in 250 μL of ethanol and sonicated at 25°C for 5 min. The lipid/ethanol solution was subsequently injected into a magnetically stirred 2.5 mL vial, which contained 100 μg of siRNA dissolved in 250 μL of 50 mM citric acid buffer, pH 4. The lipid suspension was stirred for 10 min and then extruded using a handheld extruder (Avestin, Ottawa, ON, Canada) through 80 nm polycarbonate membranes (Whatman International, Kent, UK) 5 times at room temperature. Ethanol was removed by dialysis against PBS (pH 7.4) without Ca2+ and Mg2+ for 24h. Encaspulation efficiency of siRNA was quantified by measuring the fluorescence signal using the Quant-iT RiboGreen RNA Assay Kit (Life Technologies, Carlsbad, CA) in the presence or absence of 0.4% Triton-X. Fluorescence was measured using the Fluostar fluorescence plate reader (BMG Labtech, Cary, NC) (Ex/Em = 485/520 nm). Liposome size and zeta potential were measured 3 times per sample on a Zetasizer NanoZS (Malvern, Westborough MA) and averaged.
In Vivo Mouse Factor VII Knockdown Experiments
Six-to eight-week-old, female CD-1 mice (Charles River Laboratories, Wilmington, MA) were administered ILL-siRNA formulations via tail vein injection at an siRNA concentration of 5 mg/kg (15:1 w:w ratio of ILL:siRNA) in a total volume of 200 μL. Control mice received an injection of 200 μL of phosphate buffered saline without calcium or magnesium salts (PBS). 48h after administration, animals were anesthetized with isoflurane. Blood was collected by submandibular cheek bleeding. Serum samples were obtained by allowing the blood to clot for 30 min and then centrifuging for 15 min at 15,000 rpm at 4°C. The supernatant was collected and analyzed for serum levels of Factor VII protein using the Biophen VII chromogenic assay (Aniara, Mason, OH) according to manufacturer’s instructions. A standard curve was generated using serially diluted concentrations of PBS-treated animals. Serum Factor VII levels of mice treated with ILL-siRNA formulations were expressed as percentage of PBS-control. Each group consisted of n=3 mice. DLinDMA liposomes (40:40:10:10 DLinDMA:Chol:DSPC:PEG-DMG) served as a positive control.20
RESULTS AND DISCUSSION
Chemistry and structure of Ionizable Lysine-Based Lipids
Ionizable lysine-based lipids (ILL) contain a lysine head group linked to a long-chain dialkylamine through an amide linkage at the lysine α-amine (Fig. 1). The resulting lipid structure contains a primary and tertiary amine, as well as a single carboxylate. As such, the overall net charge of ILL depends upon the ionization state of these protonatable moieties (Fig. 2). At low pH, all ionizable groups should be protonated, yielding a net charge of +2. As the pH increases, deprotonation of the carboxylate reduces the overall net charge to +1, and subsequent deprotonation of the amines results in a net charge of 0 or −1. We hypothesize that, similar to other ionizable delivery systems, this pH-dependent ionization behavior will promote siRNA encapsulation at low pH and membrane destabilization along the endocytic pathway, while preventing protein opsonization at physiological pH.12 The presence of the anionic moiety gives these lipids zwitterionic characteristics that may reduce the immunostimulatory effects that are characteristic of cationic lipid systems.14
We synthesized four distinct lipids to identify the impact of structural modifications on the biophysical and transfection behavior of these lipids (Fig. 1): LOA-LysC2 contains a linoleyloleylamine (LOA) and a two-carbon spacer between the amide-linked lysine and the LOA core; DOA-LysC2 contains the same two-carbon spacer but a dioleylamine (DOA) hydrophobic region; LOA-LysC2-OMe is identical to LOA-LysC2, except the carboxylic acid is methylated to prevent ionization of this functional group and eliminate the cationic to neutral charge transition; LOA-LysC1 is analogous to LOA-LysC2, but contains a single carbon linker between the amide and the LOA core. This focused lipid library provides a platform to study how lipid hydrophobic fluidity, net charge, and amine pKa impact the biophysical and transfection behavior of ILL.
The general ILL synthetic scheme allows for the orthoganol manipulation of lipid structure. Full details of all synthetic schemes can be found in the Supporting Information. Synthesis begins with the formation of a dialkylamine by coupling a long chain fatty acid to oleylamine, followed by reduction of the resulting amide using lithium aluminum hydride. This long-chain dialkylamine is then functionalized with either t-butylbromoacetate or t-butylacrylate to create a protected trialkylamino acid with a one- or two-carbon spacer between the tertiary amine and the carboxylate. Following deprotection, the acid group is coupled to the α-amine of a protected lysine. Global or selective deprotection of this compound yields the final ILL product. All compounds were purified by HPFC prior to biophysical characterization or transfection studies.
Liposomal formulation for biophysical characterization
ILL liposomes for biophysical characterization were prepared by rehydrating a dried lipid film of the desired composition in 10 mM Tris buffer with 150 mM NaCl, pH=7.4. Two different liposomal formulations were used; 60:40 POPC:ILL and 40:40:15:5 ILL:Chol:POPC:PEG-DMG. Both preparations formed small (80–120 nm), stable liposomes following sonication for 30 minutes at room temperature.
pH-dependent ionization of ILL containing liposomes
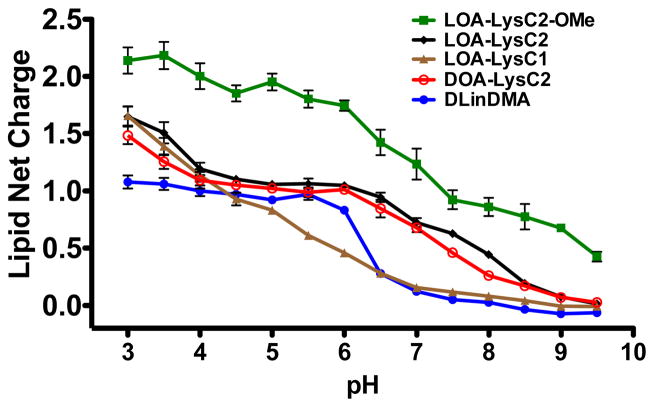
The ionization behavior of ILL liposomes (40:40:15:5, ILL:Chol:POPC:PEG-DMG) was studied by comparing their relative surface charge as a function of pH using a TNS assay.24,26 TNS is an anionic fluorophore whose fluorescence intensity increases drastically in a hydrophobic environment due to the elimination of quenching by water molecules. TNS partitions into cationic lipid membranes to a greater extent than neutral or anionic membranes due to electrostatic interactions. This behavior makes TNS an ideal candidate for exploring the surface charge of ILL containing liposomes across a range of pH. As the ILL headgroup is protonated with decreasing pH, the level of TNS fluorescence should increase due to an increasing concentration of TNS in the hydrophobic environment. The presence of multiple ionizable groups in ILL makes data normalization difficult because the shape of the ionization curve differs depending on lipid structure. As such, normalizing to the maximum and minimum values for each sample does not accurately portray their relative ionization behavior. However, normalization to a formally positive lipid (DOTAP) and a neutral lipid (DOPC) allows us to identify the relative net charge of the ILL as a function of pH. The data for each ILL can then be compared to other ILL, as well as lipids with a single ionizable amine, such as DLinDMA.20 Lipid structures for these control lipids can be found in Fig. S1, Supporting Information.
TNS fluorescence in the presence of ILL liposomes was measured in 0.5 pH unit increments from pH = 3.0–9.5 (Fig. 3). Our results indicate that all ILL show pH-dependent ionization; however, structural changes clearly impact their ionization behavior. All ILL show a maximum net charge at pH=3.0 and a minimum net charge at pH = 9.5, as expected. LOA-LysC2, which was the template for all synthesized lipids, reaches a maximum net charge of +1.7, and exhibits a broad, double sigmoidal ionization curve, eventually achieving a minimum value of 0.0 at pH = 9.5. Altering the hydrophobic region of LOA-LysC2 does not impact its ionization behavior, as seen by the matching DOA-LysC2 curve. Altering the length of the linker between the 3° amine and the amide linkage significantly impacts the shape of the ionization curve, but has no impact on the maximum or minimum lipid charge. This is illustrated by the ionization behavior of LOA-LysC1 liposomes. The shorter linker in LOA-LysC1 lowers the pKa of the 3° amine, likely due to the electron withdrawing effect of the amide in close proximity. LOA-LysC2-OMe, which has a methylated carboxylic acid moiety, exhibits a significantly different ionization profile due to the absence of the anionic group. At low pH, LOA-LysC2-OMe liposomes achieve a net charge of 2.0, indicating that both amines are fully protonated.
Figure 3.
pH-dependent ionization of ILL liposomes (40:40:15:5 ILL:Chol:POPC:PEG-DMG)
None of the ILL with an ionizable carboxylate reach a net charge of 2.0 (maximum theoretical net charge) in the tested pH range. However, methylation of the carboxylate results in a fully ionized head group. This indicates that the carboxylate moiety is either not fully protonated or is somehow interacting with the free amino groups to disrupt interactions with the TNS fluorophore. Interestingly, all of the ILL ionization curves differ significantly from that of DLinDMA, a well characterized ionizable amino lipid20, which shows a sigmoidal shape with a maximum net charge of 1.0 and a sharp transition to 0.0 between pH = 6.0 and 7.0. ILL do not follow the sigmoid shape observed for other ionizable amino lipids.12,20,24 These lipids show a broader, more complicated ionization curve, possibly due to the presence of multiple ionizable moieties with varying pKa in each lipid. Additionally, differences in amine substitution (1° versus 3°) or orientation in the bilayer (interfacial versus extended) may affect lipid ionization and alter the pH-dependent surface charge.
Lysis of biomembrane mimicking vesicles by ILL liposomes
It is hypothesized that the charge state of lipids impacts their ability to disrupt anionic biomembranes along the endocytic pathway and deliver contents to the cytosol.7,12,24 Cationic lipids can ion-pair with naturally occurring anionic lipids, inducing a phase transition from the stable Lα to the unstable HII phase, promoting endosomal escape.18,27
The efficiency of this process is believed to be essential for the effective delivery of liposomal contents to the cytosol.7,8,18,20 However, formally cationic systems are generally immunostimulatory and have short plasma half-lives due to protein opsonization and subsequent clearance by the reticuloendothelial system, making them poor candidates for systemic delivery.8 Alternatively, ionizable lipids that become cationic at pH < 7.4 exhibit limited interactions with anionic membranes in circulation, but have strong electrostatic interactions along the endocytic pathway, thereby promoting efficient delivery. Several of these ionizable systems have shown promising in vivo data in pre-clinical models.12,14
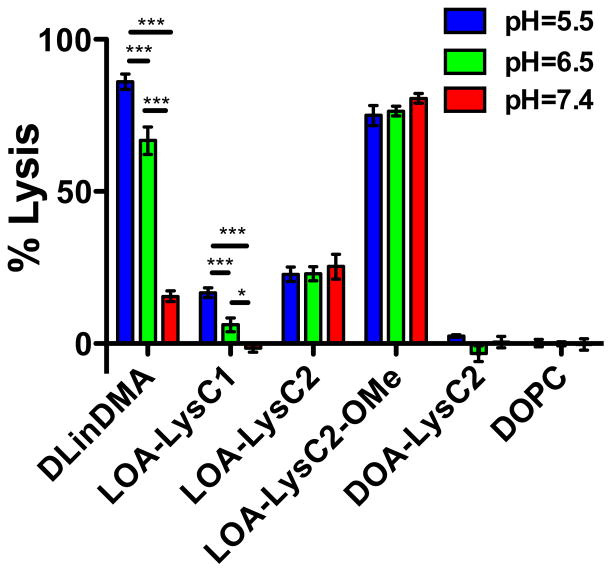
Membrane lysis activity at low-pH is expected to correlate with transfection efficiency, and may help identify lipids with favorable properties for siRNA delivery in vitro and in vivo. Understanding the pH-dependent interactions between ILL containing liposomes and anionic membranes provides insight into their expected behavior in circulation. We investigated the capacity of ILL containing liposomes to lyse biomembrane mimicking vesicles (BMV) encapsulating the fluorophore/quencher ANTS/DPX as a function of pH. BMV consisted of 45:20:20:15 DOPC:DOPE:DOPG:cholesterol28, and ILL liposomes contained 40:60 ILL:POPC. pH values of 7.4, 6.5, and 5.5 were chosen to assay lipid behavior under physiologically relevant conditions.
The synthesized ILLs provide a platform for investigating how changes in lipid net charge (LOA-LysC2-OMe), hydrophobic group fluidity (DOA-LysC2), and lipid ionization behavior (LOA-LysC1) impact membrane lysis compared to LOA-LysC2 liposomes. Additionally, we compare the behavior of ILL with DLinDMA, a well characterized ionizable lipid that is effective for in vivo siRNA delivery, and DOPC, a zwitterionic membrane phospholipid that is neutral across the tested pH range. DOPC liposomes show no lysis across the tested pH-range, as expected. DLinDMA liposomes show strong pH-dependent lysis characteristics, with a 5-fold increase in lysis at pH = 5.5 (86%) compared to pH = 7.4 (16%). Interestingly, DLinDMA still shows significantly more lysis at pH = 7.4 than DOPC, indicating that these lipids are not fully deprotonated at physiological pH (Fig. 4).
Figure 4.
Lysis of biomembrane mimicking vesicles (45:20:20:15 DOPC:DOPE:DOPG:Chol) by ILL containing liposomes (60:40 POPC:ILL)
Since all ILLs exhibit pH-dependent ionization, we anticipated that we would observe pH-dependent BMV lysis for all ILL liposomes due to changes in electrostatic interactions as a function of pH. Interestingly, we observed that only LOA-LysC1 shows pH-dependence membrane lysis (Fig. 4). LOA-LysC1 becomes cationic at a lower pH than all other ILL, possibly explaining this difference in biophysical behavior. At pH = 5.5, LOA-LysC1 shows a slightly lower extent of lysis (18%) than LOA-LysC2 (23%), our base lipid for all structural modifications. This may be due to the lower net surface charge of LOA-LysC1 at pH = 5.5. DLinDMA shows a similar pH-dependent lysis profile; however, the extent of lysis at pH=5.5 is four-fold higher, indicating it is more membrane active.
LOA-LysC2 and LOA-LysC2-OMe show pH-independent membrane lysis behavior. LOA-LysC2 promotes roughly 23% lysis, and LOA-LysC2-OMe, which is structurally analogous but has a methylated carboxylate, and therefore a higher charge density, shows 79% lysis across the tested pH range (Fig. 4). The difference in extent of lysis is likely due to differences in liposomal surface charge; LOA-LysC2-OMe liposomes are more cationic due to the absence of the anionic moiety, and therefore promote stronger electrostatic interactions with BMV. The pH-independent behavior of these two lipids was unexpected based on their ionization behavior; we predicted that the higher net surface charge at low pH would promote electrostatic interactions and increase membrane lysis. Instead, there appears to be a surface charge threshold above which there exists a constant, lipid dependent, extent of lysis. It is interesting to note that LOA-LysC2-OMe liposomes at pH = 7.4 have an identical surface charge to LOA-LysC2 liposomes at pH = 5.5, yet their extent of lysis is three-fold higher. This indicates that, independent of total surface charge, head group structure impacts the ILLs membrane lysis properties. This may be due to changes in lipid orientation, amine group presentation, or head group hydration in the bilayer caused by the presence of an anionic moiety in the LOA-LysC2 head group.
Decreased fluidity in the hydrophobic lipid tails is known to attenuate lipid mixing and fusion between lipid bilayers.20 Comparing the membrane lysis behavior of LOA-LysC2 and DOA-LysC2, which contains a more rigid dioleyl hydrophobic region, we see that decreasing the fluidity of the ILL tails prevents membrane lysis across the entire pH range (Fig. 4). In this case, the stability of the ILL liposome is likely preventing electrostatic-driven membrane fusion and lysis from occurring.
in vitro screening of ILL-siRNA complexes
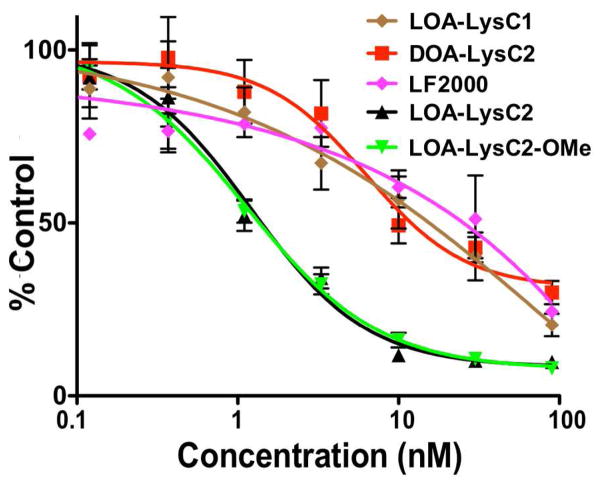
We investigated ILL lipoplexes as siRNA delivery systems in a stably transfected HeLa-Luc cell line. Anti-luciferase and non-specific siRNA was complexed with ILL liposomes (40:40:20, ILL:Chol:DOPE) at N:P = 10. This ratio showed equivalent knockdown with lower toxicity compared to higher N:P ratios (data not shown). Concentration dependent knockdown was assayed by incubating cells 0.11 – 90 nM of siRNA for 24 hrs at 37 °C. Lipofectamine 2000 (LF2000) was used as a positive control due to its well characterized, potent transfection in vitro. Our results show that all ILLs show potent, dose dependent knockdown (Fig. 5). Structural differences impact the transfection efficiency of these systems, and their efficacy generally correlates with their capacity to lyse BMV. LOA-LysC2 and LOA-LysC2-OMe, which showed the highest BMV lysis across all tested pH values, exhibited nearly identical knockdown behavior. Both ILLs exhibited an IC50 of 1.1 nM. DOA-LysC2 and LOA-LysC1, both of which showed lower BMV lysis, exhibited IC50 values of 11.5 nM and 16.6 nM respectively. All ILLs tested showed equivalent or superior transfection to LF2000, and no significant toxicity was seen in any formulation at the tested concentrations.
Figure 5.
siRNA mediate knockdown of Renilla luciferase in stably transfected HeLa-Luc cells. ILL lipoplexes (40:40:20 ILL:Chol:DOPE) at N:P = 10 were used for all experiments.
Though transfection efficiency generally correlates with the trends seen in the BMV lysis data, there are discrepancies in the overall behavior. For example, LOA-LysC2-OMe shows significantly higher BMV lysis than LOA-LysC2, but has almost identical transfection behavior. Also, DOA-LysC2 showed no BMV lysis but still showed potent transfection. These differences may be due to differences in the lipid formulation between the lysis and transfection experiments. Inclusion of fusogenic DOPE lipids in transfection formulations likely increases the transfection efficiency by promoting membrane disruption. However, the order of magnitude difference in IC50 between ILLs that effectively lyse BMV and those that do not indicates that this characteristic may be important regardless of liposomal formulation.
Factor VII knockdown IN VIVO with ILL liposomes
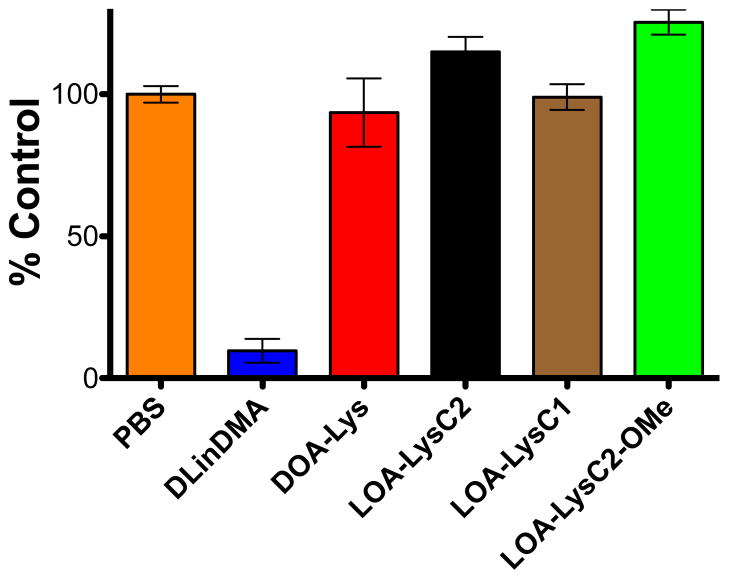
All ILL were tested for their ability to deliver siRNA in vivo. The mouse Factor VII model was chosen as the primary in vivo screen for ILL liposome mediated delivery of siRNA to hepatocytes via systemic injection.25 Stable siRNA encapsulating liposomes suitable for systemic delivery were formulated using 40:40:10:10 ILL:Chol:DSPC:PEG-DMG at a 15:1 lipid:siRNA (w:w) ratio. DSPC and PEG-DMG were used in place of DOPE in these formulations to promote liposomal stability in vivo. The encapsulation efficiency and particle sizes of the resulting particles were 83% to 93% and 86 nm to 104 nm (Table S1, Supporting Information). siRNA against the blood clotting protein Factor VII was administered at 5 mg/kg via tail vein injection. At 48 hrs post injection, blood was drawn and protein levels were assayed. Factor VII levels were unchanged compared to a PBS control for all tested ILL formulations (Fig. 6). A positive control formulation containing DLinDMA showed potent knockdown, thereby verifying the validity of the assay. Additional formulations containing LOA-LysC2 or LOA-LysC2-OMe with varying helper lipids and lipid ratios also showed no significant knockdown of Factor VII in vivo (Fig. S2 and S3, Supporting Information).
Figure 6.
siRNA-mediated knockdown of Factor VII in vivo in a murine model. ILL-siRNA formulations were administered at 5 mg/kg via tail vein injection (15:1 w:w ILL:siRNA), and Factor VII levels were assayed 48 hours following administration. ILL liposomal formulation – 40:40:10:10 ILL:Chol:DSPC:PEG-DMG
Despite their potency in vitro, ILL show no in vivo knockdown in hepatocytes. The difficulty of translating effective in vitro transfection reagents to viable in vivo siRNA delivery systems is well documented; cationic systems are rapidly cleared by the RES, preventing accumulation and transfection in hepatocytes.7,8 Additionally, structure-activity relationships for ionizable lipids have shown that slight changes in lipid structure significantly impact their in vivo efficacy.9,12,20 Though our ionization data indicates that ILL are more cationic at low pH, the two most membrane active ILL (LOA-LysC2-OMe and LOA-LysC2) show no pH dependent lysis behavior. Despite their promising in vitro transfection data, this indicates that these two ILL may be too cationic at pH = 7.4 to avoid protein or membrane interactions in circulation, stimulating RES uptake and preventing hepatocyte targeting. LOA-LysC1, which exhibits pH-dependent membrane lysis, has a lower extent of lysis at low pH that may prevent transfection in vivo. DOA-LysC2 appears to be less membrane active than all other ILL, possibly explaining its lack of efficacy. Changes to the liposomal formulation of LOA-LysC2 and LOA-LysC2-OMe did not improve results (data not shown). However, a broad, systemic manipulation of liposomal formulation, particularly the percentage of ILL as well as the choice of helper lipids or inclusion of targeting moieties may help to improve efficacy in these ILL systems.
SUMMARY
Here, we have described the synthesis and characterization of ionizable lysine-based lipids (ILL), a novel class of lipids with pH-dependent biophysical behavior. The goal of this work was to investigate how ionizable lipids with polyvalent, zwitterionic head groups behave compared to ionizable cationic systems. Four distinct ILLs were synthesized, and structural variations between ILLs impacted their biophysical behavior as well as their ability to deliver siRNA in vitro. All ILLs became increasingly cationic with a reduction in pH; however, only LOA-LysC1 was neutral at physiological pH. The ionization curves of ILL differ significantly from DLinDMA, likely due to the more complex, polyvalent head groups. All ILLs show potent siRNA mediated luciferase knockdown in vitro in a stably transfected HeLa-Luc cell line, with LOA-LysC2-OMe and LOA-LysC2 exhibiting IC50 values of roughly 1 nM. Interestingly, our in vitro data correlated well with our membrane lysis results, indicating that electrostatically driven membrane disruption promotes transfection. All ILLs form small diameter, monodisperse liposomes across multiple liposomal formulations, and efficiently encapsulate siRNA. However, ILL liposomes showed no in vivo knockdown in hepatocytes using a mouse Factor VII model. Given their potent transfection capabilities in vitro, the lack of in vivo efficacy may be due to poor targeting of liposomes to hepatocytes. Further optimization of these systems, through systemic manipulation of liposomal formulation or chemical modification of the ILL structures, may lead to higher in vivo efficacy.
Supplementary Material
Acknowledgments
This work was supported by the NSF Graduate Research Fellowship Program, NIH–EB003008, Deutsche Forschungs Gemeinschaft (DGF), and Pfizer. We thank Dr. Vincent Venditto, Dr. Emily Perttu, and Dr. Derek van der Poll for helpful discussion and technical advice.
ABBREVIATIONS
- ANTS
8-aminonophthalene-1,3,6-trisulfonate
- Chol
Cholesterol
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOPG
1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- DOTAP
1,2-dioleoyl-3-trimethylamonium-propane
- DLinDMA
1,2-dilinoleyloxy-3-N,N-dimethylaminopropane
- DPX
p-xylene-bis-pyridinium bromide
- DSPC
1,2-distearoyl-sn-glycero-3-phosphocholine
- ILL
Ionizable Lysine-based lipid
- LF2000
Lipofectamine 2000
- PBS
Phosphate buffered saline without calcium and magnesium salts
- PEG-DMG
1-(monomethoxypolyethyleneglycol)-2,3-dimyrystoylglycerol
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- TNS
Sodium 2-(p-toluidino)-6-naphthalenesulfonic acid
Footnotes
SUPPORTING INFORMATION DESCRIPTION
Supporting Information Available: Detailed synthetic schemes and chemical characterization data, additional lipid structures, siRNA encapsulation and liposomal characterization data, and additional Factor VII data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat Rev Drug Discovery. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 2.Peer DD, Lieberman JJ. Special delivery: targeted therapy with small RNAs. Gene Ther. 2011;18:1127–1133. doi: 10.1038/gt.2011.56. [DOI] [PubMed] [Google Scholar]
- 3.Puri A, Loomis K, Smith B, Lee J. Lipid-based nanoparticles as pharmaceutical drug carriers: from concepts to clinic. Crit Rev Ther Drug. 2009;26:523–580. doi: 10.1615/critrevtherdrugcarriersyst.v26.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discovery. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu JJ, Langer R, Chen J. A novel mechanism is involved in cationic lipid-mediated functional siRNA delivery. Mol Pharm. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J Intern Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanton MG, Colletti SL. Medicinal chemistry of siRNA delivery. J Med Chem. 2010;53:7887–7901. doi: 10.1021/jm1003914. [DOI] [PubMed] [Google Scholar]
- 8.Li W, Szoka FC. Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- 9.Adami RC, Seth S, Harvie P, Johns R, Fam R, Fosnaugh K, Zhu T, Farber K, McCutcheon M, Goodman TT, Liu Y, Chen Y, Kwang E, Templin MV, Severson G, Brown T, Vaish N, Chen F, Charmley P, Polisky B, Houston ME. An Amino Acid-based Amphoteric Liposomal Delivery System for Systemic Administration of siRNA. Mol Ther. 2011;19:1141–1151. doi: 10.1038/mt.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceballos C, Khiati S, Prata CAH, Zhang XX, Giorgio S, Marsal P, Grinstaff MW, Barthélémy P, Camplo M. Cationic Nucleoside Lipids Derived from Universal Bases: A Rational Approach for siRNA Transfection. Bioconjugate Chem. 2010;21:1062–1069. doi: 10.1021/bc100005k. [DOI] [PubMed] [Google Scholar]
- 11.Yang HW, Yi JW, Bang EK, Jeon EM, Kim BH. Cationic nucleolipids as efficient siRNA carriers. Org Biomol Chem. 2010;9:291–296. doi: 10.1039/c0ob00580k. [DOI] [PubMed] [Google Scholar]
- 12.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DWY, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, Maclachlan I, Cullis PR, Madden TD, Hope MJ. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 13.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DWY, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci USA. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abrams MT, Koser ML, Seitzer J, Williams SC, DiPietro MA, Wang W, Shaw AW, Mao X, Jadhav V, Davide JP, Burke PA, Sachs AB, Stirdivant SM, Sepp-Lorenzino L. Evaluation of Efficacy, Biodistribution, and Inflammation for a Potent siRNA Nanoparticle: Effect of Dexamethasone Co-treatment. Mol Ther. 2010;18:171–180. doi: 10.1038/mt.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Fan H, Levorse DA, Crocker LS. Interaction of cholesterol-conjugated ionizable amino lipids with biomembranes: lipid polymorphism, structure-activity relationship, and implications for siRNA delivery. Langmuir. 2011;27:9473–9483. doi: 10.1021/la201464k. [DOI] [PubMed] [Google Scholar]
- 16.Obata Y, Suzuki D, Takeoka S. Evaluation of cationic assemblies constructed with amino acid based lipids for plasmid DNA delivery. Bioconjugate Chem. 2008;19:1055–1063. doi: 10.1021/bc700416u. [DOI] [PubMed] [Google Scholar]
- 17.Zelphati O, Szoka FC. Mechanism of oligonucleotide release from cationic liposomes. Proc Natl Acad Sci USA. 1996;93:11493–11498. doi: 10.1073/pnas.93.21.11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Szoka FC. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 19.Hafez IM, Maurer N, Cullis PR. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 20.Heyes J, Palmer L, Bremner K, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Controlled Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann TS, Lee ACH, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, Judge AD, Lam K, Mcclintock K, Nechev LV, Palmer LR, Racie T, Röhl I, Seiffert S, Shanmugam S, Sood V, Soutschek J, Toudjarska I, Wheat AJ, Yaworski E, Zedalis W, Koteliansky V, Manoharan M, Vornlocher HP, Maclachlan I. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 22.Fricker G, Kromp T, Wendel A, Blume A, Zirkel J, Rebmann H, Setzer C, Quinkert RO, Martin F, Müller-Goymann C. Phospholipids and lipid-based formulations in oral drug delivery. Pharm Res. 2010;27:1469–1486. doi: 10.1007/s11095-010-0130-x. [DOI] [PubMed] [Google Scholar]
- 23.Walsh CL, Nguyen J, Szoka FC. Synthesis and characterization of novel zwitterionic lipids with pH-responsive biophysical properties. Chem Commun. 2012;48:5575–5577. doi: 10.1039/c2cc31710a. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Fan H, Levorse DA, Crocker LS. Ionization Behavior of Amino Lipids for siRNA Delivery: Determination of Ionization Constants, SAR, and the Impact of Lipid pKa on Cationic Lipid–Biomembrane Interactions. Langmuir. 2011;27:1907–1914. doi: 10.1021/la104590k. [DOI] [PubMed] [Google Scholar]
- 25.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Jayaprakash KN, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DWY, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DG. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nature Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey A, Cullis P. Modulation of membrane fusion by asymmetric transbilayer distributions of amino lipids. Biochemistry. 1994;33:12573–12580. doi: 10.1021/bi00208a007. [DOI] [PubMed] [Google Scholar]
- 27.Hafez IM, Cullis PR. Roles of lipid polymorphism in intracellular delivery. Adv Drug Delivery Rev. 2001;47:139–148. doi: 10.1016/s0169-409x(01)00103-x. [DOI] [PubMed] [Google Scholar]
- 28.Koynova R, Wang L, MacDonald RC. An intracellular lamellar-nonlamellar phase transition rationalizes the superior performance of some cationic lipid transfection agents. Proc Natl Acad Sci USA. 2006;103:14373–14378. doi: 10.1073/pnas.0603085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.