Abstract
The prestalk region of the Dictyostelium slug is comprised of an anterior population of pstA cells and a posterior population of pstO cells. They are distinguished by their ability to utilize different parts of the promoter of the ecmA gene. We identify, by mutational analysis and DNA transformation, CA-rich sequence elements within the ecmA promoter that are essential for pstA-specific expression and sufficient to direct pstA-specific expression when multimerised. The CA-rich region was used in affinity chromatography with nuclear extracts and bound proteins were identified by mass spectrometry. The CA-rich elements purify MrfA, a protein with extensive sequence similarity to animal Myelin-gene Regulatory Factor (MRF)-like proteins. The MRF-like proteins and MrfA also display more spatially limited but significant sequence similarity with the DNA binding domain of the yeast Ndt80 sporulation-specific transcription factor. Furthermore, the ecmA CA-rich elements show sequence similarity to the core consensus Ndt80 binding site (the MSE) and point mutation of highly conserved arginine residues in MrfA, that in Ndt80 make critical contacts with the MSE, ablate binding of MrfA to its sites within the ecmA promoter. MrfA null strains are delayed in multicellular development and highly defective in pstA-specific gene expression. These results provide a first insight into the intracellular signaling pathway that directs pstA differentiation and identify a non-metazoan orthologue of a family of molecularly uncharacterised transcription factors.
Keywords: Dictyostelium, Myelin-gene Regulatory Transcription Factor, prestalk cell
Introduction
Terminal cell type differentiation in Dictyostelium generates a mass of spores borne on a tapering cellular stalk that is embedded into a conical basal disc, also composed of stalk cells. The orchestrated movement of the prestalk cells creates this structure and the need to move differentially is, presumably, a main reason for the existence of heterogeneity in the prestalk population. There are at least six Dictyostelium prestalk (pst) sub-types, defined by their patterns of marker gene expression, by their location in the slug and by their movement patterns and fates at culmination (Gaudet et al., 2008). Three of the principal populations, pstA, pstO and pstB cells, can be distinguished from one another using reporter fusions with the promoters of ecmA and ecmB; two genes that encode related extracellular matrix proteins.
PstA cells utilize cap-site proximal ecmA promoter sequences, termed ecmA sequences, while pstO cells utilize cap-site distal sequences, the ecmO sequences (Early et al., 1993); the complete promoter of the ecmA gene is termed the ecmAO sequence. PstA cells occupy the approximate front one-half of the prestalk region and the pstO cells form a band behind them. At culmination there is an ordered, “reverse fountain” movement of these two populations to occupy, respectively, the bottom and the top halves of the stalk. PstB cells express the ecmB gene at a higher relative level than the ecmA gene (Jermyn et al., 1996). They form a cluster, tightly apposed to the substratum. This cluster is normally located towards the back of the slug but the pstB cells stain strongly with neutral red and visualisation of living slugs shows them to be highly dynamic, making occasional forays as far forward as the prestalk-prespore boundary (Dormann et al., 1996). At culmination the pstB cells move to an anterior position, apposed to the substratum, and the descending stalk impales them so that they come to form the basal disc.
Up to the slug stage differentiation occurs non-positionally, in response to diffusible signals with subsequent cell sorting acting to generate pattern (reviewed in Kay and Thompson, 2009). This is best characterised for pstO and pstB differentiation. Here the diffusible signalling molecule is DIF-1, a chlorinated hexaphenone produced by the prespore cells (Kay and Thompson, 2001). Mutants defective in DIF-1 production are defective in pstO and pstB cell differentiation but to different degrees. Such “DIF-less” mutants retain a pstO region but the cells within it display greatly reduced ecmAO:lacZ expression (Thompson and Kay, 2000). In contrast, the slugs formed by DIF-less mutants contain very little if any pstB tissue and their culminants lack, or possess only rudimentary, basal discs (Keller and Thompson, 2008; Saito et al., 2008). PstA cell differentiation appears normal in DIF-less mutants and there is evidence that another, unidentified polyketide may act to induce their differentiation (Serafimidis and Kay, 2005).
DIF-1 signaling has been characterised at the transcriptional level for both pstO and pstB differentiation. DimA, a bZIP transcription factor, is required for ecmAO:lacZ expression in pstO cells and for the induction, by DIF-1, of ecmA expression in a monolayer assay(Thompson et al., 2004). DimB, a dimerisation partner of DimA, is also required for DIF-1 inducibility of ecmA in monolayer cells and it binds to two sequence elements within the ecmO promoter sequence (Huang et al., 2006; Zhukovskaya et al., 2006). DimA and DimB translocate to the nucleus in response to DIF and ChIP analysis shows that DimB binds to the ecmA promoter. There is also strong evidence for an involvment of DimB in pstB differentiation; DimB is highly nuclear-enriched in pstB cells, the DimB null strain lacks pstB cells, the ecmB gene is not DIF-inducible in the null mutant and DimB binds to the ecmB promoter when cells are exposed to DIF-1 (Saito et al., 2008; Yamada et al., 2011).
The cap-site proximal promoter region of the ecmA gene, that directs pstA specific gene expression, is in contrast totally uncharacterized. We show that it functions, in part at least, via a novel transcription factor that contains a DNA-binding domain with sequence and functional similarity to Ndt80, a very well characterized yeast sporulation regulator. Ndt80 binds to a DNA sequence known as the middle sporulation element (MSE) and it activates a battery of genes required for progression through meiosis (Chu et al., 1998; Lamoureux et al., 2002). The sequence similarity of the novel protein to Ndt80 is restricted to the DNA binding domain but there is much more extensive similarity to vertebrate CNS Myelin-gene Regulatory Factor (MRF; Emery et al., 2009). Hence we name the Dictyostelium protein MrfA (Myelin gene regulatory factor-like A).
MRF, the first family member to be characterized, is expressed selectively in post-mitotic oligodendrocytes (Emery et al., 2009). It is required for the expression of a large number of CNS myelin genes, its over-expression stimulates expression of CNS myelin genes and mice lacking MRF in the oligodendrocyte lineage are defective in myelination. Despite its biological importance its mechanism of action is unknown and most other members of the family are completely uncharacterized. The one, highly controversial exception to this is the C. elegans orthologue, pqn-47, which is a regulator of molting (Russel et al., 2011). Based on their failure to detect any nuclear enrichment of a PQN-47::GFP fusion protein, these authors strongly question the validity of the assignment of MRF as a transcription factor. We show, in support of the original observations on murine MRF, that the closely related protein MrfA is a bona fide transcription factor.
Results
A 39-mer from within the ecmA promoter is both necessary and sufficient to direct pstA-specific expression
Previous deletion analysis of the ecmA promoter identified a region with a 5′ end point at nt −493 (numbered relative to the cap site) that directs expression selectively in pstA cells (Zhukovskaya et al., 2006). The 117nt region adjacent to this 5′ end point is, as is typical of all Dictyostelium promoters, highly AT-rich and it can be deleted, to yield construct ecmA376:lacZ, with no apparent effect on the spatial expression pattern (Fig. 1A). The adjacent 39 nucleotides are much less AT-rich and just a partial deletion of this, very CA-rich, region, to yield ecmA357:lacZ, abolishes expression. We term the sequence encompassing the entire CA-rich region the 39-mer. In order to determine whether the 39-mer contains all the sequence information necessary to direct pstA-specific expression, it was 4-fold multimerised and placed upstream of minimal promoter elements derived from an actin gene (Fig. 1B). The resultant, 4×39-mer:lacZ construct shows robust and correctly localized staining. Thus the 39-mer sequence is, when multimerised, sufficient for pstA-specific expression.
Fig. 1. Analysis of the ecmA promoter sub-region that directs expression in pstA cells.
(A) Schematic representation of the two 5′ deletions of the ecmA promoter that define the 39-mer and expression analysis of slugs expressing them. (B). Schematic representation and expression analysis of 4×39:lacZ, a lacZ construct containing actin 15 basal promoter elements coupled to a 4-fold multimerized tandem array of the 39-mer. Bar represents 200 μm.
Affinity chromatography using the 39-mer yields the CNS MRF orthologue MrfA
The 39-mer was oligomerised, bound to a matrix and used in affinity chromatography with nuclear extracts derived from slug cells. After two rounds of chromatography, SDS electrophoresis resolved many stained bands (Fig. 2A and see legend) and slices containing these were excised and subjected to mass spectrometry. The hits include DimB and MrfA. The MrfA sequence was originally determined from a random cDNA clone (rcdK for random cDNA: DDB_G0284183 in http://dictybase.org/) used in genomic mapping (Kuspa and Loomis, 1996). We propose the more meaningful name of MrfA because there is strong similarity along most of the MrfA protein length (excepting the simple repetitive sequence) with vertebrate CNS MRF (Myelin-Gene Regulatory Factor; (Emery et al., 2009). Since the DimB null is not defective in PstA differentiation (Zhukovskaya et al., 2006), we chose to determine whether MrfA plays any role in the process.
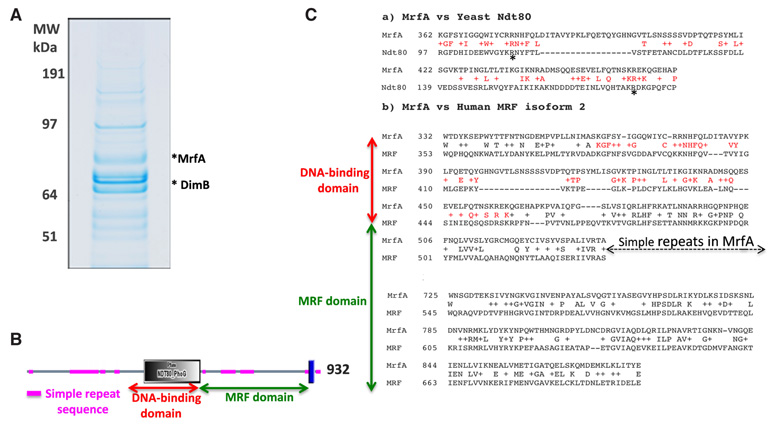
Fig. 2. Partial purification and identification of MrfA.
(A) Preparative SDS gel electrophoresis of the proteins purified by affinity chromatography on the 39-mer. The complexity of the protein mixture reflects the fact that all DNA binding proteins will bind at some level to any DNA molecule. This is kept under control by competition, exerted by including or pre-incubating with non-specific DNA, but this is a difficult balance: too stringent a competition may cause low abundance or low affinity proteins to be lost. These could still be important players, because interactions with other transcription factors may amplify their effect in vivo. Therefore we utilise intermediate competitor levels and rely upon blanket mass spectrometry analysis, followed by bio-informatic triage to select candidates for further more detailed analysis. A good example of this was the identification of DimB and MybE, the pstO regulators, from a welter of other bands in similar analyses (Fukuzawa et al., 2006; Zhukovskaya et al., 2006). The predicted size of MrfA is 105 kDa but the gel region containing it is estimated to be approximately 75 kDa. We assume therefore that we are detecting a proteolytic cleavage fragment of MrfA. (B) MrfA structure, the blue bar near the C terminus represents a predicted membrane spanning domain (Persson and Argos, 1994). (C) a) NCBI BLAST alignment at the default settings of yeast Ndt80 and MrfA showing the DNA binding domains. Asterisks highlight two arginine residues that are essential for DNA binding of Ndt80. b) NCBI BLAST alignment at the default settings of Human Myelin-gene Regulatory Factor (MRF) and Dictyostelium MrfA showing the region of sequence similarity. The gap arrowed is mainly comprised of simple repeat sequence (Fig. 2B). There is also a weaker sequence similarity with a small region of the MRF domain common to several cell surface anchor proteins of prokaryotes and to bacteriophage structural proteins but the significance of this is unclear.
Murine MRF is the founder member of a family, which we will term the MRF-like family, that is represented in vertebrate and invertebrate animals but not in fungi. All members contain a region with sequence similarity to the DNA binding domain of yeast Ndt80 (Fig. 2B and Fig. 2C section a). The defining feature of the family is the additional presence, nearer the C terminus, of an approximate 200 amino acid region of further homology which we term the MRF domain (Fig. 2C section b). In MrfA there is also a hydrophobic region very near the C terminus (Fig. 2B) that could, in principle, constitute a membrane-spanning (TM) domain of the kind found in membrane-tethered transcription factors (Brown et al., 2000). This interesting possibility is under investigation.
The predicted MrfA DNA binding domain produced in E. coli interacts with the CA-rich sequence elements in the ecmA 39-mer
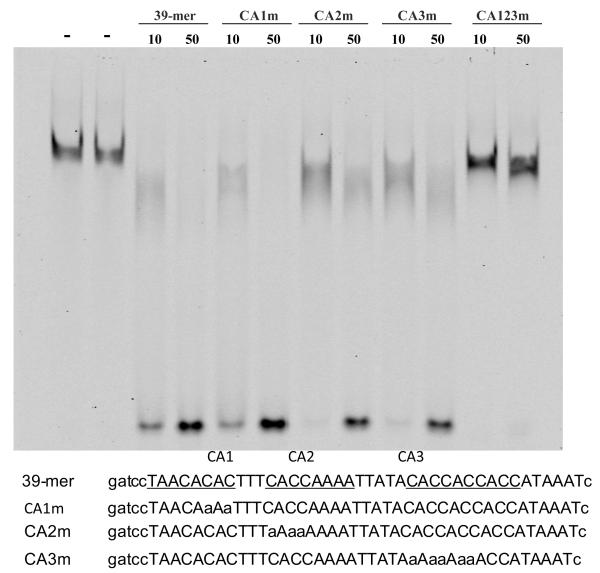
A segment of MrfA, containing amino acids 294-546 and spanning the predicted DNA binding domain, was oligo-histidine tagged and expressed in E. coli. The fusion protein was purified by metal ion chromatography and used in band shifts with the 39-mer as probe. In the absence of specific competitor there is a retarded band (Fig. 3). The core consensus binding site for the interaction of yeast Ndt80 with sporulation-specific promoters is CRCAAA (Jolly et al., 2005). The 39-mer contains three discrete regions, CA1, CA2 and CA3, that possess similarity with this sequence. These regions were mutated, separately and in combination, and the 39 nt sequences containing them were used as competitors in band shift assays. The separate mutations produce only relatively small effects on the efficiency as a competitor. However, at concentrations where the unmutated 39-mer acts as a potent competitor, mutation of all three regions eliminates competition. This suggests independent binding to the three regions and is consistent with the smearing to higher mobility of the retarded products observed with the individually mutated competitors; they presumably represent probe molecules complexed with varying numbers of MrfA molecules.
Fig. 3. In vitro binding of recombinant MrfA to the ecmA 39-mer.
Bacterially expressed MrfA protein region 294-546, a region that spans the predicted DNA-binding domain, was used in band shift assay. Protein amounts were 0.5 μg per reaction. Parental, 39-mer, and mutated 39-mer sequences are shown below. The C-rich sequences (CA1, CA2 and CA3) are underlined and C to A substitutions introduced into the mutant competitors are shown in small case. “-”, 10 and 50 represent competitor in ng.
The CA-rich sequence elements are necessary for reporter expression in pstA cells
We next determined the effect of mutations in the CA rich regions on the in vivo expression of ecmA376:lacZ (Fig. 4). Each of the three separate block mutations that were analysed by band shift, CA1m, CA2m and CA3m, render the ecmA376 promoter fragment less efficient, with CA1m and CA3m showing the lowest relative level of expression. Thus, in contrast to their behavior in band shift assay, all three CA-rich regions are needed for optimal expression in vivo.
Fig. 4. Mutational analysis of the ecmA 39-mer.
The CA-rich sequences (CA1-3; underlined) within the 39-mer region of ecmA376:lacZ were mutated, by C to A substitutions as indicated by small case letters, to generate CA1m:lacZ-CA3m:lacZ (Fig. 1A and Fig. 3). The staining patterns of parental ecmA376:lacZ and each of the three mutant constructs are shown. They were all stained for 16 hours. CA1m:lacZ and CA3m:lacZ showed almost no staining by 8 hours of incubation, at which time the wild-type control (ecmA376:lacZ) showed strong labeling in pstA cells (data not shown). Bar represents 200 μm.
Mutation of two conserved residues important for specific DNA binding by Ndt80 ablates binding of MrfA to the ecmA 39-mer
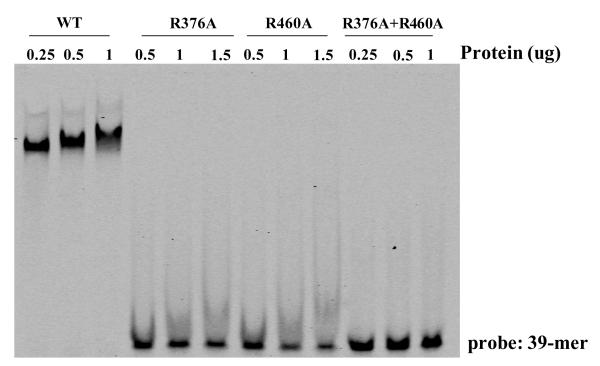
Next, a functional relationship to Ndt80 was confirmed at the protein level by inserting point mutations in MrfA, to replace residues known to be critically important in the interaction of Ndt80 with the MSE. The structure of the Ndt80 DNA binding domain, complexed with an MSE (Lamoureux et al., 2002), shows it to be a member of the Ig-fold group of transcription factors, a family that also includes p53 and NFkB. Two arginine residues, R111 and R177, respectively interact with the first and second C-G base pairs in the core consensus MSE (asterisked in Fig. 2C section a). Mutation of either of these amino acids leads to a 20-fold decrease in Ndt80-binding affinity and a greater than 100-fold decrease in biological activity (Fingerman et al., 2004). We mutated each equivalent residue in MrfA to alanine, to create R376A and R460A (Fig. 2C section a) and we also constructed the double mutant R376A+R460A. The proteins were expressed in E. coli and used in band shift assay with the 39-mer as probe. Increasing amounts of protein, normalized by concentration estimates from aliquots resolved on an SDS gel, were used in the band shift (Fig. 5). The unmutated protein retards the probe at all concentrations tested while the two single mutant proteins and the double mutant are inactive.
Fig. 5. Functional analysis of residues of MrfA essential for binding the ecmA 39-mer.
Mutant MrfA proteins that contain R to A substitutions in two residues essential for DNA binding by Ndt80 (Fig. 2C section a) were expressed as HIS fusions in E. coli and assayed for binding to the 39-mer. The fusion protein amounts were normalized by parallel gel electrophoretic analyses, followed by staining, and are expressed as the number of ug used in each reaction.
mrfA null mutants display delayed development
A null mutant for MrfA (the mrfA- strain) was generated by homologous gene disruption (Fig. 6). There is a progressive delay in the mutant’s development. By the mound stage the mutant displays an approximate five hour delay that persists through to the slug stage and then increases, with many mutant structures taking several days to complete culmination (Fig. 7). During migration, null mutant slugs frequently split along their length (Fig. 8A). The timing of these developmental defects are consistent with the expression pattern of mrfA as displayed on “dictyExpress” at http://dictybase.org/; there is little or no expression in growing cells, an approximately linear rise up to through aggregation and a gradual decrease thereafter.
Fig. 6. Generation of an mrfA- strain.
(A) At the top is a schematic representation of the knockout construct used for targeted mutagenesis of the mrfA gene. The coding region of mrfA is boxed, with two introns (gray boxes) and the Ndt80 homologous domain (black box). The knockout construct was based on a cDNA clone obtained from NBRP, Japan (clone SSA107) as a template. A blasticidin cassette was inserted at the unique EcoR1 site of SSA107, as described in methods. (B) Confirmation of gene targeting by PCR. Genomic DNA was extracted from Ax2 (lane WT) and the null strain (lane mrfA-) and PCR was performed with primer pairs P2-P3 or BsrF-P3. The P2-P3 primer pair generates a 611 bp product for WT and 2111 bp for mrfA-. This increased size is as expected if the blasticidin cassette is indeed inserted into the mrfA gene by homologous recombination. The BsrF-P3 primer pair generates a 1725 bp product for mrfA- but not for WT DNA. Since this uses the same WT DNA as is used for the P2-P3 PCR reaction the lack of signal in the WT lane cannot be due to template degradation. (C) Confirmation of gene inactivation by RT-PCR. Total RNA was extracted from the Ax2 strain (lane marked WT) and the null strain (lane marked mrfA-). After synthesis of cDNAs PCR was performed with the P1-P3 primer pair, which generates a 1545 bp product only in the WT lane. IG7 was analysed in the same reaction as a ubiquitously expressed control.
Fig. 7. Development of the mrfA- strain.
The parental Ax2 and the mrfA- strains were developed on KK2 agar at a density of 1.0×106 cell/cm2 for the times shown.
Fig. 8. Cellular differentiation in the mrfA- strain.
(A) Side views of a parental and a mrfA- mutant slug formed by neutral red stained cells. The position of the pstB cluster is indicated by multiple arrows. (B) Cellular differentiation at the slug stage was analyzed using cell type-specific lacZ constructs transformed into parental and mutant cells.
The example shown in Fig. 8A was stained with neutral red, which stains all prestalk cells and so orients the structures. Neutral red is also invaluable as a marker for pstB cells, the precursors of the basal disc cells, because there is as yet no molecular marker specific for pstB cells (Dormann et al., 1996; Saito et al., 2008). There is a pstB population in the particular null mutant slug shown here. It is located, as always, on the ventral surface and, in this particular example, it is in the anterior half of the slug. Most often, as in the control slug, the pstB cells are located in the rear of the slug and this explains why the presence of a pstB population, and of the basal disc derived from it, is sporadic in the null mutant (data not shown); because when a slug splits longitudinally a rearward pstB population will become separated from the residual, anterior slug tissue.
The expression of pstA markers is highly defective in the mrfA- strain
The other major prestalk cell sub-types were analysed using lacZ constructs (Fig. 8B). PstAB differentiation, which generates a cone of cells in the slug tip and which is assayed using ecmB:lacZ, is normal. So too are prespore and pstO differentiation, respectively assayed using pspA:lacZ and ecmO:lacZ. In agreement with the neutral red staining pattern, total anterior prestalk differentiation, assayed using ecmAO:lacZ, also appears normal in the mutant. However, analysis of two pstA-specific markers significantly modifies this conclusion. We first analysed expression of the multimerised 39-mer construct, 4×39-mer:lacZ in parental and mrfA- slugs (Fig. 9). The control slug shows strong staining, predominantly confined to the pstA region. In contrast, in the null mutant there are only a very few staining cells, scattered throughout the slug. Parental slugs expressing ecmA376:lacZ are also strongly stained in the pstA region but here there is a low level of residual staining in the null mutant, predominantly restricted to the extreme anterior (Fig. 9).
Fig. 9. Expression of pstA markers in parental, mrfA- and mrfA-/dimB- strains.
Cells with the indicated genotypes, and transformed with the ecmA376:lacZ or 4×39-mer:lacZ constructs, were developed to the slug stage and stained overnight.
We speculated that the residual expression of ecmA376:lacZ in the anterior of mrfA- slugs might be due to partial redundancy with DimB, because:
DimB is bound by the 39-mer in affinity chromatography (Fig. 2A) .
the 40-mer contains several sequence tracts which resemble the sequence of R2, the highest affinity DimB binding site in the ecmO region (Zhukovskaya et al., 2006).
an oligonucletide containing CA3 sequences is specifically retarded in band shift analysis using recombinant DimB (unpublished results).
We therefore constructed a double null strain, in which both the mrfA and dimB genes are disrupted. However, the ecmA376:lacZ construct is expressed in the double null strain at the same level as in the mrfA- strain (Fig. 9). Hence DimB is not the partially redundant partner.
Discussion
PstO-specific gene expression is relatively well understood; the extracellular signal, DIF-1, and several cognate transcription factors have been identified. PstA expression is, by comparison, much less well investigated. Here we define a 39 nucleotide region of ecmA, identified as essential by mutational analysis, that directs strong ecmA-specific expression when multimerised and juxtaposed to heterologous minimal promoter elements. The 39-mer therefore contains all the information necessary for pstA-specific expression. We also identify a transcription factor that binds the 39-mer and that is therefore a potential mediator of extracellular signalling. The protein, MrfA, has strong sequence similarity along most of its length with vertebrate MRF, including a predicted DNA binding domain with similarity to that of the yeast transcription factor Ndt80. Two lines of evidence support a role for MrfA in pstA differentiation.
Mutations in the MrfA binding sites compromise in vivo activity
The three CA-rich sequence tracts, CA1-3, constitute a major proportion of the 39-mer sequence. Simultaneous mutation of all three sequences ablates competition activity in a band-shift assay using recombinant MrfA. The core consensus sequence for binding of Ndt80 to sporulation-specific genes, CRCAAA, has sequence similarity to the CA-rich blocks. In an vivo assay mutation of either CA1 or CA3 alone greatly reduces reporter expression in the pstA region. We suggest therefore that, although the band shift assay implies independent binding of MrfA to the separate sites in vitro, in vivo functioning requires the co-operative interaction of multiple MrfA molecules.
MrfA is essential for full levels of ecmA-specific gene expression
Gene disruption provides further, more direct evidence that MrfA is a key regulator of pstA differentiation. The clearest effect is observed for the multimerised 39-mer reporter construct, where the absence of MrfA eliminates expression. For the −376 construct, there is a very low residual level of anterior expression in the null mutant. This is we surmise the result of partial redundancy with one or more other transcription factors that recognize elements downstream of the 39-mer region. We also suppose that redundancy explains why the full length, ecmAO, promoter is active in pstA cells of the MrfA null mutant. We tested the possibility that DimB might be the partially redundant partner but it is not. There are, however, several other proteins in Dictyostelium with sequence similarity to MrfA, so one of these might be the redundant partner.
The mrfA- strain also displays two morphological phenotypes that may relate to their defective pstA differentiation. The mutant cells aggregate with initially normal kinetics but then later there is a progressive delay in their development. Later still, as the mutant slugs migrate, they continuously divide in two along their length and, although DIF-1 is not the inducer of pstA differentiation, this is also a hallmark of mutants defective in DIF-1 signaling (Huang et al., 2006; Thompson et al., 2004; Zhukovskaya et al., 2006). PstA cell differentiation is, however, believed to be induced by another, as yet unidentified, polyketide (Serafimidis and Kay, 2005). If correct, then the implication is that MrfA is activated by this polyketide and MrfA offers an entry point from which to dissect the cognate signaling pathway.
Finally, we resolve a controversy over the metazoan MRFs. In the recent analysis of pqn-47, the nematode MRF orthologue, strong doubts were expressed as to the validity of the alignment of murine MRF with the DNA binding domain of Ndt80 (Russel et al., 2011). Here, the functional homology of Ndt80 and MrfA was confirmed by mutating the MrfA equivalents of two key residues needed for binding of Ndt80 to the MSE. This negated specific DNA binding in vitro. Thus the MRF-like proteins are by implication confirmed as transcription factors and, in the case of MrfA, defined DNA binding sites are identified in a known target gene This information should help guide future research into the metazoan MRFs.
Materials and Methods
Cell growth and development
D. discoideum Ax2 (Gerisch isolate) was used as the parental strain. Cells were grown in HL5 medium (Watts and Ashworth, 1970) and developed on water or KK2 (20mM K2HPO4/KH2PO4 pH 6.2) agar, or on membrane filters, at a density of 1.0×106 cells/cm2. LacZ staining was performed as described by Zhukovskaya et al., (2006). For neutral red staining cells harvested from growth were suspended in KK2 containing 0.008% neutral red and then washed with KK2 three times.
Promoter analysis
For 5′ deletions the ecmA promoter sequence was PCR amplified using a primer with a BamHI site at the 5′ end, which was paired with a reverse primer containing a start codon and a sequence that added a BglII site at the 3′ end of the PCR product. After digestion with BamHI and BglII, the fragment was cloned into BamHI/BglII-cut Actin15dΔBam:gal vector, replacing the regulatory elements of the actin15 promoter. This deletion in Actin15ΔBam:gal inactivates the promoter but it retains minimal elements that can function when upstream elements from a heterologous promoter are inserted (Ceccarelli, Mahbubani and Williams, 1991; Fukuzawa et al., 2006). For the mutations, nucleotide substitution(s) were introduced into each 5′ primer, with a BamHI cohesive end. Various deleted and mutated promoter constructs were grown in the presence of G418 (50 ug/ml) and developed either on water agar plates or nitrocellulose filters until the slug stage.
Protein purification and mass spectrometry
Nuclear extract derived from 3.5×1011 slug cells was purified by precipitation with 40% (w/v) ammonium sulfate. It was then loaded onto a DNA affinity column bearing an oligonucleotide corresponding to the 39-mer sequence (Fukuzawa et al., 2006). This was synthesized as a duplicate copy, annealed with the complementary strand, multimerized by ligation and coupled to sepharose beads. Bound proteins were eluted with 0.4 M KCl. The eluted proteins were further purified through a second round of binding on the affinity column, concentrated and loaded onto an SDS-polyacrylamide gel. After staining with Colloidal Blue, protein bands were excised from the gel and digested, in-gel, with trypsin. The resulting peptides were analysed by Q-TOF mass spectrometry.
Protein expression, purification and EMSA
The region of mrfA encoding the Ndt80 DNA binding domain orthologous region (amino acids 294-546) was cloned in pET21a(+) NdeI / BamHI site (Novagen, Ltd). It was expressed in E. coli strain BL21 Codonplus RIL (Stratagene, Ltd), as a His-fusion protein, and purified using TALON metal affinity resin (Clontech, Ltd). Point mutations were introduced using the QuikChange mutagenesis kit (Stratagene, Ltd). Band-shift analysis was performed using fluorescent labeled probe with non-labeled competitor (Yamada et al., 2010).
Construction of disruptant strains
The mrfA disruption construct was generated by insertion of a 1.5 Kb blasticidin resistance cassette into the EcoR1 site, positioned at nucleotide 2308 relative to the initiation codon in the mrfA gene, into an mrfA cDNA clone (Fig. 6). A dimB- strain was isolated using a disruption construct bearing hygromycin resistance, just as described by Zhukovskaya et al., (2006). Briefly, the DimB-coding sequences extending from +1 to +1375 was cloned into a plasmid vector and a BamHI hygromycin resistance cassette was inserted at the unique BglII site, positioned at nucleotide 600 relative to the initiation codon. Transformants were isolated clonally and screened for gene disruption by PCR and by RT-PCR. The dimB-/mrfA- double mutant was created by disruption of dimB in the mrfA- strain, again using hygromycin as the selection.
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research from the JSPS (Japan Society for the Promotion of Science) to M. Fukuzawa (no. 21570214) and by Wellcome Trust Program Grant 082579/Z to J. G. Williams.
Abbreviations used in this paper
- ECM
extracellular matrix
- MRF
myelin-gene regulatory factor
- pst
prestalk
References
- BROWN MS, YE J, RAWSON RB, GOLDSTEIN JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- CECCARELLI A, MAHBUBANI H, WILLIAMS JG. Positively and negatively acting signals regulating stalk cell and anterior-like cell differentiation in Dictyostelium. Cell. 1991;65:983–989. doi: 10.1016/0092-8674(91)90550-i. [DOI] [PubMed] [Google Scholar]
- CHU S, DERISI J, EISEN M, MULHOLLAND J, BOTSTEIN D, BROWN PO, HERSKOWITZ I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- DORMANN D, SIEGERT F, WEIJER CJ. Analysis of cell movement during the culmination phase of Dictyostelium development. Development. 1996;122:761–769. doi: 10.1242/dev.122.3.761. [DOI] [PubMed] [Google Scholar]
- EARLY AE, GASKELL MJ, TRAYNOR D, WILLIAMS JG. Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development. 1993;118:353–362. doi: 10.1242/dev.118.2.353. [DOI] [PubMed] [Google Scholar]
- EMERY B, AGALLIU D, CAHOY JD, WATKINS TA, DUGAS JC, MULINY A, WE SB, IBRAHIM A, LIGON KL, ROWITCH DH, BARRES BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FINGERMAN IM, SUTPHEN K, MONTANO SP, GEORGIADIS MM, VERSHON AK. Characterization of critical interactions between Ndt80 and MSE DNA defining a novel family of Ig-fold transcription factors. Nucleic Acids Res. 2004;32:2947–2956. doi: 10.1093/nar/gkh625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FUKUZAWA M, ZHUKOVSKAYA NV, YAMADA Y, ARAKI T, WILLIAMS JG. Regulation of Dictyostelium prestalk-specific gene expression by a SHAQKY family MYB transcription factor. Development. 2006;133:1715–1724. doi: 10.1242/dev.02327. [DOI] [PubMed] [Google Scholar]
- GAUDET P, WILLIAMS JG, FEY P, CHISHOLM RL. An anatomy ontology to represent biological knowledge in Dictyostelium discoideum. BMC Genomics. 2008;9:130. doi: 10.1186/1471-2164-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG EY, BLAGG SL, KELLER T, KATOH M, SHAULSKY G, THOMPSON CRL. bZlP transcription factor interactions regulate DIF responses in Dictyostelium. Development. 2006;133:449–458. doi: 10.1242/dev.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JERMYN KA, TRAYNOR D, WILLIAMS JG. The initiation of basal disc formation in Dictyostelium discoideum is an early event in culmination. Development. 1996;122:753–760. doi: 10.1242/dev.122.3.753. [DOI] [PubMed] [Google Scholar]
- JOLLY ER, CHIN CS, HERSKOWITZ I, LI H. Genome-wide identification of the regulatory targets of a transcription factor using biochemical characterization and computational genomic analysis. BMC Bioinformatics. 2005;6:275. doi: 10.1186/1471-2105-6-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAY RR, THOMPSON CR. Forming patterns in development without morphogen gradients: scattered differentiation and sorting out. Cold Spring Harb Perspect Biol. 2009;1:a001503. doi: 10.1101/cshperspect.a001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAY RR, THOMPSON CR. Cross-induction of cell types in Dictyostelium: evidence that DIF-1 is made by prespore cells. Development. 2001;128:4959–4966. doi: 10.1242/dev.128.24.4959. [DOI] [PubMed] [Google Scholar]
- KELLER T, THOMPSON CR. Cell type specificity of a diffusible inducer is determined by a GATA family transcription factor. Development. 2008;135:1635–1645. doi: 10.1242/dev.020883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUSPA A, LOOMIS WF. Ordered yeast artificial chromosome clones representing the Dictyostelium discoideum genome. Proc Natl Acad Sci USA. 1996;93:5562–5566. doi: 10.1073/pnas.93.11.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMOUREUX JS, STUART D, TSANG R, WU C, GLOVER JN. Structure of the sporulation-specific transcription factor Ndt80 bound to DNA. EMBO J. 2002;21:5721–5732. doi: 10.1093/emboj/cdf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANSTEIN DJ, SCHUSTER HP, MORANDINI P, HUNT DM. Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene. 1995;162:129–134. doi: 10.1016/0378-1119(95)00351-6. [DOI] [PubMed] [Google Scholar]
- RUSSEL S, FRAND AR, RUVKUN G. Regulation of the C. elegans molt by pqn-47. Dev Biol. 2011;360:297–309. doi: 10.1016/j.ydbio.2011.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO T, KATO A, KAY RR. DIF-1 induces the basal disc of the Dictyostelium fruiting body. Dev Biol. 2008;317(2):444–453. doi: 10.1016/j.ydbio.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SERAFIMIDIS I, KAY RR. New prestalk and prespore inducing signals in Dictyostelium. Dev Biol. 2005;282:432–441. doi: 10.1016/j.ydbio.2005.03.023. [DOI] [PubMed] [Google Scholar]
- THOMPSON CRL, FU Q, BUHAY C, KAY RR, SHAULSKY G. A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development. 2004;131:513–523. doi: 10.1242/dev.00939. [DOI] [PubMed] [Google Scholar]
- THOMPSON CRL, KAY RR. The role of DIF-1 signaling in Dictyostelium development. Mol Cell. 2000;6:1509–1514. doi: 10.1016/s1097-2765(00)00147-7. [DOI] [PubMed] [Google Scholar]
- WATTS DJ, ASHWORTH JM. Growth of myxamoebae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J. 1970;119:171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMADA Y, NUNEZ-CORCUERA B, WILLIAMS JG. DIF-1 regulates Dictyostelium basal disc differentiation by inducing the nuclear accumulation of a bZIP transcription factor. Dev Biol. 2011;354:77–86. doi: 10.1016/j.ydbio.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHUKOVSKAYA NV, FUKUZAWA M, YAMADA Y, ARAKI T, WILLIAMS JG. The Dictyostelium bZIP transcription factor DimB regulates prestalk-specific gene expression. Development. 2006;133:439–448. doi: 10.1242/dev.02190. [DOI] [PubMed] [Google Scholar]