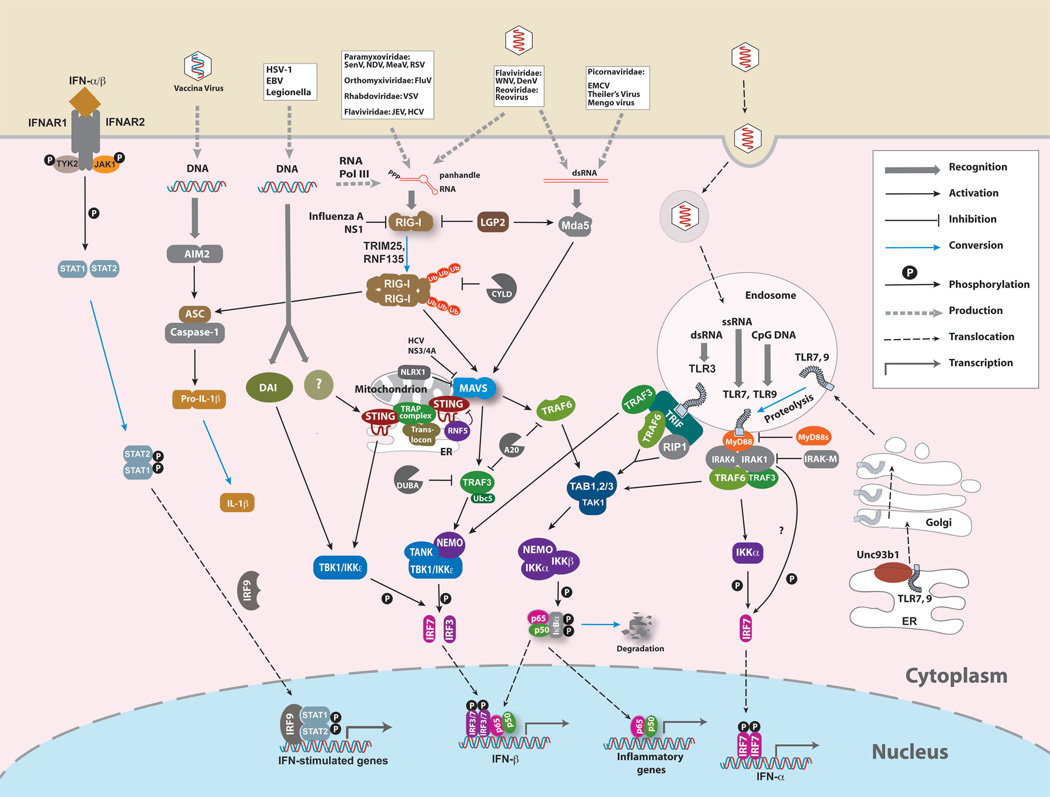
Viral diseases remain a challenging global health issue. Innate immunity is the first line of defense against viral infection. A hallmark of antiviral innate immune responses is the production of type 1 interferons and inflammatory cytokines. These molecules not only rapidly contain viral infection by inhibiting viral replication and assembly, but also play a crucial role in activating the adaptive immune system to eradicate the virus. Recent research has unveiled multiple signaling pathways that detect viral infection, with several pathways detecting the presence of viral nucleic acids. This SnapShot focuses on innate signaling pathways triggered by viral nucleic acids that are delivered to the cytosol and endosomes of mammalian host cells.
Cytosolic pathways
Many viral infections, especially those of RNA viruses, result in the delivery and replication of viral RNA in the cytosol of infected host cells. These viral RNAs often contain 5’-triphosphate (5’-ppp) and panhandle-like secondary structures composed of double-stranded segments. These features are recognized by members of the RIG-I-like Receptor (RLR) family, which includes RIG-I, MDA5, and LGP2 (Fujita, 2009; Yoneyama et al., 2004). All RLRs contain a DEAD/H-box RNA helicase domain in the middle. In addition, RIG-I and LGP2 contain a C-terminal regulatory domain (RD) that binds to 5’-pppRNA. MDA5, on the other hand, recognizes long double-stranded RNAs (dsRNAs) as well as single-stranded RNAs (ssRNAs) derived from picornaviruses. RIG-I and MDA5, but not LGP2, also contain two N-terminal CARD domains that interact with the CARD domain of the adaptor protein MAVS (also known as IPS-1, VISA or CARDIF), which resides in the mitochondrial outer membrane. MAVS interacts with STING (also known as MITA), a transmembrane protein in mitochondria and the endoplasmic reticulum (ER). On the ER membrane, STING associates with the TRAP complex, which may be involved in recruiting the protein kinases TBK1 and IKKε to phosphorylate the transcription factor IRF3. MAVS also recruits the ubiquitin ligases TRAF3 and TRAF6, which activate TBK1 and another kinase IKK, respectively. IKK phosphorylates IκB, an inhibitor of the master transcription factor NF-κB, leading to the ubiquitination of IκB and its subsequent degradation. NF-κB then enters the nucleus to turn on a plethora of proinflammatory genes. NF-κB also associates with IRF3, IRF7 and other transcription factors to induce production of IFN-β.
Infection by some DNA viruses and intracellular bacteria also leads to potent induction of type-1 interferons. The cytosolic DNAs introduced by these pathogens are detected by distinct sensors. AT-rich DNA is recognized by RNA polymerase III, which transcribes the DNA into 5’-pppRNA that triggers the RIG-I pathway. Cytosolic DNA can also induce interferons through RIG-I-independent mechanisms, including those involving other DNA sensors such as DAI and adaptor proteins such as STING. In addition to inducing production of type 1 interferons, cytosolic DNA can activate the inflammasome, which converts the IL-1β precursor protein into the mature cytokine. In this pathway, cytosolic DNA is recognized by AIM2, which forms a complex with ASC and procaspase-1, resulting in caspase-1 activation (Schroder et al., 2009). Cytosolic RNA can also activate the inflammasome through RIG-I, which engages ASC to activate caspase-1.
Endosomal pathway
When viruses enter cells through endocytosis, their nucleic acids are detected in the lumen of the endosomes by a subset of Toll-like receptors (TLRs), including TLR3, TLR7/8, and TLR9. TLR3 detects dsRNAs, TLR7/8 recognize ssRNAs, and TLR9 senses unmethylated CpG DNA (Akira et al., 2006). These TLRs are synthesized in the ER, where they associate with the transmembrane protein Unc93b1, which escorts the TLR proteins to endosomes (Kim et al., 2008; Tabeta et al., 2006). For TLR9, and possibly TLR7, their ectodomains must be cleaved by a protease in the endosome before they become functional receptors.
Binding of nucleic acids to TLR7, 8 and 9 leads to the recruitment of the cytosolic adaptor protein MyD88, which in turn recruits the protein kinases IRAK4 and IRAK1. IRAK4 phosphorylates and activates IRAK1, which binds and activates TRAF6 and TRAF3, leading to activation of IKK and TBK1, respectively. In plasmacytoid dendritic cells, the MyD88-IRAK1-TRAF6 complex recruits and activates IRF7, which induces production of IFN-α. Binding of dsRNA to TLR3 triggers recruitment of the adaptor protein TRIF, which in turn recruits TRAF3 and TRAF6 to activate downstream kinases. TRIF also recruits RIP1, a protein kinase involved in NF-κB activation.
Ubiquitin in antiviral immunity
Ubiquitination and deubiquitination play pivotal roles in both RLR and TLR pathways of antiviral defense (Bhoj and Chen, 2009). Following binding to viral RNA, RIG-I is conjugated with Lysine-63 (K63) polyubiquitin chains by the ubiquitin ligases TRIM25 and RNF135. This ubiquitination is apparently important for RIG-I to activate MAVS. Downstream of MAVS, the ubiquitin ligase activity of TRAF3 and TRAF6 also is important for activation of TBK1 and IKK, respectively. Furthermore, K63 polyubiquitination plays an important role in the activation of NF-κB and IRF7 by TRAF6 in the endosomal pathway. Several deubiquitination enzymes (DUBs) function as negative regulators of antiviral responses (Sun, 2008). The tumor suppressor protein CYLD removes polyubiquitin chains from RIG-I, whereas DUBA cleaves polyubiquitin chains from TRAF3. The anti-inflammatory protein A20 is another DUB that inhibits the RIG-I and TLR pathways at a step upstream of TBK1 and IKK.
Interferon signaling and positive feedback regulation
Type 1 interferons bind to a common receptor composed of two subunits, IFNAR1 and IFNAR2. Engagement of this receptor complex leads to recruitment and activation of the tyrosine kinases JAK1 and TYK2, which phosphorylate the transcription factors STAT1 and STAT2. The activated STAT proteins associate with IRF9 to form a complex called ISGF3, which regulates the expression of a large array of interferon-stimulated genes. The products of these genes act in concert to inhibit viral infection, replication and assembly. In addition, interferons induce the expression of several key components of the RIG-I pathway, such as RIG-I, TRIM25 and IRF7, thereby amplifying the antiviral response.
Negative regulation of antiviral immunity
After an effective immune response is launched, it must be dampened down or shut down completely in a timely manner to avoid damage to the host. In addition to the DUBs described above, a growing list of host factors has been found to inhibit the RIG-I and TLR pathways. Examples of this list include NLRX1, RNF5, ITCH, MyD88 and IRAK-M. Pathogens have also evolved strategies to counter the host immune response. For example, hepatitis C virus uses the viral protease NS3/4A to cleave MAVS from the mitochondrial membrane thus blocking induction of type 1 interferon gene expression; this immune evasion strategy assists the virus, enabling it to establish persistent infection in human liver cells. Another example is provided by the influenza A virus, which uses NS1 to inhibit RIG-I activation.
Abbreviations
- DenV
Dengue virus
- DUBA
Deubiquitination enzyme A
- EBV
Epstein-Barr virus
- EMCV
encephalomyocarditis virus
- FluV
Flu virus
- HCV
hepatitis C virus
- HSV
herpes simplex virus
- IFNAR
Interferon-alpha/beta receptor
- IKK
IκB kinase
- IRF
IFN regulatory factor
- IRAK
IL-1R-associated kinase
- ITCH
Itchy E3 ubiquitin protein ligase homolog
- ISGF3
IFN-stimulated gene factor 3
- JAK1
Janus kinase
- JEV
Japanese encephalitis virus
- LGP2
laboratory of genetics and physiology 2
- MAVS
Mitochondrial anti-viral signaling protein
- MCMV
murine cytomegalovirus
- MDA5
melanoma differentiation-associated gene 5
- MeaV
Measles virus
- MyD88
myeloid differentiation primary response gene 88
- NDV
Newcastle disease virus
- NF-κB
nuclear factor kappa B
- NLRX1
NLR family member X1
- NS1
Non-structural protein 1
- NS3/4A
Non-structural protein 3/4A
- RIG-I
retinoic acid-inducible gene I
- RIP
receptor-interacting protein
- RNF
Ring finger protein
- RSV
Respiratory syncytial virus
- SenV
Sendai virus
- STAT
Signal transducer and activator of transcription
- STING
stimulator of interferon genes
- TANK
TRAF family member-associated NF-κB activator
- TBK1
TANK-binding kinase
- TRAF
TNF receptor-associated factor
- TRAP
translocon associated protein
- TRIF
TIR-containing adaptor inducing IFNβ
- TRIM25
Tripartite motif-containing 25
- TYK2
Tyrosine kinase 2
- VSV
Vesicular stomatitis virus
- WNV
West Nile virus.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Fujita T. A nonself RNA pattern: tri-p to panhandle. Immunity. 2009;31:4–5. doi: 10.1016/j.immuni.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotidesensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- Schroder K, Muruve DA, Tschopp J. Innate immunity: cytoplasmic DNA sensing by the AIM2 inflammasome. Curr Biol. 2009;19:R262–265. doi: 10.1016/j.cub.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, Crozat K, Mudd S, Mann N, Sovath S, Goode J, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3-7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]