Abstract
Though iron and oxygen are required to sustain essential biological processes, an excess of either can result in oxidative stress. Therefore, mammals tightly regulate cellular and systemic iron and oxygen homeostasis. At the cellular level, the hypoxia-inducible transcription factors (HIFs) are key mediators of oxygen homeostasis through their regulation of genes involved in anaerobic metabolism and oxygen delivery, among others. Iron regulatory proteins (IRPs) largely govern cellular iron homeostasis through their effects on the translation and stability of mRNAs involved in iron uptake, utilization, export, and storage. Here, we describe regulatory factors for each pathway that sense both iron and oxygen availability and coordinate the maintenance of mammalian iron and oxygen homeostasis at both the cellular and systemic levels.
Keywords: iron, oxygen, hypoxia, iron regulatory protein (IRP), hypoxia-inducible factor (HIF), FBXL5, iron- and 2-oxoglutarate-dependent dioxygenase, hemerythrin
Mammalian cells employ iron and oxygen in processes that are essential for life, including oxidative phosphorylation, synthesis of metabolites and cofactors, and posttranslational modifications.1–3 In contrast, excess amounts of iron and oxygen result in cytotoxic oxidative stress,4,5 necessitating tight regulation of iron and oxygen availability. The hypoxia-inducible factors (HIFs), and the iron regulatory proteins (IRPs) are key mediators of cellular oxygen and iron homeostasis, respectively. Because iron and oxygen are often intimately connected in their metabolism, it is not surprising that their levels are coordinately regulated in cells. Such cross-talk is achieved in part by cellular regulatory factors that sense and respond to both iron and oxygen, and it is reinforced by overlap in the gene targets regulated by each pathway.
Overview of Mammalian Cellular Oxygen Homeostasis
Appropriate responses to changes in cellular oxygen tension are largely mediated through transcriptional activation of genes by HIFs.6 HIF is a dimer of basic helix-loop-helix (bHLH) and Per-Arnt-Sim (PAS) domain transcription factors, HIF-α and HIF-β.1 TheHIF-β subunit, also known as aryl-hydrocarbon-nuclear transporter (ARNT), is a constitutively expressed partner for multiple bHLH-PAS transcription factors.6 The HIF-α subunit, however, is regulated in response to changes in cellular oxygen availability.
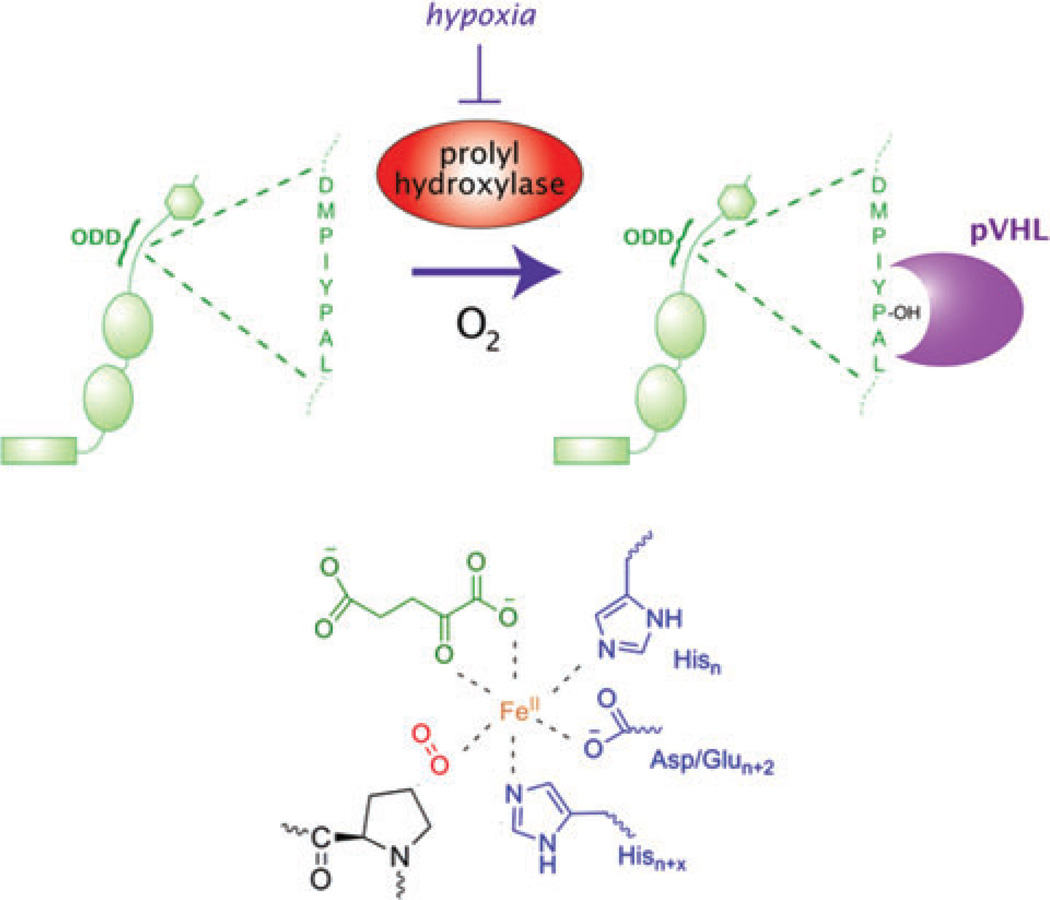
Under normoxic conditions, HIF-α is ubiquitinated by an E3 ligase complex containing the product of the von Hippel–Lindau tumor suppressor gene (VHL) and targeted for degradation by the proteasome (Fig. 1).7,8 A specific region in HIF-α, termed the oxygen degradation domain (ODD), is necessary and sufficient for VHL recognition of HIF-α. A family of three iron and 2-oxoglutarate-dependent dioxygenases hydroxylate proline residues in the ODD, thereby recruiting HIF-α to the VHL E3 ligase complex.9–11 These dioxygenases hydroxylate the HIF-α ODD under normoxic conditions but are inhibited in hypoxia, thus sparing HIF-α from VHL-mediated degradation. The recruitment of transcriptional co-activators to HIF-α is also inhibited in an oxygen-dependent manner by hydroxylation of an asparagine residue in its C-terminal transactivation domain by another iron and 2-oxoglutarate-dependent dioxygenase, factor inhibiting HIF-1 (FIH-1).12
Figure 1.
HIF regulation by iron and oxygen levels. Under normoxic conditions, HIF-α is targeted for degradation by the VHL-containing E3 ubiquitin ligase complex. VHL recognizes HIF-α following hydroxylation of proline residues by Fe(II)- and 2-oxoglutarate-dependent dioxygenases (top). These enzymes use iron in their active sites (bottom) to activate molecular oxygen. Under conditions of low oxygen and/or iron, HIF-α cannot be hydroxylated and is spared from degradation.
The biochemical properties of these dioxygenases allow them to serve as both iron and oxygen sensors in the cell. These enzymes require iron to activate dioxygen to hydroxylate residues on protein substrates as well as the co-substrate 2-oxoglutarate (Fig. 1). By virtue of their substrate requirements, these dioxygenases are believed to function as direct oxygen sensors in the pathway, with in vitro Km values for oxygen that appear to correlate with relative in vivo sensitivities to oxygen.1 However, the relationship between dioxygenase activity and changes in oxygen availability is likely to be further modulated by other mechanisms such as feedback loops and mitochondria-derived signals.1 Because of their iron requirement, these enzymes may also function as iron sensors, as evidenced by low dioxygenase activity seen in cells deficient in iron.13
Induction of HIF-α results in transcription of genes that confer adaptation to hypoxia. For example, at the cellular level HIF induces the transcription of genes involved in glycolysis, promoting adaptation to anaerobic metabolism.6 HIF can also regulate systemic hypoxic responses, for example, by inducing erythropoietin (EPO) and vasoendothelial growth factor (VEGF) gene expression to promote greater delivery of oxygen to tissues.6
Overview of Iron Homeostasis
In mammals, iron is primarily absorbed from the diet in the form of insoluble ferric salts through the proximal part of the small intestine. Intestinal epithelial cells (IECs) absorb iron through the apical membrane protein divalent metal transporter 1 (DMT1) and then export it into the bloodstream through the basolateral membrane protein, ferroportin (FPN).5,14,15 Exported iron binds to its serum carrier, transferrin (Tf), and is then transported throughout the body to be utilized by all cells, particularly erythroid precursors in the bone marrow.16
Since mammals have no active means to secrete excess iron from the body, iron levels are primarily regulated through control of dietary absorption in the intestine, mobilization of iron stores from the liver, and recycling in the reticuloendothelial system.16 A major effector hormone that regulates this systemic homeostasis is hepcidin.13 In response to high iron concentrations, hepcidin is transcriptionally upregulated in the liver and secreted into the bloodstream, where it binds to its receptor, FPN, causing this iron exporter to be internalized and degraded.17 As a result, export of iron into the blood stream from the intestine and splenic macrophages decreases, and Tf-bound iron levels decrease. In states of iron deficiency, hepcidin transcription is repressed, and FPN levels increase, promoting greater iron availability throughout the body.16
At the cellular level, iron homeostasis occurs largely through coordinated regulation of iron import, utilization, and storage. Cells acquire iron mostly through receptor mediated endocytosis of Tf through transferrin receptor 1 and 2 (TfR 1 and 2).5 Cells utilize much of this iron by incorporation into enzyme prosthetic groups such as iron sulfur clusters or heme, while excess iron is sequestered by ferritin, an iron storage protein.4,16 These processes are coordinately regulated as a function of cellular iron bioavailability at the level of posttranscriptional regulation.
Posttranscriptional regulation of iron metabolism was first described in ferritin mRNA, where in states of low iron, a cis-acting iron response element (IRE)18 in the 5′ untranslated region (UTR) promotes the binding of IRPs.2,5 IRP-binding results in suppression of ribosome-binding and decreases synthesis of the ferritin protein.2 IREs alternatively function to stabilize mRNAs when they are located in the 3′ UTR.5 In the case of TfR1 mRNA, IRP-binding protects the transcript from endonucleolytic degradation.19 IREs have since been identified in other genes (Table 1), though functional roles have not been validated for all. Thus, in the low iron state, IRPs bind to IREs to increase bioavailable iron levels by upregulation of iron uptake and downregulation of iron utilization and storage. When cells have excess iron, IRPs fail to bind IREs, reducing bioavailable iron by a decrease in iron uptake, and an increase in utilization and storage (Fig. 2).
TABLE 1.
Genes Containing Iron Response Elements
| Gene symbol | UTR | Protein name | Proposed function |
|---|---|---|---|
| ALAS25 | 5′ | 5′ aminolevulinic acid synthase | Heme synthesis |
| APP41 | 5′ | Amyloid precursor protein | Alzheimer’s disease pathogenesis |
| CDC14A18 | 3′ | Cell division cycle homolog A | Phosphatase; cell cycle |
| DMT15 | 3′ | Divalent metal transporter 1 | Iron import |
| EPAS127 | 5′ | HIF-2α | Hypoxia-induced transcription |
| FTH5 | 5′ | Ferritin heavy chain | Iron storage |
| FTL5 | 5′ | Ferritin light chain | Iron storage |
| mACO18 | 5′ | Mitochondrial aconitase | Citrate-isocitrate conversion |
| SLC40A15 | 5′ | Ferroportin | Iron export |
| SNCA42 | 5′ | Alpha-synuclein | Synucleinopathy pathogenesis |
| TFRC5 | 3′ | Transferrin receptor 1 | Cellular iron uptake |
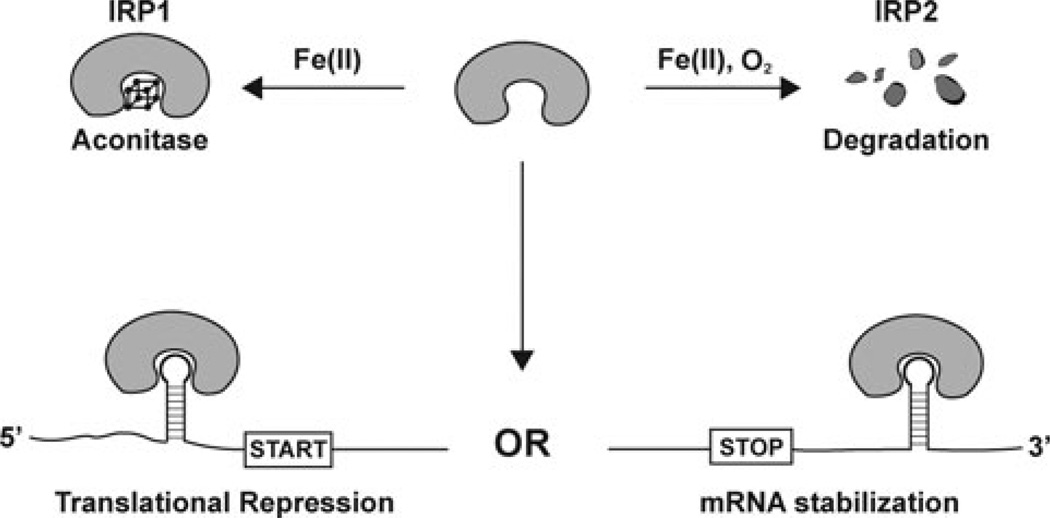
Figure 2.
Iron dependent regulation of the IRPs. In states of low iron, IRPs are capable of binding IREs in the 5′ or 3′ UTR in mRNAs of genes contributing to iron homeostasis. IRP-binding in turn blocks translation initiation or mRNA degradation, respectively. In states of high iron, IRP1 loses its IRE-binding activity following assembly of an iron sulfur cluster and conversion to a cytosolic aconitase. In contrast, IRP2 is polyubiquitinated and degraded by the proteasome under iron- and oxygen-replete conditions.
Iron governs the IRP1 and 2 binding activity through distinct mechanisms (Fig. 2). In states of high iron, IRP1 assembles an iron sulfur cluster, gaining aconitase activity but losing its ability to bind RNA in the process.2 IRP2, despite being 60% identical to IRP1, cannot assemble an iron sulfur cluster. Instead, in the high iron state IRP2 is ubiquitinated and degraded by the proteasome, thus removing its RNA-binding activity from cells.15
Together, IRP1 and 2 are essential regulators of mammalian iron homeostasis, though they can largely compensate for one another, as deletion of either gene results in viable and fertile mice.20 IRP2 null mice do, however, display microcytic anemia and iron overload in the intestine and liver.21–23 While global IRP deficiency results in misregulation of iron homeostasis, an inappropriate excess of IRP-binding activity has also been shown to affect systemic iron management. For example, the Shiraz phenotype of microcytic anemia and iron overload is due to a mutation in glutaredoxin 5, a critical component of iron sulfur cluster biogenesis.24 In this setting, the IRE-binding form of IRP1 is upregulated even in the iron replete state, resulting in repression of heme synthesis by inhibition of erythroid 5′ aminolevulinic acid synthase (eALAS).24,25
Cross-Talk Between O2 and Fe Homeostasis Regulatory Pathways
The bioavailability of iron affects the cellular hypoxic response in several ways. As noted in cell culture studies, manipulation of iron levels in the media alters HIF-α induction through effects on dioxygenase activity.1 As a result, it is proposed that dioxygenases may function as iron sensors, though it remains to be seen whether these enzymes play an extensive role in physiological iron sensing. Iron’s role in the hypoxic response pathway has been further underscored by the recent discovery of an IRE within the 5′ UTR of HIF-2α.27 Consequently, HIF-2α translation is downregulated in states of iron deficiency and upregulated in iron excess.27 HIF-2α function can also be altered by small molecules that modulate the binding of IRP1 to IREs.28 The physiological importance of this additional layer of regulation remains to be studied, though it could serve to limit hypoxia-induced erythropoiesis when body iron stores are limited.27
Oxygen or iron deficiency can in turn, alter cellular iron homeostasis following HIF induction. HIF binds to HREs found within several genes involved in iron metabolism (Table 2), promoting iron uptake from serum, iron scavenging, and iron retention. HIF is also known to impact systemic iron homeostasis. For example, HIF-2α directly mediates dietary iron absorption through transcription of genes involved in IEC iron transport,30,31 underlying an observation made nearly 50 years ago that hypoxia increases dietary iron absorption through the intestine.29 The hypoxia response pathway was also found to be crucial to the regulation of the systemic hormone, hepcidin.32,33 HIF-1 induction, resulting from hypoxia or reduced iron availability, represses hepcidin expression in the liver upon binding toHREs within the hepcidin promoter. Reduced hepcidin levels should, in turn, enhance absorption of dietary iron to compensate.
TABLE 2.
HIF Target Genes Involved in Iron Homeostasis
| Gene symbol | Protein name | Proposed function |
|---|---|---|
| CP13 | Ceruloplasmin | Iron oxidation in bloodstream |
| DMT130,31 | Divalent metal transporter 1 | Iron import |
| HAMP32,a | Hepcidin | Systemic iron effector |
| HMOX113 | Heme oxygenase-1 | Heme catabolism |
| TFRC13 | Transferrin receptor | Cellular iron uptake |
| TF43,44 | Transferrin | Serum iron transport |
HIF represses transcription of hepcidin.
In addition to mediating HIF-dependent induction of genes involved in iron homeostasis, oxygen levels have also been known to directly influence cellular iron levels through the IRP pathway. Oxygen can influence IRP1 activity in cells by affecting the stability of its iron sulfur cluster.34 With respect to IRP2, hypoxia promotes its RNA-binding activity through increased stability, even under iron-replete conditions that usually favor its degradation.35 Though several models have been proposed to explain how cellular iron and oxygen levels are sensed and translated to changes in IRP2 regulation, the responsiblemechanisms remain poorly understood.15
A Candidate Fe and O2 Sensor Governing IRP Stability
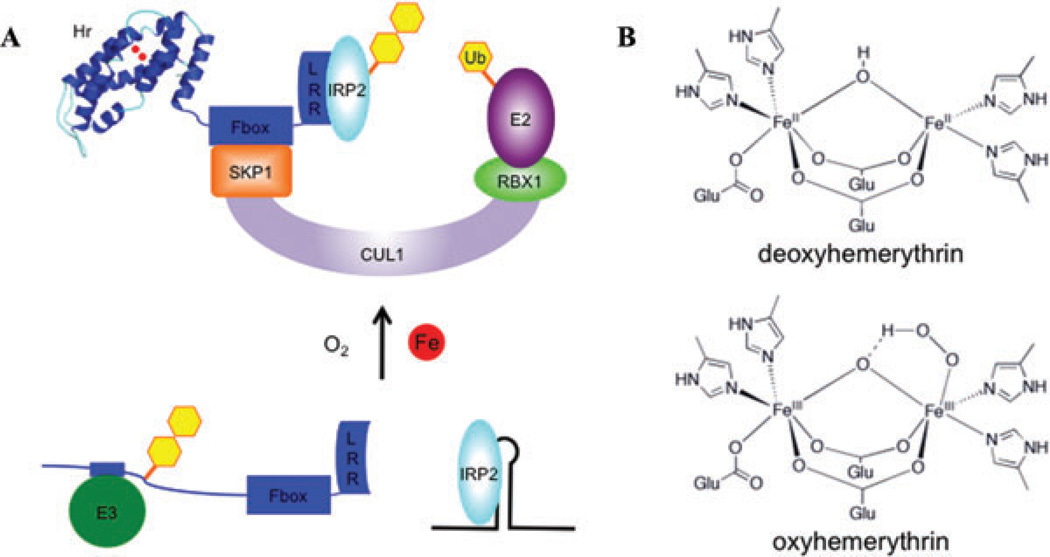
Our laboratory has recently identified an iron-regulated E3 ligase that mediates the iron-dependent degradation of IRP2.36 This E3 ligase, a member of the Skp-Cullin-F-box (SCF) complex class of E3 ligases, interacts with IRP2 and can catalyze its polyubiquitination in vitro. The F-box-containing subunit of this SCF complex, FBXL5, is regulated by iron at the level of its own protein stability: FBXL5 is stabilized under iron-replete conditions and degraded when cellular iron availability is low. Iron-dependent regulation is conferred by an iron- and oxygen-binding domain located within FBXL5’s N-terminus. In the presence of iron, this region folds into a hemerythrin domain, previously only found in marine invertebrates and bacteria, and frequently serve as oxygen carriers.37,38 Structurally, the hemerythrins form an alpha helical bundle, wherein they directly bind to two iron atoms through carboxylate and imidazole side chains of its amino acids (Fig. 3). The two iron atoms form a μ-oxo di-iron center contained within the hydrophobic core of the alpha helical bundle, and can reversibly bind oxygen (Fig. 3).37
Figure 3.
Model of FBXL5’s proposed role in cellular iron homeostasis. (A) In states of low iron bioavailability, FBXL5’s hemerythrin domain is likely destabilized, allowing for a putative E3 ubiquitin ligase to recognize a degron and promote FBXL5’s degradation. Limiting levels of FBXL5 result in IRP2 stabilization and binding to IREs. In contrast, when iron bioavailability increases, FBXL5’s hemerythrin domain forms a stable alpha helical bundle by directly binding iron and oxygen. In this state, FBXL5 is able to mask its degron and assemble into a SCF complex that promotes the polyubiquitination and degradation of IRP2. (B) Model of hemerythrin iron- and oxygen-binding site. Imidazole and carboxylate residues from conserved amino acid side chains bind a diiron center connected by a bridging oxygen atom (μ-oxo). Oxygen can be reversibly bound onto the penta-coordinate iron and forms a peroxo radical species that is stabilized by protonation and coordination to the bridging oxygen atom.
In the setting of iron sensing and IRP2 ubiquitination, FBXL5 may sense iron and oxygen concentrations through the stability of its hemerythrin domain. In the absence of iron and oxygen, the hemerythrin domain is unstable. Unfolding of the hemerythrin domain appears to unmask a degron for an as-yet unidentified E3 ligase to promote FBXL5 degradation. This decrease in FBXL5 protein accumulation results in a corresponding increase in IRP2 stability. Conversely, in settings of high iron and oxygen the hemerythrin domain is stabilized, concealing the FBXL5 degron. The ensuing increase in its stability allows FBXL5 to assemble into a SCF complex capable of catalyzing the polyubiquitination and degradation of IRP2 (Fig. 3).
Summary
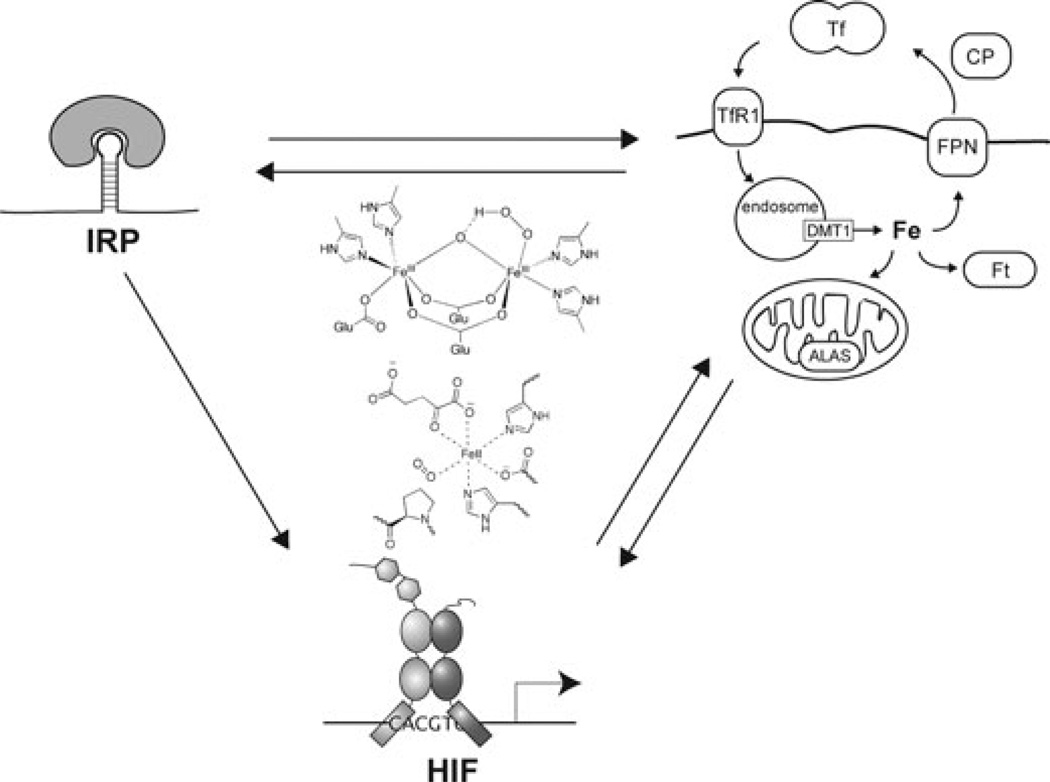
Coordinate regulation of iron and oxygen homeostasis facilitates appropriate responses to perturbations of either of these metabolites at the cellular level. In cells, the HIF and IRP pathways respond to changes in both iron and oxygen levels. We now know of enzymes in each pathway that are capable of directly sensing and signaling changes in cellular iron and oxygen levels: dioxygenases for HIF and the hemerythrin containing FBXL5 for IRPs (Fig. 4). By binding both iron and oxygen, these sensors provide a basis for cross-talk between these pathways. Furthermore, cells exhibit cross-talk between the HIF and IRP pathway through overlapping control of gene expression. This cross-talk between iron and oxygen homeostasis pathways at the cellular level may also be extended to the systemic level. In addition, coordinate regulation of oxygen and iron homeostasis may be achieved indirectly through a variety of other mechanisms not discussed here.39,40 Together, these observations provide new insights and opportunities to address diseases stemming from misregulation of oxygen and iron homeostasis, including anemia, iron overload disorders, susceptibility to infection, ischemic damage, and cancer.
Figure 4.
Cross-talk between pathways mediating iron and oxygen homeostasis. At the cellular level, both iron and oxygen are sensed by dioxygenases and hemerythrin to regulate HIF-α and IRP2 stability, respectively. In addition, the HIF and IRP pathways are intertwined at the level of target gene expression, as the HIF-2α 5′ UTR contains an IRE, while both HIF and IRPs mediate expression of genes involved in cellular and systemic iron homeostasis.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 2007;3:144–153. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- 2.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 3.Wallander ML, Leibold EA, Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta. 2006;1763:668–689. doi: 10.1016/j.bbamcr.2006.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galaris D, Pantopoulos K. Oxidative stress and iron homeostasis: mechanistic and health aspects. Crit. Rev. Clin. Lab. Sci. 2008;45:1–23. doi: 10.1080/10408360701713104. [DOI] [PubMed] [Google Scholar]
- 5.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 6.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 7.Ivan M, Kondo K, Yang H, et al. HIF-alpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 8.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 9.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 10.Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 11.Ivan M, Haberberger T, Gervasi DC, et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lando D, Peet DJ, Gorman JJ, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peyssonnaux C, Nizet V, Johnson RS. Role of the hypoxia inducible factors HIF in iron metabolism. Cell Cycle. 2008;7:28–32. doi: 10.4161/cc.7.1.5145. [DOI] [PubMed] [Google Scholar]
- 14.Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu. Rev. Nutr. 2008;28:197–213. doi: 10.1146/annurev.nutr.28.061807.155521. [DOI] [PubMed] [Google Scholar]
- 15.Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N. Y. Acad. Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- 16.Wrighting DM, Andrews NC. Iron homeostasis and erythropoiesis. Curr. Top. Dev. Biol. 2008;82:141–167. doi: 10.1016/S0070-2153(07)00006-3. [DOI] [PubMed] [Google Scholar]
- 17.Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 18.Piccinelli P, Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–966. doi: 10.1261/rna.464807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koeller DM, Casey JL, Hentze MW, et al. A cytosolic protein binds to structural elements within the iron regulatory region of the transferrin receptor mRNA. Proc. Natl. Acad. Sci. USA. 1989;86:3574–3578. doi: 10.1073/pnas.86.10.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyron-Holtz EG, Ghosh MC, Iwai K, et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaVaute T, Smith S, Cooperman S, et al. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 2001;27:209–214. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- 22.Smith SR, Ghosh MC, Ollivierre-Wilson H, et al. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol. Dis. 2006;36:283–287. doi: 10.1016/j.bcmd.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Galy B, Ferring D, Minana B, et al. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2) Blood. 2005;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- 24.Wingert RA, Galloway JL, Barut B, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- 25.Pondarre C, Antiochos BB, Campagna DR, et al. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron_sulfur cluster biogenesis. Hum. Mol. Genet. 2006;15:953–964. doi: 10.1093/hmg/ddl012. [DOI] [PubMed] [Google Scholar]
- 26.Knowles HJ, Raval RR, Harris AL, et al. Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res. 2003;63:1764–1768. [PubMed] [Google Scholar]
- 27.Sanchez M, Galy B, Muckenthaler MU, et al. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat. Struct. Mol. Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 28.Zimmer M, Ebert BL, Neil C, et al. Small-molecule inhibitors of HIF-2a translation link its 5′UTR iron-responsive element to oxygen sensing. Mol. Cell. 2008;32:838–848. doi: 10.1016/j.molcel.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendel GA. Studies on iron absorption. I. The relationships between the rate of erythropoiesis, hypoxia and iron absorption. Blood. 1961;18:727–736. [PubMed] [Google Scholar]
- 30.Shah YM, Matsubara T, Ito S, et al. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152–164. doi: 10.1016/j.cmet.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mastrogiannaki M, Matak P, Keith B, et al. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J. Clin. Invest. 2009;119:1159–1166. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peyssonnaux C, Zinkernagel AS, Schuepbach RA, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J. Clin. Invest. 2007;117:1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolas G, Chauvet C, Viatte L, et al. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J. Clin. Invest. 2002;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyron-Holtz EG, Ghosh MC, Rouault TA. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306:2087–2090. doi: 10.1126/science.1103786. [DOI] [PubMed] [Google Scholar]
- 35.Hanson ES, Rawlins ML, Leibold EA. Oxygen and iron regulation of iron regulatory protein 2. J. Biol. Chem. 2003;278:40337–40342. doi: 10.1074/jbc.M302798200. [DOI] [PubMed] [Google Scholar]
- 36.Salahudeen AA, Thompson JT, Ruiz JC, et al. An E3 ligase possessing an iron responsive hemerythrin domain is a regulator of iron homeostasis. Science. doi: 10.1126/science.1176326. Submitted. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stenkamp RE. Dioxygen and hemerythrin. Chem. Rev. 1994;94:715–726. [Google Scholar]
- 38.French CE, Bell JM, Ward FB. Diversity and distribution of hemerythrin-like proteins in prokaryotes. FEMS Microbiol. Lett. 2008;279:131–145. doi: 10.1111/j.1574-6968.2007.01011.x. [DOI] [PubMed] [Google Scholar]
- 39.Hentze MW, Kuhn LC. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor CT. Mitochondria and cellular oxygen sensing in the HIF pathway. Biochem. J. 2008;409:19–26. doi: 10.1042/BJ20071249. [DOI] [PubMed] [Google Scholar]
- 41.Lee DW, Andersen JK. Role of HIF-1 in iron regulation: potential therapeutic strategy for neurodegenerative disorders. Curr. Mol. Med. 2006;6:883–893. doi: 10.2174/156652406779010849. [DOI] [PubMed] [Google Scholar]
- 42.Olivares D, Huang X, Branden L, et al. Physiological and pathological role of alpha-synuclein in Parkinson disease through iron mediated oxidative stress; the role of a putative iron-responsive element. Int. J. Mol. Sci. 2009;10:1226–1260. doi: 10.3390/ijms10031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manalo DJ, Rowan A, Lavoie T, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 44.Xia X, Lemieux EM, Li W, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. USA. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]