Abstract
While it is well known that individual integrins are critical mediators of cell behavior, recent work has shown that when multiple types of integrins simultaneously engage the ECM, cell functions are enhanced. However, it is not known how integrins spatially coordinate to regulate cell adhesion because no reliable method exists to segregate integrins on the cell membrane. Here, we use a microcontact printing-based strategy to pattern multiple ECMs that bind distinct integrins in order to study how integrins might interact. In our technique, proteins are first adsorbed uniformly to a poly(dimethyl siloxane) stamp, and then selectively “de-inked.” Our strategy overcomes several inherent limitations of conventional microcontact printing, including stamp collapse and limited functionality of the surface patterns. We show that integrins spatially segregate on surfaces patterned with multiple ECMs, as expected. Interestingly, despite spatial segregation of distinct integrins, cells could form adhesions and migrate across multicomponent surfaces as well as they do on single component surfaces. Together, our data indicate that although cells can segregate individual integrins on the cell surface to mediate ECM-specific binding, integrins function cooperatively to guide cell adhesion and migration.
Introduction
Adhesive interactions between cells and their underlying substrate are critical to many cell functions including growth factor signaling,1 differentiation,2–5 survival,6,7 and migration.8–10 Cells adhere specifically to extracellular matrix (ECM) ligands on their substrate.11 The principal transmembrane receptors that bind to the ECM, recruit additional proteins to sites of ECM binding, bind the actin cytoskeleton, and therefore transduce ECM ligand binding into cellular events, are the heterodimeric integrin proteins.12,13 24 distinct integrins have been identified to date, many of which bind to different types of ECM.
The concept that specific integrins are critical to specific cell behaviors ranging from differentiation to migration is based on classic experiments using substrates coated with a single type of ECM, or a promiscuous ligand combined with integrin-specific blocking antibodies.2,14 Interestingly, many cells co-express multiple integrins that bind to distinct ECM ligands. For instance, endothelial cells and mesenchymal stem cells express β1-containing heterdimers such as α1β1 and α2β1, and also express the αvβ5 heterodimer; many β1-containing integrin heterodimers bind to type-I collagen but not vitronectin, whereas αvβ5 binds to vitronectin but not type-I collagen.15
Studies using a variety of cells and stimulation methods suggest ‘cross-talk’ between specific integrins.16–18 For instance, an integrin specific for collagen type I (α2β1) actively represses activation of integrins specific for fibronectin (α5β1 and αvβ3) when endothelial cells are exposed to shear stress while adhering to collagen type I, and vice-versa when the cells are adhering to fibronectin.18 Moreover, substrates composed of mixtures of ECM proteins that engage multiple integrins in concert can have synergistic effects, such as enhancing embryonic stem cell differentiation5,19 and endothelial cell survival.20 However, experiments to date were performed on substrates presenting either individual, purified ECM proteins, or homogenous mixtures of ECM proteins. A lack of techniques with which to present multiple adhesive ligands in spatially organized patterns has prevented studies of spatial interactions between integrins.
Spatial control of cell behavior by integrins has been demonstrated by subcellularly patterning integrin ligands, including purified ECM proteins and adhesive ligands such as the arginine-glycine-aspartic acid (RGD) tripeptide sequence. The majority of current patterning techniques rely on microcontact printing of proteins.21,22 This simple method uses a poly(dimethyl siloxane) (PDMS) stamp inked with ECM proteins to pattern a conventional cell culture substrate, then coats the remainder of the substrate with materials that prevent cell adhesion.23 Patterning approaches have enabled the discovery that the spatial pattern of adhesion guides cell structure and function from the molecular24,25 to whole-cell8,26,27 scales. However, these patterning approaches currently print only a single adhesive ligand. Multiple ligands can be patterned by simply printing multiple times, but spatial registration between successive printing steps is not trivial.28
Here, we utilize a simple extension of conventional microcontact printing of proteins to encode a surface with distinct patterns of multiple ECMs to spatially segregate integrin receptors. The technique is based on cyclic inking and patterned de-inking of a PDMS stamp, and allows the generation of microscale, sparse, multicomponent surface patterns. By using the technique to generate surfaces presenting either patterned vitronectin (VN) or collagen type I (CI), or VN and CI simultaneously, we segregated integrin receptors on a cell membrane. In concordance with the literature, cells use αvβ5 but not β1 to bind to VN, and β1 but not αvβ5 to bind to CI. Strikingly, we observed that cells spanning VN and CI regions can assemble yet segregate co-existing αvβ5 and β1 adhesions, and single adhesions spanning both the VN and CI regions are formed with αvβ5 and β1 segregated within the adhesion. Moreover, cells can decipher a directional migration cue that is only evident from using the union of both VN and CI adhesions provided on the substrate. These results indicate that integrins function in concert to guide cell adhesion and migration.
Results
Development of the stamp-off method
We first set out to design a strategy to pattern surfaces with protein, keeping in mind that the strategy should be compatible with patterning multiple proteins. In our approach, a featureless, poly(dimethyl siloxane) (PDMS) stamp is first inked with protein (For initial studies, we used fluorescently-tagged bovine serum albumin; Fig. 1a, i). In parallel, a PDMS template, cast against a photolithographically-generated master to generate features in relief, is cleaned and activated by ultraviolet (UV) ozone treatment23 (Fig. 1a, ii). The inked stamp is then rinsed, dried, and placed in conformal contact with the template, transferring protein from the stamp surface to the template where contact is made (Fig. 1a, iii). Thereby, the previously featureless stamp surface encodes a pattern corresponding to the features of the template. Finally, this pattern is transferred to a cell culture substrate by microcontact printing (Fig. 1a, iv). Using fluorescent microscopy to track this method before and after stamp-off and stamping indicates this is a reasonable method to generate protein surface patterns. Importantly, as we directly show shortly, this strategy is compatible with patterning multiple proteins.
Fig. 1. The stamp-off process.
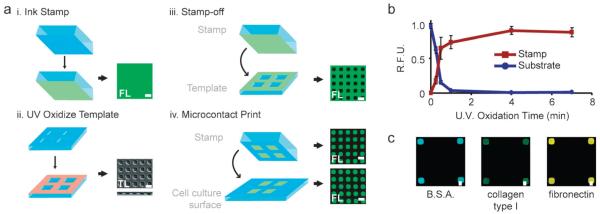
(a) Schematic representation of the stamp-off process. For certain substrates, either fluorescent (FL) or transmitted light (TL) images are shown. (b) Relative fluorescence intensity versus time of ultraviolet oxidation on the stamp (red), and substrate (blue). R.F.U., relative fluorescence units. Means ± s.e.m. are from three independent experiments. (c) Either bovine serum albumin conjugated to AlexaFluor-488, collagen type I, or fibronectin were loaded onto a flat stamp, stamped-off, transferred to a cell culture surface, immunolabeled and imaged. Note the lack of fluorescence intensity outside of the patterned squares. All scale bars, 10 μm.
Complete protein transfer from the stamp surface during contact between the stamp and template is key to the stamp-off approach. We previously reported that cleaning and rendering a PDMS template hydrophilic by UV ozone permits protein transfer from stamp to template.23 To characterize protein transfer effciency here, we measured the fluorescent intensity of AlexaFluor 488-tagged bovine serum albumin on the stamp and template after stamping-off as a function of UV ozone treatment time. Consistent with prior results, no protein transfer from stamp to template occurs without UV ozone treatment (Fig. 1b). In contrast, UV ozone treatment for 7 min permitted complete protein transfer from stamp to template. This effect is not specific to bovine serum albumin, as detection of type I collagen and fibronectin by immunofluorescence similarly indicated complete protein removal from the stamp following 7 min of UV ozone treatment time (Fig. 1c). Thus, UV ozone treatment of the PDMS template permits complete protein removal from the stamp, enabling a topographically featureless stamp to encode a protein pattern.
Advantages of stamp-off over conventional microcontact printing
Conventional microcontact printing suffers from the key limitation that PDMS stamps bearing small, sparse features are prone to deformation and collapse during printing.29 Stamp collapse depends both on the geometry of the features, and the pressure applied during stamping.30 Because the stamp-off technique uses a topographically featureless stamp, we hypothesized that it could overcome the deformation and collapse problems of conventional microcontact printing. To test this idea, conventional PDMS stamps bearing 20 μm tall, 20 μm × 20 μm square features with variable spacing were loaded with AlexaFluor 488-tagged bovine serum albumin (BSA) and stamped onto a substrate. When stamps with spacing of features similar to the characteristic feature size and height were used (25 μm spacing between 20 μm features sizes with 20 μm height), no collapse occurred and pattern fidelity was high (Fig. 2a i). However, significant collapse occurred when the spacing greatly exceeded the height of the stamp, as evidenced by pattern fouling from stamps bearing >100 μm spacing (Fig. 2a ii, iii). In our stamp-off technique, in contrast, identical but inverse features on a template were used to remove AlexaFluor 488-tagged bovine serum albumin from a topographically featureless stamp. The stamp bearing the pattern was then placed in conformal contact with a substrate. Regardless of the feature spacing, no pattern fouling occurred (Fig. 2a iv–vi). These data directly indicate that the stamp-off technique obviates the risk of pattern fouling associated with stamp deformation and collapse during conventional microcontact printing.
Fig. 2. Advantages of stamp-off.
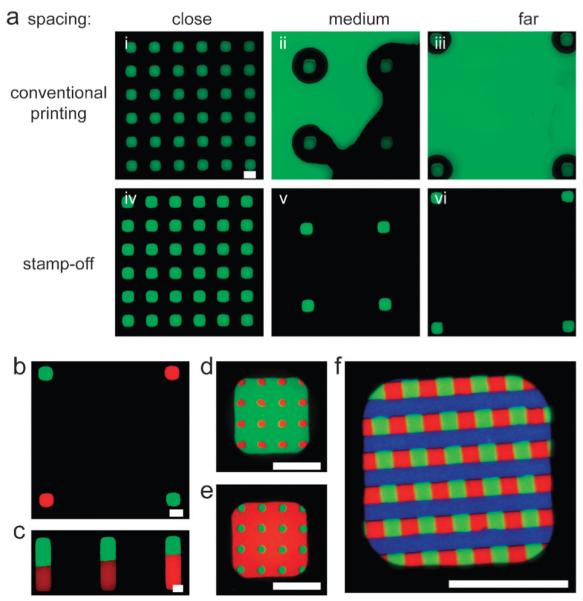
(a) Conventional microcontact printing (top row), and stamp-off (bottom row), for 20 μm features that are spaced 20, 110, or 200 μm apart (edge-to-edge). (b)–(e) A flat stamp was loaded with BSA conjugated to AlexaFluor-594 (red), stamped-off, reloaded with BSA conjugated to AlexaFluor-488 (green), stamped-off, and imaged. (f) A flat stamp was loaded with BSA conjugated to AlexaFluor-594 (red), stamped-off, reloaded with BSA conjugated to AlexaFluor-488 (green), stamped-off, reloaded with BSA conjugated to AlexaFluor-647 (blue), stamped-off, and imaged. All scale bars, 20 μm.
Despite this limitation, conventional microcontact printing has illuminated the role that geometry has in regulating cell differentiation, proliferation, and polarity.4,6,26,31,32 These insights have been gained from studies on dual-component patterned surfaces generated by conventional microcontact printing, wherein one protein is patterned on a surface, and the remainder of the surface is coated to prevent adhesion. In contrast, through iteration of the steps outlined in Fig. 1A i–iii, we generated discrete (Fig. 2b) and adjacent (Fig. 2c–e) 3-color patterns, as well as higher-order patterns (Fig. 2f). In general, n deinking and re-inking steps can create n+1-component surface patterns. We used fluorescently-conjugated BSA for these examples; so long as the iterative inking steps do not involve proteins that bind to one another, the pattern remains segregated. If two inking proteins did bind each other, then the second inking step would lead to adsorption of the second protein on both the bare (stamped-off) region and the region containing the first protein. The stamp-off process allows in some cases for positioning of features at substantially higher spatial resolutions than the actual resolution of stamp placement. For example, to generate the pattern shown in Fig. 2b via stamp-off, we first generated a checkerboard (no uncolored region), then stamped-off with an array of holes whose periodicity matched the periodicity of the checkerboard. The resolution of stamp placement in the second step only needed to be good enough to land each hole entirely within a square in the checkerboard. To generate the pattern shown in Fig. 2b by sequential conventional microcontact printing, the resolution of stamp placement must be equal to the precision required for the edge-to-edge spacing. As such, one can with proper design position features with high resolution using low resolution manual stamp placement via our technique. Thus, whereas conventional microcontact printing is limited to generation of dual-component surfaces, the stamp-off technique enables easy preparation of multicomponent surface patterns.
Integrin segregation by stamp-off
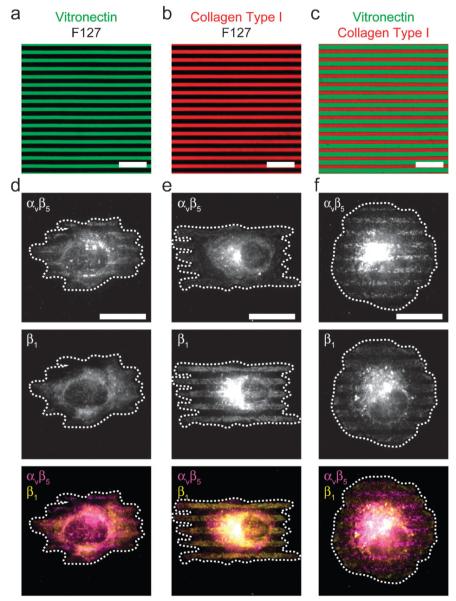
Since stamp-off enables multifunctional surface patterns, we used it to ask whether we could localize integrin receptors on a cell to spatially distinct regions of ECM proteins. Human umbilical vein endothelial cells (HUVECs) express both β1 and αvβ5 integrins, and the coordinated use of these integrins by endothelial cells is essential to angiogenesis.33,34 Importantly, β 1 integrin (in a heterodimer with an α subunit, usually α1 or α2) binds to collagen type I (CI) but not vitronectin (VN), while αvβ5 integrin recognizes VN but not CI. To investigate this segregation further, we leveraged the ability of stamp-off to generate surfaces patterned with CI, VN, or CI and VN simultaneously (Fig. 3a–c). When we plated HUVECs on these substrates and immunostained for specific integrins, we observed that adhesion receptors localized in a predictable and reproducible manner. On surfaces composed of alternating, 3 μm stripes of VN and non-adhesive F127 Pluronics, cells spread across many stripes and used αvβ5, but not β1, to form adhesions on the VN (Fig. 3d). On surfaces composed of alternating, 3 μm stripes of CI and F127 Pluronics, cells used β1, but not αvβ5 to form adhesions on the CI (Fig. 3e). Cells did not form adhesions onto the non-adhesive stripes on either surface. On surface patterns displaying alternating, 3 μm stripes of CI and VN, cells spread isotropically as expected, but used β1 and αvβ5 to bind to CI and VN, respectively (Fig. 3f). Another integrin known to bind VN but not CI, αvβ3, localized very similarly (Supplementary Fig. 1†). 2 h after plating, the cells adopted polarized morphology, but the integrins remained segregated (Fig. 4a). Interestingly, distinct integrins composed an adhesion complex that spanned multiple ECMs (Fig. 4b,c). Similar integrin segregation was seen in human mesenchymal stem cells (hMSCs), which also express αv- and β1-based integrin heterodimers (data not shown). These data directly indicate that cells can co-express yet spatially segregate integrin receptors on the cell surface when they encounter distinct ECMs simultaneously.
Fig. 3. Integrin segregation on multicolor surface patterns.
(a)–(c) Fluorescent micrographs of BSA conjugated to AlexaFluor-488 (green) to represent vitronectin, BSA conjugated to AlexaFluor-594 (red) to represent collagen type I and black to represent non-adhesive F127. (d)–(f) Micrographs of human umbilical vein endothelial cells (HUVECs), seeded on substrates patterned as in (a)–(c), fixed after 1 h, and immunolabeled. Note the colocalization of αvβ5 integrin to vitronectin, and β1 integrin to collagen type I. All scale bars, 20 μm.
Fig. 4. Continuous adhesions composed of compositionally and spatially segregated types of integrins.
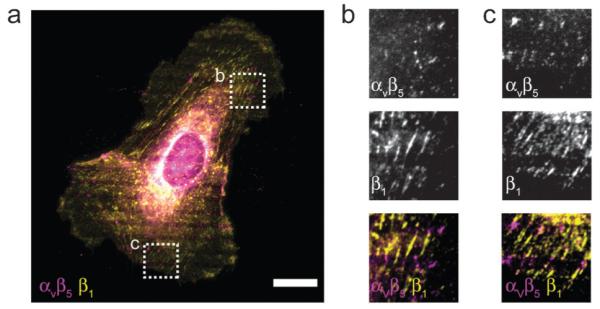
(a–c) Micrograph of a HUVEC seeded on a pattern with alternating lines of vitronectin and collagen type I as in Fig. 3c, fixed 2 h after seeding and immunolabaled. Panels (b) and (c) correspond boxed regions in (a). Note the segregation of αvβ5 and β1 integrins throughout the cell (a), and within single adhesions (b, c). Scale bar, 20 μm.
Coordinate regulation of migration by distinct integrin types
Cells express distinct integrins yet segregate αvβ5 from β1 on multicomponent substrates, suggesting that multiple integrins may coordinate to guide migratory direction on such substrates. To test this hypothesis, we measured the migratory trajectories of HUVECs on surfaces composed of patterned, symmetric VN or CI, versus the same VN and CI patterns positioned adjacently (Fig. 5a–c). Cells spread and migrated without directional bias on surfaces displaying patterned and symmetric VN or CI (Fig. 5a, b). We used the stamp-off technique to generate trifunctional surfaces that presented VN, CI and non-adhesive regions to the cells. Interestingly, cells spread and migrated parallel to the adhesive co-pattern of VN and CI, and did not show a bias for either VN or CI (Fig. 5c). To characterize cell migration, we measured the cell speed (rate of displacement) and persistence (the average time between significant direction changes) from the migration trajectories. Although cell speed was similar on the three surfaces (Fig. 5d), migration persistence was significantly higher on the surfaces displaying both VN and CI than the surfaces displaying either VN or CI (Fig. 5e). Consistently, these patterned substrates directed hMSC migration in the same way as the HUVECs: hMSC migration was not directional on patterned VN or CI alone, but highly directional on patterned VN and CI (data not shown).
Fig. 5. Multicolor surface patterns direct cell migration.
(a–c, top) Fluorescent micrographs of BSA conjugated to AlexaFluor-488 (green) to represent vitronectin, BSA conjugated to AlexaFluor-594 (red) to represent collagen type I and black to represent non-adhesive F127. (a–c, middle) HUVECs were seeded on patterned substrates, and recorded via time-lapse, phase-contrast microscopy. Representative cells are shown. (a–c, bottom) Representative migration trajectories of cells on patterned substrates. Each black line represents a single cell, originating from (0,0), tracked every 15 min for 12 h. 15 cells are plotted per graph. (d, e) Migration speed (d) and persistence (e) as determined from the trajectory of at least 60 cells from three independent experiments. Trajectories were fit to the persistent random walk model to calculate the persistence time.49 See Materials and Methods for details. Means ± s.e.m. are from 3 independent experiments. All scale bars, 20 μm.
These differences in cell shape and migration were due to either the combined presence of VN and CI on the surface, or the adhesive geometry of the parallel stripes. The latter possibility was likely since adhesive geometry has previously shown to direct migration.9,35 To directly distinguish between these possibilities, we examined the migration responses of cells on single-component stripes of identical geometry to that of Fig. 5c. Parallel stripes of VN alone, CI alone, and the combined VN and CI stripes resulted in similar spreading and migration patterns (Fig. 5c, Supplementary Fig. 2†), indicating that adhesive geometry indeed directed migration. Together, these data therefore suggest that cells can assemble a composite picture from distinct ECMs whose ensemble pattern conveys directional information even though the individual patterns do not convey directional information. Because cells used at least two distinct integrin heterodimers to bind to these surfaces (αv-based heterdimers to bind vitronectin, and β1-based heterodimers to bind type-I collagen; Fig. 3 and 4, Supplementary Fig. 1†), cells appear to be able to use distinct integrin receptors together, perhaps even indiscriminately, to guide migration direction.
Discussion
The stamp-off technique offers clear advantages over conventional microcontact printing even for single-protein stamping. To overcome stamp deformation and collapse investigators have used PDMS stamps backed with glass,22 PDMS stamps coated with a rigid material,36 and stamps made from a material more rigid than conventional PDMS.37 In contrast, stamp-off can easily achieve small, sparse features with unlimited distance between features without the risk of deformation and collapse. Several investigators have devised creative methods to generate multifunctional surface patterns using techniques other than iterative, conventional microcontact printing.38 Unfortunately, the limited accessibility and reproducibility of these methods has prevented their widespread adoption and application to biologic assays. In contrast, the stamp-off method is capable of easily generating robust, multifunctional surface patterns without any additional specialized equipment. A similar method to stamp-off based on cyclic inking and de-inking was recently described, except that the method used silicon wafers as the stamp-off template.39 However, the method was not applied to the interaction of cells with a surface. The use of PDMS molds instead of silicon wafers as stamp-off templates presented herein will enable wider adoption of the approach to biological researchers that do not have access to cleanroom facilities. Thus, the stamp-off technique presented here provides a facile mechanism to generate multifunctional, ECM-based surface patterns for studying cell-material interactions.
Directed migration is thought to be guided by soluble signals in many settings, including embryonic development,40 wound healing,41 and angiogenesis.42 Emerging evidence suggests that non-soluble, adhesive signals can also be important to directing migration in vivo.43,44 It was recently demonstrated that adhesive interactions may polarize cells during directed migration of neural crest cells prior to establishment of a soluble gradient of the chemokine Sdf1 in developing Xenopus.43 In vitro, many lines of evidence suggest that the ECM can impinge strongly on cell polarity and migration.8,9,26,32
Cells migrate up a gradient of immobilized ECM, in a process known as haptotaxis.45 Similarly, cells orient and migrate in the direction of closer spacing between integrinligand bonds.24 Surfaces displaying patterned ECM in a non-adhesive background can direct cell migration by constraining cell adhesion to patterned regions.9,35 Moreover, the spatial pattern of adhesion can direct cell polarity, and the degree of traction force generation.8,26,46 Results here suggest that cells can also coordinate spatial information by generating a composite picture of the geometry of the ECM, even if that involves distinct integrins and multiple ECMs, to decode such geometric cues. Although the mechanism by which this coordination occurs remains unclear, it is likely that cytoplasmic interactions between integrins and the cytoskeleton are suffciently universal that cells can use different integrins interchangeably to spread and migrate with an integrated cytoskeleton on a complex ECM.
A single focal adhesion involves up to hundreds of bound integrin heterodimers.47 When cells bind to multiple ECMs simultaneously they use different types of integrins.16,18 However, it is not known whether each focal adhesion is a cluster of a single type of integrin, or a mixture of different types. Our data indicate that a single focal adhesion can be composed of different types of integrins. Moreover, the integrins within a focal adhesion clearly segregate based on the underlying ECM: αv localized to VN, while β1 localized to CI even when a single adhesion spanned both ECMs. Although some studies suggest differences in focal adhesion dynamics and composition depending on the bound integrin, our studies suggest that there is suffcient sharing of the structural components that lie between the cytoskeleton and the integrin that these adhesions can flexibly reorganize as they cross ECM boundaries. How a cell regulates, and is regulated by, adhesions composed of spatially and compositionally distinct integrins bonded to distinct ECM proteins remains an open question that necessitates further study.
Studies demonstrating functional overlap between integrins during physiological processes suggest that distinct integrin receptors may regulate the actin cytoskeleton in a coordinated manner.13,16,33 Detailed in vitro studies of the integrin-actin cytoskeleton linkage during cell migration48 have focused on one integrin receptor subtype. Thus, the view that cells use distinct integrin receptors to cooperatively regulate the actin cytoskeleton has been controversial. We present direct evidence that (1) cells use distinct integrin receptors to bind to different kinds of ECM simultaneously, and (2) cells migrate along adhesive paths regardless of the type of ECM that composes the path. These data together suggest that cells distinguish between ECM ligands by using different integrin receptors, and likely use spatial information from these integrins as an ensemble to regulate spatial decisions such as the direction of migration. Given the importance of cell adhesion and migration, it is critical to identify mechanisms that mediate crosstalk between integrin receptors during migration. Surfaces that display multiple types of ECMs such as those employed here will be key to this line of study.
Experimental
Cell culture and reagents
Human umbilical vein endothelial cells were obtained from, and cultured as prescribed by the manufacturer (Lonza, Walkersville, MD). Other biological reagents included: bovine serum albumin conjugated to AlxeaFluor 488, 594 or 647 (Invitrogen, Carlsbad, CA), fibronectin from human plasma (BD Biosciences, Bedford, MA), rat tail collagen type I (BD Bioscience, Bedford, MA), vitronectin from human plasma (Sigma-Aldrich, St. Louis, MO), anti-active-β1 (clone 9EG7, BD Biosciences, Bedford, MA), anti-αvβ5 (clone P1F6, Santa Cruz Biotechnology, Santa Cruz, CA), anti-αvβ3 (clone LM609, Millipore, Billerica, MA), anti-collagen type I (polyclonal, Meridian Life Science, Saco, ME), anti-human fibronectin (polyclonal, MP Biomedicals, Santa Ana, CA) and Pluronics F127 (Sigma-Aldrich, St. Louis, MO). poly(dimethyl siloxane) (PDMS; Sylgard 184, Dow Corning, Midland, MI) was used at 10 : 1 (w : w) base : curing agent.
Soft lithography
Patterned PDMS stamps were cast from a photoresist-patterned silicon wafer, as previously described.23 Stamp-off templates were cast similarly, but from negative photoresist patterns on the silicon wafer. Flat PDMS stamps were cast from an unpatterned silicon wafer. For microcontact printing, PDMS stamps were inked with protein at 50 μg ml−1 in H2O (for fibronectin and vitronectin), 50 μg ml−1 in PBS (for bovine serum albumins), or 100 μg ml−1 in 1% (v/v) acetic acid (for collagen type I), all for 1 h at room temperature. The stamps were then thoroughly rinsed in H2O and blown dry with a stream of N2. In parallel, the target substrate (a stamp-off template or PDMS cell culture substrate) was treated with ultraviolet ozone for specified times (Jelight Company, Irvine, CA). The stamp was then placed in conformal contact with the target substrate for ~1 s. F127 Pluronics was adsorbed to PDMS surfaces from a 0.2% (w/v) solution for 1 h at room temperature to prevent protein adsorbtion to non-functionalized portions of the PDMS.
Immunofluorescence and microscopy
Substrates were fixed in chilled acetone for 3 min at −20 °C, and blocked and immunolabeled in 10% goat serum. Samples were imaged on a Zeiss AxioVert 200M, and images were acquired with an AxioCam HRm using AxioVision software (Carl Zeiss, Thornwood, NY). Fixed samples were imaged using a 63×, N.A. 1.4 Plan Apochromat objective, and live samples were imaged with a phase contrast, 10×, N.A. 0.25 A-plan objective. An environmental chamber was used to control temperature and CO2 during live experiments (In Vivo Scientific, St. Louis, MO).
Conclusion
As investigators delve deeper into molecular mechanisms that underlie adhesive cell behaviors, it is increasingly important to dissect pathways mediating cell adhesion. In this spirit, we believe the surface patterning method presented herein will lead to a greater understanding of mechanisms of fundamentally adhesive events, such as cell migration.
Supplementary Material
Insight, innovation, integration.
Many types of integrins can mediate cell adhesion but have distinct effects on cell functions ranging from differentiation to migration. It is not known, however, whether cells can coordinate the simultaneous engagement of multiple types of integrins to regulate cellular behaviors. To engage multiple types of integrins in a spatially controlled manner, we created a technique to simultaneously pattern multiple integrin ligands, or extracellular matrix proteins (ECMs), on a surface. In this setting, distinct types of integrins segregate to their corresponding ECM spots. Interestingly, cells were able to coordinate simultaneous engagement of spatially segregated αvβ3 and β1-based integrins to assemble focal adhesions and guide migration. This technique may shed light on interactions between adhesion receptors in a variety of settings.
Acknowledgements
The authors wish to thank S. Raghavan for helpful discussions, and C. K. Choi, G. L. Lin, and E. Toro for critically reading the manuscript. This work was supported by grants from the National Institutes of Health (EB00262, EB08396, HL73305, GM74048), and Center for Engineering Cells and Regeneration of the University of Pennsylvania, the National Science Foundation Graduate Research Fellowship (RD), the National Science Foundation Louis Stokes Alliances for Minority Participation Program (MK), and the Institute of Regenerative Medicine at the University of Pennsylvania (SG).
Footnotes
Electronic supplementary information (ESI) available: Supplementary Fig. 1 and 2. See DOI: 10.1039/c0ib00129e
References
- 1.Schwartz MA, Assoian RK. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J. Cell Sci. 2001;114:2553–60. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]; Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nat. Cell Biol. 2002;4:E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 2.Spiegelman BM, Ginty CA. Fibronectin modulation of cell shape and lipogenic gene expression in 3T3-adipocytes. Cell. 1983;35:657–666. doi: 10.1016/0092-8674(83)90098-3. [DOI] [PubMed] [Google Scholar]
- 3.Adams JC, Watt FM. Changes in keratinocyte adhesion during terminal differentiation: reduction in fibronectin binding precedes alpha 5 beta 1 integrin loss from the cell surface. Cell. 1990;63:425–435. doi: 10.1016/0092-8674(90)90175-e. [DOI] [PubMed] [Google Scholar]; Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE. Engineering cell shape and function. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]; Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 4.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 5.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 6.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 7.Ingber DE. Fibronectin controls capillary endothelial cell growth by modulating cell shape. Proc. Natl. Acad. Sci. U. S. A. 1990;87:3579–3583. doi: 10.1073/pnas.87.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]; Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–349. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]; Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J. Theor. Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, Bruzewicz DA, Wong AP, Piel M, Whitesides GM. Directing cell migration with asymmetric micropatterns. Proc. Natl. Acad. Sci. U. S. A. 2005;102:975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia N, Thodeti CK, Hunt TP, Xu Q, Ho M, Whitesides GM, Westervelt R, Ingber DE. Directional control of cell motility through focal adhesion positioning and spatial control of Rac activation. FASEB J. 2008;22:1649–1659. doi: 10.1096/fj.07-090571. [DOI] [PubMed] [Google Scholar]
- 10.Davidson LA, Marsden M, Keller R, Desimone DW. Integrin alpha5beta1 and fibronectin regulate polarized cell protrusions required for Xenopus convergence and extension. Curr. Biol. 2006;16:833–844. doi: 10.1016/j.cub.2006.03.038. [DOI] [PubMed] [Google Scholar]; Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay ED. Extracellular matrix. J. Cell Biol. 1981;91:205s–223s. doi: 10.1083/jcb.91.3.205s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neff NT, Lowrey C, Decker C, Tovar A, Damsky C, Buck C, Horwitz AF. A monoclonal antibody detaches embryonic skeletal muscle from extracellular matrices. J. Cell Biol. 1982;95:654–666. doi: 10.1083/jcb.95.2.654. [DOI] [PMC free article] [PubMed] [Google Scholar]; Berrier AL, Yamada KM. Cell-matrix adhesion. J. Cell. Physiol. 2007;213:565–573. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 13.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 14.Menko AS, Boettiger D. Occupation of the extracellular matrix receptor, integrin, is a control point for myogenic differentiation. Cell. 1987;51:51–57. doi: 10.1016/0092-8674(87)90009-2. [DOI] [PubMed] [Google Scholar]; Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 15.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J. Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wayner EA, Orlando RA, Cheresh DA. Integrins alpha v beta 3 and alpha v beta 5 contribute to cell attachment to vitronectin but differentially distribute on the cell surface. J. Cell Biol. 1991;113:919–929. doi: 10.1083/jcb.113.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roca-Cusachs P, Gauthier NC, Del Rio A, Sheetz MP. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Danen EH, Sonneveld P, Brakebusch C, Fassler R, Sonnenberg A. The fibronectin-binding integrins alpha5beta1 and alphavbeta3 differentially modulate RhoA-GTP loading, organization of cell matrix adhesions, and fibronectin fibrillogenesis. J. Cell Biol. 2002;159:1071–1086. doi: 10.1083/jcb.200205014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orr AW, Ginsberg MH, Shattil SJ, Deckmyn H, Schwartz MA. Matrix-specific suppression of integrin activation in shear stress signaling. Mol. Biol. Cell. 2006;17:4686–4697. doi: 10.1091/mbc.E06-04-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, Cho SW, Son SM, Hudson SP, Bogatyrev S, Keung L, Kohane DS, Langer R, Anderson DG. Combinatorial extracellular matrices for human embryonic stem cell differentiation in 3D. Biomacromolecules. 2010;11:1909–1914. doi: 10.1021/bm100357t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perruzzi CA, de Fougerolles AR, Koteliansky VE, Whelan MC, Westlin WF, Senger DR. Functional overlap and cooperativity among alphav and beta1 integrin subfamilies during skin angiogenesis. J. Invest. Dermatol. 2003;120:1100–1109. doi: 10.1046/j.1523-1747.2003.12236.x. [DOI] [PubMed] [Google Scholar]
- 21.Bernard A, Delamarche E, Schmid H, Michel B, Bosshard HR, Biebuyck H. Printing patterns of proteins. Langmuir. 1998;14:2225–2229. [Google Scholar]
- 22.James CD, Davis RC, Kam L, Craighead HG, Isaacson M, Turner JN, Shain W. Patterned protein layers on solid substrates by thin stamp microcontact printing. Langmuir. 1998;14:741–744. [Google Scholar]
- 23.Tan JL, Liu W, Nelson CM, Raghavan S, Chen CS. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004;10:865–872. doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- 24.Arnold M, Hirschfeld-Warneken VC, Lohmuller T, Heil P, Blummel J, Cavalcanti-Adam EA, Lopez-Garcia M, Walther P, Kessler H, Geiger B, Spatz JP. Induction of cell polarization and migration by a gradient of nanoscale variations in adhesive ligand spacing. Nano Lett. 2008;8:2063–2069. doi: 10.1021/nl801483w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell shape provides global control of focal adhesion assembly. Biochem. Biophys. Res. Commun. 2003;307:355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]; Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lehnert D, Wehrle-Haller B, David C, Weiland U, Ballestrem C, Imhof BA, Bastmeyer M. Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J. Cell Sci. 2004;117:41–52. doi: 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- 26.Thery M, Racine V, Piel M, Pepin A, Dimitrov A, Chen Y, Sibarita JB, Bornens M. Anisotropy of cell adhesive micro-environment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goffn JM, Pittet P, Csucs G, Lussi JW, Meister JJ, Hinz B. Focal adhesion size controls tension-dependent recruitment of alpha-smooth muscle actin to stress fibers. J. Cell Biol. 2006;172:259–268. doi: 10.1083/jcb.200506179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers JA, Paul KE, Whitesides GM. Quantifying distortions in soft lithography. J. Vac. Sci. Technol., B. 1998;16:88–97. [Google Scholar]
- 29.Ruiz SA, Chen CS. Microcontact printing: a tool to pattern. Soft Matter. 2007;3:168–177. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]; Xia Y, Whitesides GM. Soft lithography. Annu. Rev. Mater. Sci. 1998;28:153–184. [Google Scholar]
- 30.Hui CY, Jaogta A, Lin YY, Kramer EJ. Constraints on microcontact printing imposed by stamp deformation. Langmuir. 2002;18:1394–1407. [Google Scholar]
- 31.Ruiz SA, Chen CS. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desai RA, Gao L, Raghavan S, Liu WF, Chen CS. Cell polarity triggered by cell-cell adhesion via E-cadherin. J. Cell Sci. 2009;122:905–911. doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dupin I, Camand E, Etienne-Manneville S. Classical cadherins control nucleus and centrosome position and cell polarity. J. Cell Biol. 2009;185:779–786. doi: 10.1083/jcb.200812034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eliceiri BP, Cheresh DA. Adhesion events in angiogenesis. Curr. Opin. Cell Biol. 2001;13:563–568. doi: 10.1016/s0955-0674(00)00252-0. [DOI] [PubMed] [Google Scholar]
- 34.van der Flier A, Badu-Nkansah K, Whittaker CA, Crowley D, Bronson RT, Lacy-Hulbert A, Hynes RO. Endothelial alpha5 and alphav integrins cooperate in remodeling of the vasculature during development. Development. 2010;137:2439–2449. doi: 10.1242/dev.049551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odom TW, Love C, Wolfe DB, Paul KE, Whitesides GM. Improved pattern transfer in soft lithography using composite stamps. Langmuir. 2002;18:5314–5320. [Google Scholar]
- 37.Schmid H, Michel B. Siloxane polymers for high-resolution, high-accuracy soft lithography. Macromolecules. 2000;33:3042–3049. [Google Scholar]
- 38.Ghosh M, Alves C, Tong Z, Tettey K, Konstantopoulos K, Stebe KJ. Multifunctional surfaces with discrete functionalized regions for biological applications. Langmuir. 2008;24:8134–8142. doi: 10.1021/la8006525. [DOI] [PMC free article] [PubMed] [Google Scholar]; Raghavan S, Desai RA, Kwon Y, Mrksich M, Chen CS. Micropatterned Dynamically Adhesive Substrates for Cell Migration. Langmuir. 2010;26:17733. doi: 10.1021/la102955m. DOI: 10.1021/la102955m. [DOI] [PubMed] [Google Scholar]; Tien J, Nelson CM, Chen CS. Fabrication of aligned microstructures with a single elastomeric stamp. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1758–1762. doi: 10.1073/pnas.042493399. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hui EE, Bhatia SN. Microscale control of cell contact and spacing via three-component surface patterning. Langmuir. 2007;23:4103–4107. doi: 10.1021/la0630559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyer SR, Garcia AJ, Delamarche E. Facile preparation of complex protein architectures with sub-100-nm resolution on surfaces. Angew. Chem., Int. Ed. 2007;46:6837–6840. doi: 10.1002/anie.200700989. [DOI] [PubMed] [Google Scholar]
- 40.Jessell TM, Melton DA. Diffusible factors in vertebrate embryonic induction. Cell. 1992;68:257–270. doi: 10.1016/0092-8674(92)90469-s. [DOI] [PubMed] [Google Scholar]
- 41.Werner S, Grose R. Regulation of wound healing by growth factors andcytokines. Physiol. Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- 42.Jain RK. Molecular regulation of vessel maturation. Nat. Med. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 43.Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasad M, Montell DJ. Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev. Cell. 2007;12:997–1005. doi: 10.1016/j.devcel.2007.03.021. [DOI] [PubMed] [Google Scholar]; Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rorth P. Two distinct modes of guidance signalling during collective migration of border cells. Nature. 2007;448:362–365. doi: 10.1038/nature05965. [DOI] [PubMed] [Google Scholar]; Bronner-Fraser M. Neural crest cell migration in the developing embryo. Trends Cell Biol. 1993;3:392–397. doi: 10.1016/0962-8924(93)90089-j. [DOI] [PubMed] [Google Scholar]; Friedl P, Hegerfeldt Y, Tusch M. Collective cell migration in morphogenesis and cancer. Int. J. Dev. Biol. 2004;48:441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]; Hammerschmidt M, Wedlich D. Regulated adhesion as a driving force of gastrulation movements. Development. 2008;135:3625–3641. doi: 10.1242/dev.015701. [DOI] [PubMed] [Google Scholar]; Marsden M, DeSimone DW. Integrin-ECM interactions regulate cadherin-dependent cell adhesion and are required for convergent extension in Xenopus. Curr. Biol. 2003;13:1182–1191. doi: 10.1016/s0960-9822(03)00433-0. [DOI] [PubMed] [Google Scholar]; Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dickinson RB, Tranquillo RT. A stochastic model for adhesion-mediated cell random motility and haptotaxis. J. Math. Biol. 1993;31:563–600. doi: 10.1007/BF00161199. [DOI] [PubMed] [Google Scholar]; Smith JT, Tomfohr JK, Wells MC, Beebe TP, Jr, Kepler TB, Reichert WM. Measurement of cell migration on surface-bound fibronectin gradients. Langmuir. 2004;20:8279–8286. doi: 10.1021/la0489763. [DOI] [PubMed] [Google Scholar]; McCarthy JB, Palm SL, Furcht LT. Migration by haptotaxis of a Schwann cell tumor line to the basement membrane glycoprotein laminin. J. Cell Biol. 1983;97:772–777. doi: 10.1083/jcb.97.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker KK, Brock AL, Brangwynne C, Mannix RJ, Wang N, Ostuni E, Geisse NA, Adams JC, Whitesides GM, Ingber DE. Directional control of lamellipodia extension by constraining cell shape and orienting cell tractional forces. FASEB J. 2002;16:1195–1204. doi: 10.1096/fj.02-0038com. [DOI] [PubMed] [Google Scholar]
- 47.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev. Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]; Brown CM, Hebert B, Kolin DL, Zareno J, Whitmore L, Horwitz AR, Wiseman PW. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J. Cell Sci. 2006;119:5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]; Choi CK, Vicente-Manzanares M, Zareno J, Whitmore LA, Mogilner A, Horwitz AR. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]; Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Dobereiner HG, Freund Y, Borisy G, Sheetz MP. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; Galbraith CG, Yamada KM, Galbraith JA. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 2007;315:992–995. doi: 10.1126/science.1137904. [DOI] [PubMed] [Google Scholar]
- 49.Harms BD, Bassi GM, Horwitz AR, Lauffenburger DA. Directional persistence of EGF-induced cell migration is associated with stabilization of lamellipodial protrusions. Biophys. J. 2005;88:1479–88. doi: 10.1529/biophysj.104.047365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.