Abstract
Surface- and tip-enhanced Raman spectroscopy (SERS and TERS) are modern spectroscopic techniques, which are becoming widely used and show a great potential for the structural characterisation of biological systems. Strong enhancement of the Raman signal through localised surface plasmon resonance enables chemical detection at the single-molecule scale. Enhanced Raman spectra collected from biological specimens, such as peptides, proteins or microorganisms, were often observed to lack the amide I band, which is commonly used as a marker for the interpretation of secondary protein structure. The cause of this phenomenon was unclear for many decades. In this work, we investigated this phenomenon for native insulin and insulin fibrils using both TERS and SERS and compared these spectra to the spectra of well-defined homo peptides. The results indicate that the appearance of the amide I Raman band does not correlate with the protein aggregation state, but is instead determined by the size of the amino acid side chain. For short model peptides, the absence of the amide I band in TERS and SERS spectra correlates with the presence of a bulky side chain. Homo-glycine and -alanine, which are peptides with small side chain groups (H and CH3, respectively), exhibited an intense amide I band in almost 100% of the acquired spectra. Peptides with bulky side chains, such as tyrosine and tryptophan, exhibited the amide I band in 70% and 31% of the acquired spectra, respectively.
Introduction
Raman spectroscopy is steadily gaining importance for the structural characterisation of biological systems.1, 2 The relatively low sensitivity of Raman spectroscopy can be improved by utilising the electromagnetic field enhancement caused by rough metallic surfaces, such as silver or gold, through localized surface plasmon resonances.3 This phenomenon, known as surface-enhanced Raman scattering (SERS), enables chemical detection down to the single-molecule level.4, 5 In addition to high sensitivity, this label-free technique also provides information regarding the structural organisation of the specimen surface.6, 7 These and other advantages have made SERS one of the most extensively explored techniques in areas ranging from surface chemistry to biological chemistry and biomedical analysis.8, 9 There are two proposed mechanisms for Raman signal amplification.3 The first mechanism involves physical enhancement (GPhys), which is caused by the plasmonic excitation of the metal surface induced by the external electromagnetic field.10,11-13 In the theoretical models proposed for this enhancement, the chemistry of surface-specimen interactions is neglected.14 Alternatively, several studies have demonstrated that the experimentally observed enhancement is often substantially stronger than the theoretically predicted enhancement.15, 16 Thus, it was suggested that the chemical contribution (GChem), which includes chemical bonding and light-induced charge transfer between the adsorbed molecule and the substrate, must be considered. Recent experimental studies and mathematical calculations suggest that both GPhys and GChem should be considered simultaneously to explain the SERS phenomenon.17 However, the application of SERS is limited by lower spatial resolution. Consequently, important topographical information for the studied surfaces cannot be obtained. This disadvantage can be overcome by combining a Raman spectrometer with a scanning tunnelling microscope (STM) or an atomic force microscope (AFM). In this experimental setup, widely known as tip-enhanced Raman spectroscopy (TERS), a single gold or silver particle or edge at the apex of an AFM or STM tip greatly enhances the Raman signal, similar to SERS, in a small area around the tip.18 Unlike SERS, site-dependent, semi-quantitative conclusions can be drawn because the active enhancing particle is always the same. The AFM/STM component allows the tip position to be controlled relative to the sample, and consequently, high spatial resolution can be attained.19, 20 Working on a nanometer-scale resolution, which depends mainly on the TERS tip diameter, is critically important for studying biological samples, such as eukaryotic cells, bacteria, DNA, RNA, viruses and amyloid fibrils.21-23 It was recently demonstrated that TERS could potentially resolve the nucleotide sequence of a single DNA strand and reveal structural information of insulin fibrils on the nanometer scale.23-25 The ability of the electromagnetic field to enhance the Raman signal is comparable to the SERS effect mentioned above and consequently decays quickly as the tip-specimen distance increases. As a result, only molecules within 3-4 nm from the tip apex experience signal enhancement.26, 27
A protein Raman spectrum is typically composed of contributions from three major types of vibrational modes originating from the polypeptide backbone (amide bands) and aromatic and non-aromatic amino acid side chain residues. The positions of the amide bands depend on the conformation of the polypeptide backbone and intra- and intermolecular hydrogen bonds. Consequently, these bands can be correlated to the protein secondary structure.1, 28 The amide II band (1520-1570 cm−1) is primarily related to C-N stretching, N-H bending and C-C stretching and is very weak in non-resonance Raman spectra, which causes it to be nearly undetectable.29 Amide III bands (1230-1270 cm−1) overlap with vibrational bands of CH2 and C-C vibrations, which significantly complicates and limits their interpretation with regard to the secondary structure.1, 28 The amide I band (1640-1678 cm−1) does not overlap with the vibrational bands of other functional groups and can be directly used for the characterisation of protein secondary structure.30 For example, the position and intensity of the amide I band has been utilised to describe the structural organisation of insulin fibrils30 and globular proteins.31
In some SERS spectra acquired from peptides,32, 33 proteins,34 and microorganisms,35 it has been observed that the amide I band is dramatically suppressed. It has been hypothesised that this “silence” of the amide I band is very unlikely to be caused by a parallel orientation of all the peptide bonds in the analysed protein to the electrochemically roughened metal surface or the incoming laser light.32 Two groups proposed that the absence of the amide I band in SERS spectra is a consequence of poor specific adsorption of the peptide bonds to the metal particles.8, 32 It was proposed that no enhancement occurs even if a peptide bond is in close proximity to the metal surface if a peptide bond is not directly adsorbed on a metal particle. Lack of experimental evidence currently limits the interpretation of such SERS and TERS spectra, and the systematic study presented herein aims to clarify the nature of suppressed amide I bands in SERS and TERS spectra of proteins and protein aggregates.
We hypothesized that the silence of amide I bands in surface-enhanced Raman spectra is a consequence of the distancing of peptide bonds from the metal surface. The proximity could depend on the length (bulkiness) of the amino acid side chains, which act as spacers between the metal particles and the peptide bonds. Based on this hypothesis, amino acid sequences with small side chain groups, such as H and CH3, would be expected to consistently exhibit spectra with an amide I band. Conversely, peptides with bulky side chain groups, such as Trp or Tyr, would be expected to frequently exhibit spectra with a suppressed or absent amide I band. In this study, SERS experiments were performed on selected homo-peptides with various side chains. We found that homo-glycine and -alanine peptides with small side chain groups (H and CH3, respectively) exhibited the amide I band in nearly 100% of the acquired spectra. This value decreased dramatically as the steric hindrance of the substituents increased. For example hexa-tyrosine and tryptophan-rich peptides exhibited the amide I band in only approximately 70% and approximately 31% of the recorded SERS spectra, respectively. These results confirmed our hypothesis that small amino acid substituents, such as H and CH3, allow the peptide bonds to be in a close proximity to the surface of metal particles, whereas bulky substituents, such as phenyl and indole, shield this bond from the particle evanescent field. Notably, the SERS experiments conducted here were performed independently on different Raman setups with different SERS substrates and laser wavelengths, which strengthens the validity of our results.
Experimental
Gold colloid nano-particles
DXR/SERS Analysis package, containing 70 nm gold nano-particles and gold-coated glass slides, was purchased from Thermo Scientific, Madison, WI. According to the Certificate of Analysis, the nano-particle colloid solution has a SPR absorption peak at 541 nm, pH 6.8 and zeta potential of −28 mV. The standard deviation of the particles seize as measured by DLS is 7%. Prior to mixing with the analyzed specimen, gold nono-particles were additionally dialyzed against distilled water.
Insulin protein
Bovine insulin (Sigma-Aldrich, St. Louis, MO) was dissolved (2 mg/ml) in distilled water. The protein solution was mixed with 70 nm spherical gold colloid nano-particles (DXR/SERS Analysis Package, Thermo Scientific, Madison, WI) in 1:1 V/V ratio. After being mixed with an analyzed specimen, the solution was deposited onto a gold-coated glass slide, which was also provided by DXR/SERS Analysis package (DXR/SERS Analysis Package kit, Thermo Scientific, Madison, WI). There is an indication in the literature that the gold surface may enhance the Raman scattering when a nano-particle is in a close proximity to the surface.36, 37 However, understanding the exact enhancement mechanism of a specific SERS system is beyond the scope of this manuscript. The solution was allowed to air dry prior to spectra acquisition.
For TERS experiments on native insulin, the protein was dissolved in water (c = 0.5μM) and 1.5 μL was dropped on a clean glass slide. After drying under argon 190 TERS spectra were acquired (t = 2 s) on a grid with a point-to-point distance of 1 nm. λ = 530,9 nm, P = 450 μW). Topology of the TERS spectra acquisition is shown in the Fig. S1.
Insulin fibrils
Bovine insulin (Sigma-Aldrich, St. Louis, MO) was dissolved (60 mg/ml) in HCl, pH 2.5. The protein solution was heated at 70 °C for 2.5 hours without stirring. An aliquot of a fibril gel was diluted 1:10 V/V with HCl, pH 2.5 and mixed with 70 nm spherical gold colloid nano-particles (DXR/SERS Analysis Package, Thermo Scientific, Madison, WI) followed by deposition onto a gold-coated surface (DXR/SERS Analysis Package, Thermo Scientific, Madison, WI) or a 6 nm evaporated silver island films that was annealed for 60 s at 180 °C. The solution was allowed to air dry prior to spectra acquisition. Silver island films were examined with AFM (data not shown) and the changes in surface morphology was noticed as a result of film exposure to a low pH fibril solution. These changes could affect the SERS signal including the observed reduction in SERS intensity while Raman band positions remained unchanged.
For TERS measurements an aliquot of the fibril gel was resuspended in HCl (pH 2.5) solution with a 1:100 dilution factor (v/v). A drop of this solution was placed onto a precleaned glass. After the surface was exposed to the solution for 2–3 min, the solution excess was gently removed, and the surface was rinsed with HCl (pH 2.5) solution and dried under an argon flow. AFM topography image is shown in Fig. S2.
Peptides
Penta-glycine and penta-alanine were purchased from Bachem Bioscience (King of. Prussia, PA). Hexa-tyrosine and tryptophan-rich peptide with the sequence Trp-Arg-Trp-Trp-Trp-Trp were purchased from American Peptides (Sunnyvale, CA). Peptides were dispersed in distilled water and mixed 1:1 V/V with 70 nm spherical gold colloid nano-particles (DXR/SERS Analysis Package, Thermo Scientific, Madison, WI). The solution was placed onto gold-coated surface (DXR/SERS Analysis Package, Thermo Scientific, Madison, WI) and air dry prior to spectra acquisition.
Alternatively, hexa-Gly (c = 0.1μM), penta-Ala (0.02 μM) and tri-Tyr (c= 0.1 μM), purchased from Sigma-Aldrich, St. Louis, MO, were dissolved in water and 1.5-3 μL were placed on a 6 nm evaporated silver island film that was annealed for 60 s at 180 °C. The samples were dried under argon and used within hours.
SERS instrumentation
SERS spectra of analyzed specimens mixed with gold nano-particles were recorded on Renishaw in Via confocal Raman spectrometer coupled with Leica microscope, 20x long-range objective using WiRE 2.0 software. A 785 nm laser light was utilized for the excitation. For the spectral acquisition, automatic mapping stage (Nanonics AFM MultiView 1000 system) was used. For each sample 400-1600 SERS spectra were collected with 1 micron step from each other. Multiple consecutive spectral acquisitions showed no evidence of photodegradation. The obtained spectra were further evaluated with GRAMS/AI 7.01 software. For all reported spectra no smoothing or a background substation was performed.
SERS spectra of analyzed specimens deposited onto silver island films were acquired on an inverted Raman microscope (XploRA INV, Horiba Scientific, France) using 532 nm excitation with a laser power of 90 μW. Spectral acquisition time was 1s. As mentioned previously, the raw spectra were further evaluated with using GRAMS/AI 7.01 software. SERS spectra were recorded on the substrate surface with a point-to-point distance of 200 nm.
TERS instrumentation
Tip-enhanced Raman spectra were acquired on a Nanowizard I AFM (JPK Instruments AG, Germany) mounted on an inverted microscope (Olympus IX70, Japan). Experimental conditions were as follows: oil immersion objective, 60x, NA=1,45 (Olympus, Japan), mounted on a Piezo (PIFOC, Physik Instrumente, Germany) enabling a focus control in z-direction of the TERS tip to the laser. The amplitude of the tip movement in AC-mode was ~10 nm.
A confocal Raman spectrometer (LabRam HR, Horiba Jobin Yvon, France) with a liquid nitrogen-cooled CCD camera (ISA Spectrum One, Horiba Jobin Yvon, France) was coupled to the AFM. A Krypton ion laser (Innova 300c, λ = 530,9 nm, USA) was used as the excitation source with a power on the sample of 1.1 mW. The illumination light was linearly polarized. However, in a TERS experiment this is not necessarily important, as the polarization at the sample is likely to be dominated by effects of the tip/antenna, hence a polarization orthogonal to the sample surface is to be expected.38 TERS tips were prepared by evaporation of 20 nm silver on a commercial non-contact silicon AFM tip (NT-MDT) and stored under argon till used.
Each TERS spectrum was accumulated for 10 sec. Spectra were collected from the fibril surface with a lateral distance step that varied from 0.5 to 10 nm. The lateral steps were controlled by an additional 100×100 μm sample scanning stage (P-734, Physik Instrumente, Germany). To confirm that the TERS tip was not contaminated during the acquisition of spectra of the fibril surface, a reference spectrum from the glass slide was recorded at the end of each measurement. Again the untreated spectra were evaluated as previously mentioned.
Results and discussion
Insulin fibrils
Insulin fibrils were immobilised on a glass slide, and TERS spectra were measured at randomly chosen regions of the fibril surfaces based on the experimental conditions described previously.23 Assessment of the data indicated that 88 spectra (Fig. 1) from a total of 162 acquired spectra displayed a suppressed amide I band, while 74 spectra displayed an intense amide I band.39 The absence of amide I bands in the TERS spectra of insulin fibrils could originate from a perpendicular orientation of the peptide bonds relative to the main axis of the TERS tip. This explanation is rather unlikely because suppressed amide I bands can be observed in β-sheet structures and in α-helix structures,22 whereas peptide bonds should face towards the tip randomly.
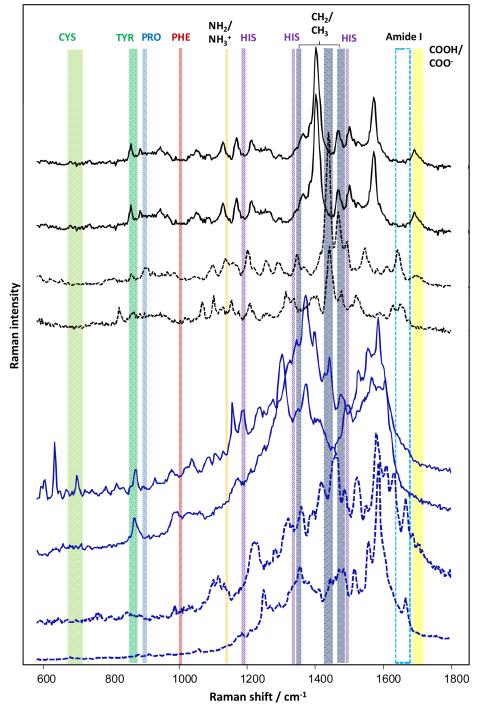
Fig. 1.
Selected TERS spectra (black) and SERS spectra (blue) of insulin fibrils with suppressed amide I bands (—) and with evident amide I bands (---). Insulin fibrils were immobilized on a glass slide for TERS measurements. An AFM tip covered by silver nanoparticles was used. A silver island film evaporated on a glass slide18 was used in the SERS experiments. For both TERS and SERS measurements, the excitation laser light with l = 532 nm was used.
The insulin fibrils were also deposited onto an evaporated silver island film. In the collected SERS spectra, almost 50% of the spectra displayed a suppressed amide I band, as shown in selected spectra in Fig. 1. In Fig. 1, the variations between the SERS and TERS spectra are quite apparent. The SERS spectra are similar, whereas the TERS spectra are different from the SERS spectra and also vary among other TERS spectra. The difference between the SERS and TERS spectra can be explained by the distinctive effects of sample and substrate preparation. Due to the nature of sample preparation, an equilibrium between the specific SERS substrate and the sample will form, and preferential binding of the specimen to the substrate is expected.
In TERS, the specimens are measured “as is” because the tip can access chemically non-preferred sites in the molecule. Consequently, the TERS spectra probe provides a slightly different but possibly more natural organization of the specimen. Furthermore, the SERS spectra reflect an average of many specimen sites and hot spots.40, 41 The TERS spectra originate from local, distinct sites, which is reflected in their spectral appearance and their variation.42, 43
In our experiments, a similar proportion of spectra for insulin fibrils exhibited suppressed amide I bands using both TERS and SERS. An individual TERS spectrum was measured from a single fibril using a single nano-size metal tip. In SERS experiments, each Raman spectrum was collected from a sample area of approximately 1 μm2 containing a large number of fibrils and metal particles. The increase in the number of nanoparticles/fibrils contributing to a single (averaged) SERS spectrum was expected to cause a significant decrease in the number of spectra with a silent amide I band. Therefore, we concluded that each SERS spectrum should be dominated by the contribution from a single hot spot. This conclusion is further supported by the fact that only one in ten SERS measurements from different spots (locations) on the silver film with deposited fibrils yielded an enhanced SERS signal. The remainder of the locations were SERS-inactive (data not shown). In the experimental setup in which gold nanoparticles were utilised for SERS characterisation of the insulin fibrils (Fig. S3), every acquisition spot was SERS-active (hot spot). The latter result indicates that more than one active spot could contribute to an individual SERS spectrum. As a result, only a third of the SERS spectra displayed a suppressed amide I band (discussed below).
Native insulin
To demonstrate that the presence or absence of the amide I band in enhanced Raman spectra depends on the amino acid sequence rather than a specific property of the amyloid fibril surface or the protein aggregation state, we studied native insulin using SERS and TERS. Insulin is a small protein hormone that consists of 51 amino acids organised in two peptide chains linked by disulphide bridges. The short protein sequence of insulin would be expected to provide equal accessibility to the 70-nm gold or silver nanoparticles.
Similar to the spectra acquired for the insulin fibrils, the SERS spectra of native insulin are very heterogeneous. Approximately 50% of the acquired spectra displayed a suppressed amide I band (Fig. 2). The TERS spectra of insulin are less diverse and predominantly exhibited a clear baseline. We often observed tyrosine (856 cm−1), cysteine (approximately 674 cm−1) and phenylalanine (1000 cm−1) bands and strong peaks approximately 1392 cm−1, which most likely originate from aliphatic groups. One can expect that for the direct verification the acquired TERS and SERS spectra originate from the analyzed protein specimen, a comparison with a normal Raman spectrum would be helpful (Fig. S4). However, this kind of information might be even misleading. First of all, the Raman cross-section of particular bands most likely will be different in enhanced and normal Raman spectra. Thus, no valuable information can be extracted from the comparison of relative intensities normal and enhanced spectra. It has been reported that specific bands can be missing in normal Raman spectra, while they are visible in the SERS/TERS spectra. This observation can be explained with the different selection rules in enhanced Raman spectroscopy (change of polarizability). Such examples are well documented in the literature. For example, amide II bands are invisible in the normal Raman spectra acquired from protein specimens and often observed in TERS and SERS protein spectra.29, 39 It has been also documented that the position of vibrational modes can change in the enhanced Raman spectra relative to that in normal Raman spectra depending on the interaction of the molecule and the metal nanoparticle.38, 44
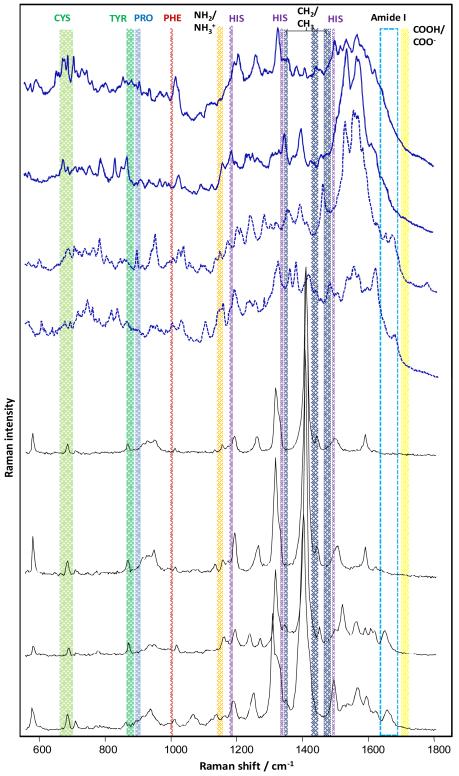
Fig. 2.
SERS (black) and TERS (blue) spectra of native insulin with suppressed amide I bands (—) and with detectable amide I bands (---). For SERS, insulin was mixed with gold nano-particles and laser excitation at λ =785 nm was used. For TERS a silver island coated AFM tip and excitation at λ =532 nm was used.
Our TERS data indicate that similar to SERS experiments, almost 50% of all the collected TERS spectra exhibited a silent amide I band (Fig. 2). Hence, we conclude that the suppression of the amide I band is not determined by the aggregation state of the protein but is due to the protein amino acid sequence.
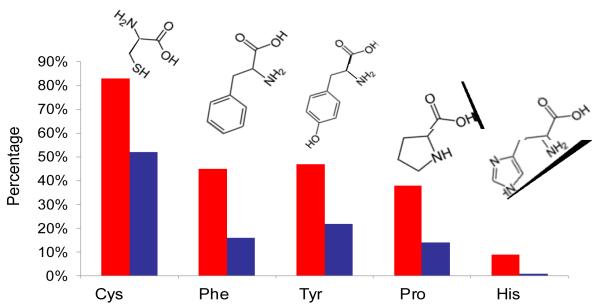
If surface plasmons preferentially enhance Raman bands of bulky side chains over the amide I band, the respective vibrational modes should be found more often in SERS/TERS spectra with a suppressed amide I band than in spectra with an intense amide I band. To confirm this hypothesis, 100 SERS spectra of native insulin without an amide I band and 100 spectra with the amide I band were assessed with respect to the occurrence of specific amino acids (Fig. 3). Amino acid residues with characteristic marker bands, such as cysteine (Cys), phenylalanine (Phe), tyrosine (Tyr), proline (Pro) and histidine (His), were chosen.29 As evident from Fig. 3, SERS spectra with suppressed amide I bands contain Cys contribution 1.5 times more often than spectra with intense amide I bands. The ratio for sequences with Tyr was nearly 2:1 and further increased for Phe and Pro (almost 3:1). His contributions resulted in a maximum with a ratio of 9:1. These results support the assumption that bulky substituents on the amino acid side chains shield the peptide bonds from the enhancing field more effectively with increasing size.
Fig. 3.
Propensities of Cys, Phe, Tyr, Pro and His in SERS spectra of native insulin with a silent Amide I band (red), and intense Amide I (blue).
Short model peptides
To further confirm that the size of the side chain determines the presence/absence of the amide I Raman band, the SERS spectra of short homo-peptides, including penta-glycine (Gly), penta-alanine (Ala), hexa-tyrosine and a tryptophan (Tpr)-rich peptide (Trp-Arg-Trp-Trp-Trp-Trp), were recorded. All the peptides were mixed with gold nanoparticles at equal concentrations and deposited onto a gold surface. In total, 411 SERS spectra were collected for each peptide using an excitation wavelength of 785 nm (Fig. 4a).
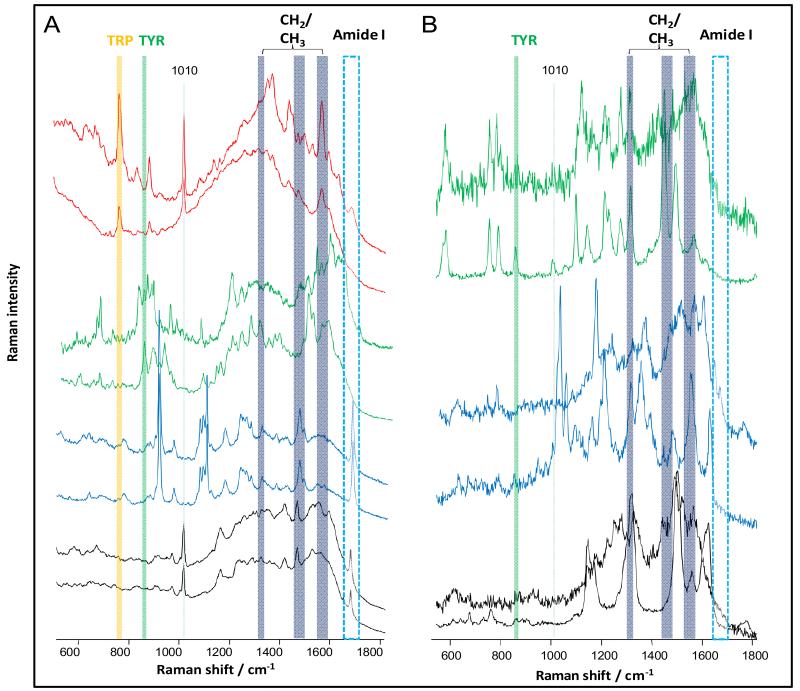
Fig. 4.
A) Selected SERS spectra of a Tpr-rich peptide (Trp-Arg-Trp-Trp-Trp-Trp) (red), hexa-Tyr (green), penta-Ala (blue) and penta-Gly (black), measured at 785 nm on gold colloids, B) selected SERS spectra of tri-Tyr, penta-Ala and penta-Gly, measured at 530 nm on silver island films.
The SERS spectra acquired from the penta-Gly homo-peptide exhibited a high degree of similarity. All the spectra displayed intense amide I bands. In addition to intense CH2 modes (1455 cm−1),45 a sharp and intense band was observed at 1010 cm−1, which can most likely be attributed to a C-C stretch.46
Similar to penta-Gly, the SERS spectra of penta-Ala also exhibit a high degree of similarity. All the spectra displayed intense amide I bands, indicating that the plasmon enhancement can access the peptide bond through the distance of the −CH3 group.45 In addition, intense vibrational modes attributed to CH2 and CH3 groups were detected.
In contrast, the SERS spectra of hexa-Tyr and a Trp-rich peptide are quite heterogeneous (Fig. 4). One cause of this heterogeneity is the different vibrational modes of the ring and =HN+ (Trp).45, 46 Each of the SERS spectra acquired from hexa-Tyr exhibited characteristic vibrational bands at 826 cm−1 and 856 cm−1 (ring breathing).45 The SERS spectra for the Trp-rich peptide also show this band (ring breathing) at 760 cm−1 and a band at 1010 cm−1, similar to penta-Gly.
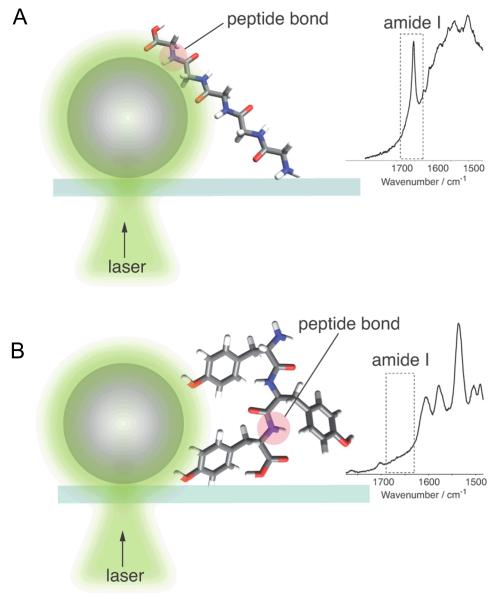
The percentages of SERS spectra with silent and intense amide I bands for each peptide were evaluated, and the results are summarised in Table 1. While all 411 SERS spectra for penta-Ala and penta-Gly display intense amide I bands, only 76% of hexa-Tyr and 31% of Trp-rich peptide SERS spectra exhibit intense amide I bands. These data confirm that the presence or absence of amide I bands in enhanced Raman spectra is apparently dependent on the proximity of the peptide bond to the nanoparticle surface. Bulky side chain groups orient the peptide bonds too far from the evanescent field of the surface plasmon and impede the enhancing effect, which suppresses amide I bands in the spectra.
Table 1.
Percentage of SERS spectra with suppressed amide I bands for each of the analyzed peptides.
| Peptide | Average % of SERS spectra without Amide I |
|
|---|---|---|
| 785/Au | 532/Ag | |
| Penta-Gly | 0 | - |
| Hexa-Gly | - | 6±1.5 |
| Penta-Ala | 0 | 7±2 |
| Tri-Tyr | - | 21±2 |
| Hexa-Tyr | 24±3 | - |
| Trp-Arg-Trp-Trp- Trp-Trp |
69±5 | |
Similar results were obtained from independent SERS measurements of similar homo-peptides (hexa-Gly, penta-Ala and tri-Tyr) using silver island films and a laser line at 532 nm (Fig. 4b). Silent amide I bands were observed in the same number of spectra for both hexa-Gly and penta-Ala, which are peptides with small side chain groups (6±1.5% and 7±2%, respectively). These values are relatively high compared with the values observed for gold colloids at 785 nm (0%), which might be due to the unique characteristics of the different metals (silver vs. gold). The dependence of the number of spectra with silent amide I bands on the nature of the metal colloids is discussed below. Nevertheless, the trend observed in our results, in which an increasing number of spectra do not display the amide I band as the size of the amino acid side chain increases, is clearly evident. The probability of silent amide I bands is three times higher in the tri-Tyr SERS spectra (21±2%). Approximately the same number of spectra without amide I bands were collected for the hexa-Tyr and tri-Tyr peptides (see Table 1). Thus, these data confirm that the number of spectra with a suppressed amide I band correlates strongly with the bulkiness of the amino acid side chain (Scheme 1).
Scheme 1.
Amino acid side chain length determines the silence of the amide I Raman band in surface enhanced spectra. Localization of the peptide bond in close proximity (A) to the surface of a 70 nm metal nano-particle leads to the enhancement of the vibrational mode and the appearance of amide I band in the SERS or TERS spectra. Distancing of the peptide bond (B) by bulky side chains in amino acids dramatically diminishes its enhancement, which causes a suppression of the amide I band. For better representation of the amide I silence phenomenon, a proper scaling of the peptide bond to the metal particle was avoided.
Intriguingly, Tyr ring vibrational modes (at 856 cm−1 and/or at 823 cm−1) were observed in 100% of the SERS spectra acquired from homo-Tyr peptide (Figure 4) while amide I bands were absent in 24±3% of these spectra. This clearly indicates that the enhancement affects one part of the molecule leaving another branch untouched. Presumably this is due to a specific orientation of the molecule on the metal surface. It is noteworthy, that the Tyr amino acid residue including the side chain and the peptide bond is not bigger than 1-nm. The fact that the Tyr side chain (phenol ring) can distance the peptide bond from the signal enhancement indicates in our opinion that the peptide bond should be in a direct contact with the metal surface to detect amide I vibrational modes in SER spectra. From this it can be concluded that a direct contact with the surface is an essential requirement for chemical interactions in SERS. Of course, more work is necessary to determine the distance dependence of the amide I mode enhancement but this is beyond the scope of this manuscript. One can hypothesize that the metal-peptide bond distance plays a crucial role in the enhancement and varies for different chemical groups and/or vibrational modes. The determination of the distance dependence of SERS for the peptide bond and Tyr will be a target of future research.
An estimation of the number of enhanced Raman spectra with suppressed amide I bands collected from native insulin and insulin fibrils is shown in Table 2. From these data, it is evident that the number of enhanced spectra with a suppressed amide I band varies between 35% and 63%. The geometrical configuration of a metal nanostructure in addition to its electromagnetic properties defines the enhancement activity.47, 48 In the homo-peptide experiments, the proportion of spectra with suppressed amide I bands varies with the excitation wavelength/metal identity. Spectra with suppressed bands are observed more frequently in SERS with silver island films (530 nm excitation) than with gold nanoparticles (785 nm excitation) (Table 2).
Table 2.
Amount of spectra with suppressed Amide I bands in TERS and SERS studies of native insulin and insulin fibrils.
| Method | Metal of nano- particles |
Excitation wavelength, nm |
Native Insulin, average, % |
Insulin fibrils, average, % |
|---|---|---|---|---|
| TERS | silver | 532 | 53±4 | 58±26 |
| SERS | silver | 532 | - | 63±9 |
| SERS | gold | 785 | 40±3.5 | 35±4.5 |
The use of gold colloids (785 nm) yields 35±4.5% of spectra without amide I bands for insulin fibrils (selected spectra in Fig. S1), while the use of silver island films (532 nm) yields 63±9% of spectra without amide I bands (selected spectra in Fig. 1). The TERS of native insulin using AFM tip covered with silver (532 nm) result in 53±4.5% of spectra with suppressed amide I bands, while the use of gold particles (785 nm) results in 40±3.5% of spectra with suppressed amide I bands (Fig. 2 and Table 2). As mentioned previously, 88 of 162 total TERS spectra displayed suppressed amide I bands. For our statistical analysis, we considered all the TERS spectra acquired from one fibril as a separate group. The large standard deviation reported in Table 2 for the TERS experiments (58±26%) indicates that some of the fibrils yielded only a few spectra with suppressed amide I bands, while some fibrils exhibited a large number of these spectra. A better understanding of the factors affecting these spectral results requires additional studies and is beyond the scope of this article.
As indicated in the Introduction, surface enhanced Raman spectra with and without amide I band have been reported in the literature for many different protein containing systems and various SERS substrates. In this study, we utilized two different SERS substrates, gold colloids and silver island films for generating SERS of insulin fibrils, native insulin and several short peptides. As evident from Table 2, a significant number of surface enhanced Raman spectra of insulin in native and fibrilar form contain no amide I band. The latter is true for all tested SERS substrates and conditions as well as for TERS of insulin fibrils. We believe that the appearance and disappearance of amide I band in surface enhanced Raman spectra is a general phenomenon and could be observed at various SERS and TERS conditions for protein containing systems.
Conclusions
The application of SERS and TERS for studying biological systems, such as globular proteins and amyloid fibrils, revealed that a substantial number of the collected spectra display a suppressed amide I band.32-35, 45, 49 Approximately 50% of all the acquired spectra from insulin fibrils do not exhibit amide I bands. This result was independent of the applied technique (TERS or SERS), nanoparticle material and instrument and raised the question as to whether this is a characteristic of the fibrillar state of insulin molecules. The study of native insulin using both SERS and TERS revealed the same result: almost 50% of the acquired spectra had a silent amide I band, suggesting that this phenomenon is not caused by the protein aggregation state. Moreover, it was shown that the absence of this band could not be attributed to a specific experimental setup or an aggregation state of the protein, but rather to the bulkiness of the amino acid side chain. SERS studies of different homo-peptides consisting of Gly-, Ala-, Tyr- and Trp-rich chains under a variety of experimental conditions clearly demonstrated that the absence of amide I bands in the spectra depended on the size of the amino acid side chain. The side chain increases the distance between the peptide bond and the metal nanoparticle preventing their immediate contact. Based on these results, we suggest that the surface enhancement of the amide I Raman band occurs due to the short-range chemical mechanism in contrast to the physical mechanism. This investigation enhances our ability to assign and better understand TERS and SERS spectra of protein samples.
A new study published after the submission of this manuscript
A research paper has been recently published by C. Blum, T. Schmid, L. Opilik, N. Metanis, S. Weidmann and R. Zenobi in J. Phys. Chem. C. (2012, 116, 23061-23066), which addresses the problem of a silent amide I band in protein SERS and TERS spectra. It is interesting that the authors report no TERS or SERS spectra of peptides and proteins, which would show the presence of amide I band. Further investigation is required to understand the difference in experimental results obtained in these two studies.
Supplementary Material
Acknowledgements
This work was supported by Award Number R01AG033719 from the National Institutes of Health (I.K.L.).
Footnotes
Electronic Supplementary Information (ESI) available: Fig S1. See DOI: 10.1039/b000000x/
Notes and references
- 1.Lednev IK. Uversky VN, Permyakov EA, editors. Protein Structures, Methods in Protein Structures and Stability Analysis. Nova Sci. 2007:1–26. [Google Scholar]
- 2.Oladepo SA, Xiong K, Hong Z, Asher SA, Handen J, Lednev IK. Chem. Rev. 2012;112:2604–2628. doi: 10.1021/cr200198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes CL, Yonzon CR, Zhang X, Van Duyne RP. J. Raman Spectrosc. 2005;36:471–484. [Google Scholar]
- 4.Kneipp J, Kneipp H, Kneipp K. Chem. Soc. Rev. 2008;37:1052–1060. doi: 10.1039/b708459p. [DOI] [PubMed] [Google Scholar]
- 5.Hering K, Cialla D, Ackermann K, Dorfer T, Moller R, Schneidewind H, Mattheis R, Fritzsche W, Rosch P, Popp J. Anal. Bioanal. Chem. 2008;390:113–124. doi: 10.1007/s00216-007-1667-3. [DOI] [PubMed] [Google Scholar]
- 6.Rasmussen T, Tantipolphan R, van de Weert M, Jiskoot W. Pharm. Res. 2010;27:1337–1347. doi: 10.1007/s11095-010-0116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qian XM, Nie SM. Chem. Soc. Rev. 2008;37:912–920. doi: 10.1039/b708839f. [DOI] [PubMed] [Google Scholar]
- 8.Pavan Kumar GV, Ashok Reddy BA, Arif M, Kundu TK, Narayana C. J. Phys. Chem. 2006;110:16787–16792. doi: 10.1021/jp063071e. [DOI] [PubMed] [Google Scholar]
- 9.Stacy AM, Van Duyne RP. Chem. Phys. Lett. 1983;102:365–370. [Google Scholar]
- 10.Jensen L, Aikens CM, Schatz GC. Chem. Soc. Rev. 2008;37:1061–1073. doi: 10.1039/b706023h. [DOI] [PubMed] [Google Scholar]
- 11.Rojas R, Claro F. J. Chem. Phys. 1993;98:998–1006. [Google Scholar]
- 12.Etchegoin PG, Le Ru EC. Phys. Chem. Chem. Phys. 2008;10:6079–6089. doi: 10.1039/b809196j. [DOI] [PubMed] [Google Scholar]
- 13.Jeanmaire DL, Van Duyne RP. J. Electroanal. Chem. 1977;84:1–20. [Google Scholar]
- 14.Moskovits M. J. Raman Spectrosc. 2005;36:485–496. [Google Scholar]
- 15.Nikoobakht B, Wang B, El-Sayed MA. Chem. Phys. Letters. 2002;366:17–23. [Google Scholar]
- 16.Alfimov MV, Fedorov YV, Fedorova OA, Gromov SP, Hester RE, Lednev IK, Moore JN, Oleshko VP, Vedernikov AI. Spectrochim. Acta A. 1997;53:1853–1865. [Google Scholar]
- 17.Duan S, Xu X, Luo Y, Tian ZQ. Chem. Commun. 2011;47:11438–11440. doi: 10.1039/c1cc14962h. [DOI] [PubMed] [Google Scholar]
- 18.Stöckle RM, Suh Y, Deckert V, Zenobi R. Chem. Phys. Lett. 2000;318:131–136. [Google Scholar]
- 19.Anderson MS. Appl. Phys. Lett. 2000;76:3130–3132. [Google Scholar]
- 20.Hayazawa N, Inouye Y, Sekkat Z, Kawata S. Opt. Commun. 2000;183:333–336. [Google Scholar]
- 21.Deckert-Gaudig T, Deckert V. Current Opin. Chem. Biol. 2011;15:719–724. doi: 10.1016/j.cbpa.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 22.Deckert-Gaudig T, Deckert V. Phys. Chem. Chem. Phys. 2010;12:12040–12049. doi: 10.1039/c003316b. [DOI] [PubMed] [Google Scholar]
- 23.Deckert-Gaudig T, Kämmer E, Deckert V. J. Biophotonics. 2012;5:215–219. doi: 10.1002/jbio.201100142. [DOI] [PubMed] [Google Scholar]
- 24.Treffer R, Deckert V. Current Opin. Biotech. 2010;21:4–11. doi: 10.1016/j.copbio.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Treffer R, Lin X, Bailo E, Deckert-Gaudig T, Deckert V. Beilstein J. Nanotech. 2011;2:628–637. doi: 10.3762/bjnano.2.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiles PL, Dieringer JA, Shah NC, Van Duyne RP. Annu. Rev. Anal. Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- 27.Pettinger B, Domke KF, Zhang D, Picardi G, Schuster R. Surface Sci. 2009;603:1335–1341. [Google Scholar]
- 28.Fabian H, Anzenbacher P. Vibr. Spectrosc. 1993;4:125–148. [Google Scholar]
- 29.Ortiz C, Zhang D, Ribbe AE, Xie Y, Ben-Amotz D. Biophys. Chem. 2007;128:150–155. doi: 10.1016/j.bpc.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Dong J, Wan Z, Popov M, Carey PR, Weiss MA. J. Mol. Biol. 2003;330:431–442. doi: 10.1016/s0022-2836(03)00536-9. [DOI] [PubMed] [Google Scholar]
- 31.Sane SU, Cramer SM, Przybycien TM. Anal. Biochem. 1999;269:255–272. doi: 10.1006/abio.1999.4034. [DOI] [PubMed] [Google Scholar]
- 32.Podstawka E, Ozaki Y. Biopolymers. 2008;89:941–950. doi: 10.1002/bip.21040. [DOI] [PubMed] [Google Scholar]
- 33.David C, Guillot N, Shen H, Toury T, de la Chapelle ML. Nanotechnology. 2010;21:475501. doi: 10.1088/0957-4484/21/47/475501. [DOI] [PubMed] [Google Scholar]
- 34.Chumanov G, Efremov R, Nabiev IR. J. Raman. Spectr. 1990;21:43–48. [Google Scholar]
- 35.Jarvis RM, Law N, Shadi IT, O'Brien P, Lloyd JR, Goodacre R. Anal. Chem. 2008;80:6741–6746. doi: 10.1021/ac800838v. [DOI] [PubMed] [Google Scholar]
- 36.Deckert-Gaudig T, Deckert V. J. Raman Spectrosc. 2009;40:1446–1451. [Google Scholar]
- 37.Deckert-Gaudig T, Deckert V. Small. 2009;5:432–436. doi: 10.1002/smll.200801237. [DOI] [PubMed] [Google Scholar]
- 38.Deckert-Gaudig T, Rauls E, Deckert V. J. Phys. Chem. C. 2010;114:7412–7420. [Google Scholar]
- 39.Kurouski D, Deckert-Gaudig T, Deckert V, Lednev IK. J. Am. Chem. Soc. 2012;134:13323–13329. doi: 10.1021/ja303263y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. Phys. Rev. Lett. 1997;78:1667–1670. [Google Scholar]
- 41.Nie S, Emory SR. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 42.Etchegoin PG, Le Ru EC. In: Surface Enhanced Raman Spectroscopy: Analytical, Biophysical and Life Science Applications. Schlücker S. Edt., editor. Wiley-VCH; New York: 2011. pp. 1–37. [Google Scholar]
- 43.Camden JP, Dieringer JA, Wang Y, Masiello DJ, Marks LD, Schatz GC, Van Duyne RP. J. Am. Chem. Soc. 2008;130:12616–12617. doi: 10.1021/ja8051427. [DOI] [PubMed] [Google Scholar]
- 44.Deckert-Gaudig T, Bailo E, Deckert V. Phys. Chem. Chem. Phys. 2009;11:7360–7362. doi: 10.1039/b904735b. [DOI] [PubMed] [Google Scholar]
- 45.Stewart S, Fredericks PM. Spectrochim. Acta Part A. 1999;55:1615–1640. [Google Scholar]
- 46.Stewart S, Fredericks PM. Spectrochim. Acta A. 1999;55:1641–1660. [Google Scholar]
- 47.Vitol EA, Orynbayeva Z, Friedman G, Gogotsi Y. J. Raman Spectrosc. 2012;43:817–827. [Google Scholar]
- 48.Zhang X, Hicks EM, Zhao J, Schatz GC, Van Duyne RP. Nano letters. 2005;5:1503–1507. doi: 10.1021/nl050873x. [DOI] [PubMed] [Google Scholar]
- 49.Blum C, Schmid T, Opilik L, Weidmann S, Fagerer SR, Zenobi R. J. Raman. Spectr. 2012 DOI: 10.1002/jrs.4099. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.