Abstract
CudA, a nuclear protein required for Dictyostelium prespore-specific gene expression, binds in vivo to the promoter of the cotC prespore gene. A 14 nucleotide region of the cotC promoter binds CudA in vitro and ECudA, an Entamoeba CudA homologue, also binds to this site. The CudA and ECudA DNA-binding sites contain a dyad and, consistent with a symmetrical binding site, CudA forms a homodimer in the yeast two-hybrid system. Mutation of CudA binding sites within the cotC promoter reduces expression from cotC in prespore cells. The CudA and ECudA proteins share a 120 amino acid core of homology, and clustered point mutations introduced into two highly conserved motifs within the ECudA core region decrease its specific DNA binding in vitro. This region, the presumptive DNA-binding domain, is similar in sequence to domains in two Arabidopsis proteins and one Oryza protein. Significantly, these are the only proteins in the two plant species that contain an SH2 domain. Such a structure, with a DNA-binding domain located upstream of an SH2 domain, suggests that the plant proteins are orthologous to metazoan STATs. Consistent with this notion, the DNA sequence of the CudA half site, GAA, is identical to metazoan STAT half sites, although the relative positions of the two halves of the dyad are reversed. These results define a hitherto unrecognised class of transcription factors and suggest a model for the evolution of STATs and their DNA-binding sites.
Keywords: Dictyostelium, CudA, Amoeboza, Plant STATs, SH2 domains
INTRODUCTION
Upon starvation, individual Dictyostelium amoebae aggregate together in groups of up to 100,000 cells that, at culmination, differentiate into either stalk cells or spore cells. Initially, the aggregating cells form themselves into a smooth hemispherical mound, but then symmetry is broken by the extrusion of an apical tip. This extends, so that the tipped aggregate is transformed into a vertical cylinder of cells: the standing slug or first finger. Under environmental conditions unfavourable for immediate culmination, the standing slug falls onto its side and migrates away, ‘searching’ for a more favourable culmination site.
cudA was discovered in an insertional mutagenesis screen as a gene that regulates the transition from slug migration to culmination (Fukuzawa et al., 1997). CudA is present in the nuclei of the subset of prestalk cells that form the slug tip, the tip organiser region of the slug (http://dictybase.org/Dicty_Info/dicty_anatomy_ontology.html), and is also present in prespore cell nuclei. Its expression in prespore cells utilises an activator element with the potential to direct expression in both the prestalk and prespore cells, and a separate repressor element that prevents expression in prestalk cells (Fukuzawa and Williams, 2000). The prespore repressor and activator proteins have not been identified, but expression in tip cells is regulated by a well-understood signalling pathway. Symmetry is broken when the ACA adenylyl cyclase becomes confined in its expression to the tip region (Verkerke-van Wijk et al., 2001) and the extracellular cAMP that is produced locally activates STATa (Fukuzawa and Williams, 2000; Dormann et al., 2001). STATa then binds to a site in the cudA promoter and induces cudA expression (Fukuzawa and Williams, 2000). Ammonia regulates the choice between slug migration and culmination, and tip-specific cudA expression is also dependent upon the AmtC ammonia transporter (Kirsten et al., 2005).
Although its expression is understood in some detail, and despite it being the only genetically characterised marker for tip cells (Fukuzawa et al., 1997), the function of CudA is unknown. The cudA-null strain is defective in biological functions ascribed to the tip and also in the expression of certain prespore genes, including the cotC spore coat protein gene (Fukuzawa et al., 1997). The fact that CudA is nuclear and that it is required for efficient cotC expression suggest that it might be a transcription factor that directly regulates prespore gene expression. By ChIP analysis, and by defining a binding site in the cotC promoter and showing that the site is essential for optimal expression in prespore cells, we confirm that CudA plays such a role. Combined with a bioinformatic analysis, the results also give insights into the evolution of STAT proteins.
STAT proteins contain an SH2 domain, a DNA-binding domain and a site of tyrosine phosphorylation (Bromberg and Darnell, 2000). In response to receptor activation, STAT monomers become tyrosine phosphorylated and there is a reciprocal interaction between the SH2 domains and phosphotyrosine residues of two STAT molecules that leads to their dimerisation. The dimer translocates to the nucleus where it regulates gene expression. This mode of ‘fast track’ signalling to the nucleus hinges upon the regulatable interaction of an SH2 domain with a specific site of tyrosine phosphorylation. Metazoa utilise this signalling mechanism prolifically; there are well over a hundred SH2 domain-containing proteins in humans (Liu, 2006). By contrast, there are only thirteen SH2 domain-containing proteins in Dictyostelium (Eichinger et al., 2005), two in Arabidopsis and just one in Oryza (Williams and Zvelebil, 2004; Gao et al., 2004). The three plant proteins, Arabidopsis (At) AtSHA and AtSHB and Oryza (Os) OsSHA, are similar in sequence and of unknown function. The SH2 domain is the only readily identifiable domain, but one of the two studies that described these plant proteins and that showed them to be functional (Gao et al., 2004) detected some sequence similarity to the linker region of STAT proteins: the sequence that joins the DNA-binding domain to the SH2 domain. Neither study could discern a DNA-binding domain.
We show that the presumptive DNA-binding domain of CudA has sequence similarity to a domain present in all three plant SH2-domain proteins and that the half site for DNA binding is conserved with metazoan STAT binding sites. This leads us to conclude that plants encode STAT-like proteins and to propose a mechanism for the co-evolution of STATs and their DNA-binding sites.
MATERIALS AND METHODS
Cell culture, development and ChIP analysis
Dictyostelium cells, strain A×2 (Gerisch isolate), were developed to the standing slug/first-finger stage on water agar plates and ChIP experiments were performed as described previously (Zhukovskaya et al., 2006). PCR was performed on immunoprecipitated chromatin or control genomic DNA using the following promoter primers: cotC, 5′-CCCATACTACATTAAAATATTTG-3′ and 5′-TCATATGCTTGTGTGTTGGG-3′ (−657 to −377 relative to the cap site); gpbA, 5′-TAAACAAACACACACCCAAC-3′ and AGGACTTACTAAAATTACAGG (−566 to −281 relative to the ATG initiation codon).
DNA affinity chromatography
Slug nuclear extracts were obtained by sonicating nuclei in DB buffer (50 mM KPO4 pH 7.5, 10% glycerol, 0.5 mM EDTA, 0.1 mM ZnCl2, 0.1 mM MgCl2, 0.01% Brij 35) containing 0.1 M NaCl, 2 mM benzamidine hydrochloride, complete protein inhibitor cocktail, 10 mM NaF and 1 mM sodium pyrophosphate. Wild-type cotC 4×14-mer sequence element (5′-gatcTGAGAATTTTCTATTGAGAATTTTCTATTGAGAATTTTCTATTGAGAATTTTCTAT-3′; the gatc overhang at the 5′ end of the oligonucleotides is added to allow labelling) or its indicated mutated form was concatemarised and coupled to CNBr-Sepharose 4B. The slug nuclear extract was precleared with blocked CNBr-Sepharose 4B and then incubated with Sepharose 4B bearing oligonucleotide. After washing with DB buffer containing 0.1 M NaCl, bound protein was extracted with DB buffer containing 0.4 M NaCl. Protein was concentrated by precipitating with 13% TCA and analysed by western blot using anti-CudA antibody.
Generation of recombinant ECudA protein and band shift analysis
The entire ECudA coding region was amplified using primers 5′-TCATATGAATAATAATACACCACTTTCTGTTAC-3′ and 5′-TGGATCCTTAAATTCTTTGTGTTTGAGGAAGTG-3′. A histidine-tagged ECudA fusion construct in pET15b (Novagen) was expressed and purified over TALON metal affinity resin (BD Biosciences). Band shift analysis was performed as described previously (Kawata et al., 1996). Oligonucleotides were labelled either with [α-32P]dATP (6000 Ci/mmol) or with Cy5-dCTP (Amersham). Gels bearing products with a Cy5-dCTP-labelled probe were scanned at 700 nm wavelength using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Yeast two-hybrid analysis
The reporter yeast strain EGY48 carrying pSH18-34 [LexAop x4-lacZ] was transformed with the different baits cloned in pEG202, and also with the CudA prey cloned in pJG4-5 (Russel et al., 1996). Each of the cloned transformants was tested by monitoring β-galactosidase expression using either galactose-containing (Gal+) medium (SD/Glu/−Ura/−His/−Trp +X-Gal +BU salts) or control (Gal−) medium (SD/Gal/Raf/−Ura/−His/−Trp +X-Gal +BU salts) to induce expression of the baits (Russel et al., 1996).
RESULTS
CudA is bound at the cotC promoter in vivo
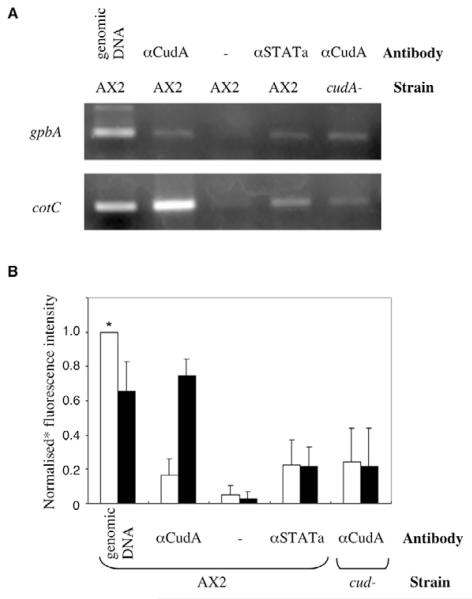
The Dictyostelium cotC spore coat protein gene shows reduced levels of expression in cudA-null slugs (Fukuzawa et al., 1997). In order to determine whether CudA acts directly, as a transcription factor that binds to the cotC promoter, we performed ChIP analysis on extracts from slug stage cells. The PCR analysis was performed using primers for cotC and gbpA, a reference gene used to normalise the data (Fig. 1A,B). When ChIP was performed using the CudA monoclonal antibody, the cotC gene was enriched relative to gbpA. Using an anti-STATa monoclonal antibody, there was no enrichment. When ChIP was performed using the CudA monoclonal antibody in a cudA-null strain, there was also no enrichment. Thus, CudA is bound at the promoter of the cotC gene.
Fig. 1. CudA binding to the cotC promoter as determined by ChIP analysis.
(A) Chromatin from A×2 and cudA-null (cudA−) Dictyostelium cells was immunoprecipitated with or without anti-CudA (aCudA) or anti-STATa (aSTATa) antibody and analysed by PCR. (B) Fluorescence intensities from three ChIP experiments performed as above were normalised against the sample marked with an asterisk; bars indicate ±s.d.
A binding site for ECudA in the cotC promoter is an interrupted dyad
In order to identify CudA binding sites within the cotC promoter, we selected the region from −549 to −319, which has previously been shown to direct prespore-specific gene expression (Powell-Coffman et al., 1994). This was divided into three sub-regions, A-C, that were synthesised as oligonucleotides (Fig. 2A). The oligonucleotides were used in affinity chromatography with slug cell extracts and only oligonucleotide B consistently bound CudA at high (0.4 M) salt concentrations (data not shown), providing preliminary evidence for a specific binding site. However, repeated attempts to map the CudA binding site in oligonucleotide B, by band shift analysis of slug nuclear extracts, were unsuccessful; a similar spectrum of retarded products was obtained using parental and cudA-null extracts. This is presumably owing to the presence of one or more CudA homologues, as described below, that share a similar DNA-binding site with CudA.
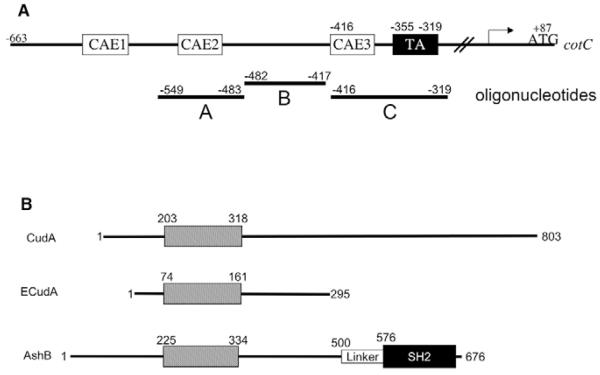
Fig. 2. Promoter structure of cotC and domain structure of CudA and CudA-like proteins.
(A) Schematic of the Dictyostelium cotC promoter. The positions of the three oligonucleotides (A, B and C), the three CA-rich elements (CAEs; binding sequences for the transcription factor GBF) and the regulatory TA-rich region are indicated. (B) Domain structures of Dictyostelium CudA, Entamoeba ECudA and Arabidopsis AtSHB.
We attempted to circumvent the problem by expressing CudA as a fusion protein in E. coli, but four different N-terminally tagged CudA fusion proteins all proved inactive in DNA binding. The conserved domain of the CudA family extends from amino acids 201 to 318 of CudA (Fig. 2B, Fig. 9A) and the constructs were MBP (maltose binding protein)-CudAaa1-802, GST (glutathione S-transferase)-CudAaa1-486, His (oligo-histidine)-CudAaa112-383 and His-CudAaa195-326; this last construct encodes a protein approximately the size of the conserved domain. We therefore turned to the much smaller Entamoeba homologue ECudA (Fig. 2B, Fig. 9A), which is only 284 aa in length.
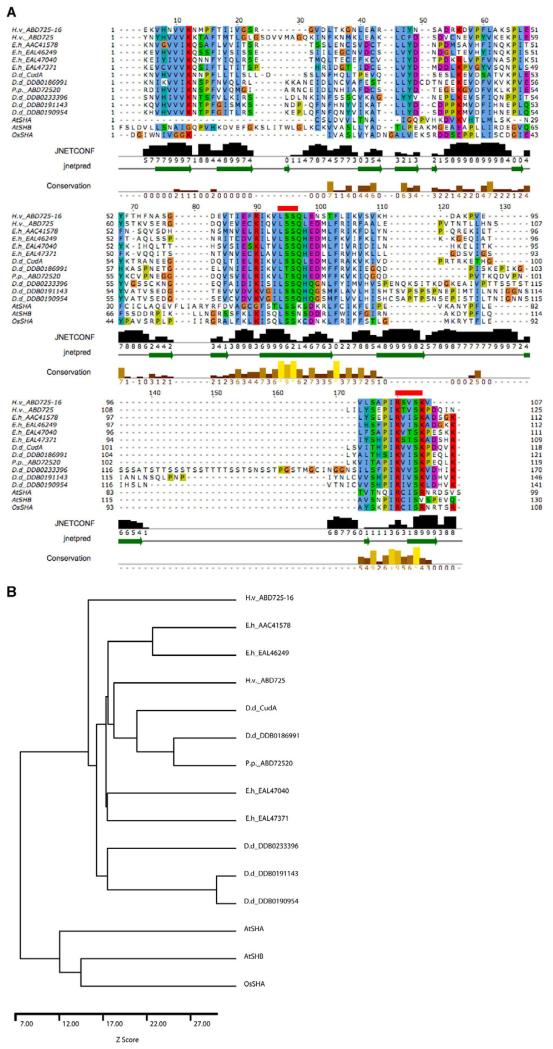
Fig. 9. CudA homologues and the evolution of STAT proteins.
(A) The core region of the Dictyostelium CudA protein compared with related proteins. Amoebozoan species of origin: D.d., Dictyostelium discoideum; E.h., Entamoeba histolytica; H.v., Hartmanella vermiformis; P.p., Physarum polycephalum. The alignment of the core domain of CudA with proteins of similar sequence was produced by AMPS (Barton, 1994), followed by manual adjustment in Jalview (Clamp et al., 2004). Accession numbers are shown only for those proteins for which no publication is available; ECudA is AAC41578. The alignment is coloured according to the ClustalX colour scheme (Thompson, et al., 1997). Residues are coloured by their physico-chemical properties as well as by how frequently they occur at each position. Thus, residues are only coloured if they show similarity to a notional ‘consensus’. Negatively charged residues are in purple; hydrophobic residues in blue; positively charged residues in red; and polar residues in green. Since glycine and proline have special properties, they are separately coloured in orange and mustard, respectively. The secondary structure prediction produced by JPred/JNet (Cuff and Barton, 2000) is shown below the alignment. The green arrows within the ‘jnetpred’ line represent predicted β-strands. The bar chart and numbers labelled ‘JNETCONF’ show the prediction confidence on a scale of 0-9. The ‘Conservation’ line highlights positions in the alignment where the physico-chemical properties of the amino acids are most highly conserved. The two red lines above the sequence show the positions of the mutations introduced to assess the importance of the two regions in DNA binding. (B) Dendrogram for the sequences shown in A. The sequences were compared pairwise and a Z-score calculated from 100 randomisations using AMPS (Barton and Sternberg, 1987).
A His-tagged form of the ECudA protein was used in a band shift assay with oligonucleotide B as the probe. This yielded a strong retardation product that was absent from the band shift performed using a control E. coli extract (Fig. 3) and that was competed by unlabelled probe. It should be noted that when increasing amounts of unlabelled oligonucleotide B were added as competitor, there was an increase in the mobility of the band shift product that paralleled the loss of probe binding. We suggest that binding of the specific competitor has two effects: direct competition that prevents binding of the probe and an added, continuing effect of transient competitor binding that causes a conformational change resulting in increased mobility of the complex. This latter effect can be explained by theoretical studies of gel retardation which posit a ‘cage effect’, whereby weakly interacting ligands are maintained in close proximity to the binding protein during the run (Cann, 1989).
Fig. 3. Analysis of in vitro binding of ECudA to oligonucleotide B.
A band shift analysis was performed using a control E. coli extract (−) and an extract of E. coli cells expressing ECudA (+), with oligonucleotide B as the probe and with the indicated amounts of unlabelled oligonucleotide B as competitor.
In order to map the ECudA binding site, a mutant scanning set was generated in which sequential blocks of six nucleotides, within the 66 nucleotides of oligonucleotide B, were replaced with the presumptively neutral sequence GCGCGC (Fig. 4). These mutant forms, M1-M11, were used as competitors in a band shift assay with oligonucleotide B as probe. Only scanning mutants M7 and M8 showed a strongly retarded band. Contained within this 12 nucleotide region is an interrupted dyad, AGAATTTTCT.
Fig. 4. Scanning mutation analysis of in vitro binding of ECudA to oligonucleotide B.
Scanning mutation analysis of ECudA binding was performed as described in the text using oligonucleotide B. The sequence of the inserted mutation was GCGCGC. Each of the 11 mutant forms was used as competitor in a band shift assay with oligonucleotide B as the probe.
Mutational analysis of the dyad element
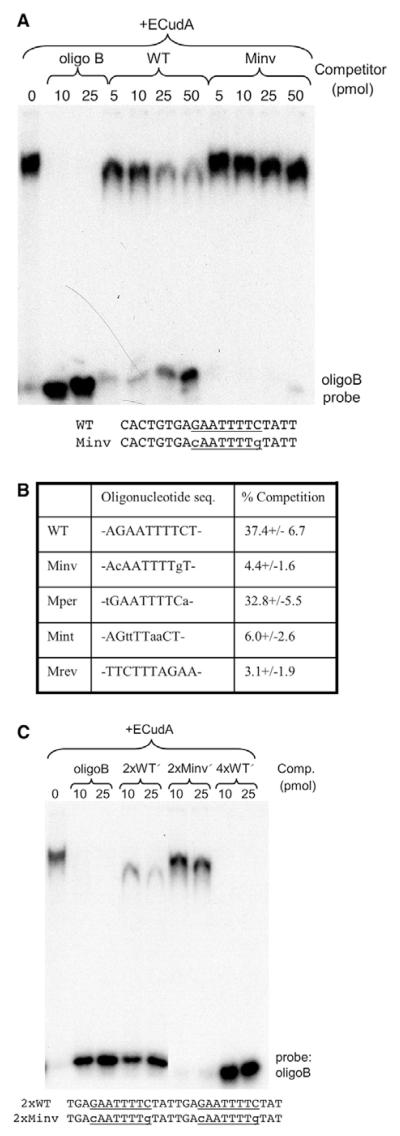
We first analysed the dyad within the context of the 20-mer that encompasses it. The unmutated 20-mer (WT) was a less effective competitor than oligonucleotide B, but there was significant competition when amounts of competitor greater than 25 pmol were added (Fig. 5A). The Minv mutant form of the 20-mer, in which the positions of the G and C residues in the dyad are swapped, was a much less effective competitor than the WT 20-mer (Fig. 5A).
Fig. 5. Mutational analysis of ECudA binding to the cotC dyad.
(A) The WT 20-mer competitor oligonucleotide (CACTGTGAGAATTTTCTATT) encompasses the interrupted dyad (underlined); the C and G within the dyad sequence are swapped in the Minv competitor. WT and Minv were used in band shift assays with oligonucleotide B and ECudA. The assays were performed using Cy5-labelled probes and the intensity of the unshifted probe bands and of the shifted (retarded) bands was measured. (B) The competition efficiency with 100 pmol of competitor is calculated as the ratio, in percentage terms, of the retarded signal to the total (retarded + unretarded) signal. This is a compilation of data from three experiments and the percentage is expressed ±s.d. (C) Point mutation analysis of ECudA binding to multimeric forms of the dyad using the dimeric competitor oligonucleotides, WT′ and Minv′, that contain tandem repeats of the 14-mer encompassing the dyad sequence. A tetramer of the WT′ form (4×WT′) was also used that contains two tandemly arrayed copies of the dimer.
Fig. 5B presents compiled data, for 100 pmol of competitor, comparing WT, Minv and several other mutant forms. When the A and T residues at the periphery of the dyad are mutated, to yield Mper, there was no effect on competition efficiency. By contrast, mutating the internal AA and TT residues, to yield Mint, greatly reduced competition efficiency. We also determined the effect of reversing the relative positions of the two arms of the dyad, to yield Mrev. This greatly reduced competition activity. In conjunction, these experiments mapped the ECudA binding site to the central eight nucleotides of the dyad and suggest that the half site is GAA.
Transcription factors often show co-operative binding to their targets (e.g. John et al., 1999), so we analysed multimeric forms of the dyad. These comprised direct repeats of the 14-mer sequence that encompasses the dyad (Fig. 5C). The 2×WT′ oligonucleotide was a significantly more effective competitor than WT, and a 4×WT′ oligonucleotide was as effective as oligonucleotide B (Fig. 5C). These data imply a co-operative interaction. Again, swapping the G and C residues within the dyad, in 2×Minv′, greatly reduced its effectiveness as a competitor of ECudA binding.
CudA also binds selectively to the ECudA dimer
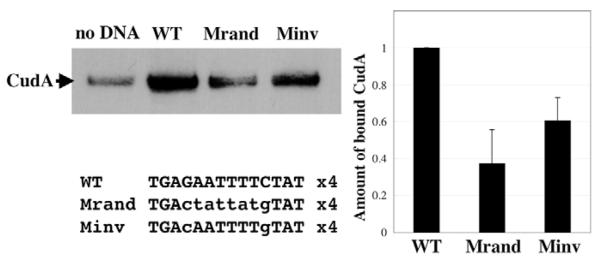
Having identified a ECudA binding site within cotC promoter oligonucleotide B, we tested whether the same site would bind CudA itself, by performing affinity chromatography and western transfer analysis. Three affinity matrices were used, each bearing a different 4-fold multimerised, 14-mer sequence. These were the WT and the Minv sequences, described above, and Mrand, a mutant form of WT in which the sequence was randomly scrambled. We also performed the binding reaction using control matrix with no DNA (Fig. 6). There was a certain level of CudA binding to the control matrix, but a 4- to 5-fold higher level of binding to the matrix bearing multimerised WT DNA. This binding was significantly reduced when the relative positions of the G and C residues were reversed, as in Minv. Binding was reduced even further when the entire sequence was randomised, as in Mrand. There was a finite level of CudA binding to the randomised DNA, presumably because CudA, like many transcription factors, has a relatively low degree of specificity to its target sequence but achieves specificity by associating with other transcription factors bound to other sites. We conclude that the dyad, defined using ECudA, also forms part or all of a CudA binding site.
Fig. 6. Demonstration that CudA binds to the ECudA dyad element.
CudA purified from Dictyostelium nuclear extracts using wild-type and mutant versions of the 4-fold multimerised 14-mer element, defined using ECudA, was analysed by western blotting. Non-specific binding was determined in a parallel purification using beads (no DNA). Results from three experiments are compiled in the bar chart. The amount of bound CudA is shown as amount bound, after subtracting non-specific binding and relative to the WT sequence with the indicated s.d. values.
Yeast two-hybrid analysis suggests that CudA forms a constitutive homodimer
The fact that ECudA and CudA bind to a site that contains an interrupted dyad would be readily explicable if the CudA protein family form homodimers. In order to determine whether CudA has this capacity, it was inserted into a yeast two-hybrid ‘bait’ vector and also into a compatible ‘prey’ vector. The read-out for interaction in the two-hybrid system utilised is β-galactosidase activity and, when bait and prey were both present, the resultant yeast colony stained strongly with X-Gal (Fig. 7). Secondary modifications, such as phosphorylation, are unlikely to occur correctly in a yeast cell. Hence, CudA appears to have the intrinsic ability to form a homodimer. This conclusion is supported by the chromatographic behaviour of ECudA, which runs at the size expected for a dimer (S. Cameron and W. Hunter, personal communication). It is also of course possible that, within the context of a Dictyostelium cell, CudA forms heterodimers with other CudA family members.
Fig. 7. Yeast two-hybrid analysis of CudA homodimerisation.
The CudA prey is constitutively expressed and in the presence of galactose as an inducer (in Gal+ medium) the bait is also expressed. The test bait is CudA and the STATa SH2 domain is used as a negative control bait. Because there is an interaction between bait and prey, i.e. homodimerisation of CudA, lacZ expression is activated and is detected with X-Gal. Staining was for 3 days at 30°C. The same bait and prey combinations were tested by growth selection using a leu marker and gave identical results (data not shown).
The dyad element is necessary for efficient expression from cotC in prespore cells
The biological function of the dyad element was tested by fusing cotC promoter fragments to the lacZ gene, transforming the constructs into parental and cudA-null cells and analysing β-galactosidase expression at the slug stage. All constructs share the same coding-region-proximal fusion point with lacZ (see legend to Fig. 8A), but are differently deleted distally. The informative constructs contain varying portions of the 41 nucleotide, 3′ proximal segment of oligonucleotide B, i.e. the region that contains the dyad (Fig. 4).
Fig. 8. Mutational analysis of the cotC promoter.
(A) Deletion analysis. A 5′ to 3′ deletion series with a start point at nucleotide −659 (relative to the cap site) was constructed and fused to the lacZ gene at a point 30 nucleotides downstream from the cotC initiation codon. The constructs were used to prepare stable transformants in A×2 and cudA-null cells and analysed as pooled populations. Note the shorter staining time used for the −659 construct. (B) Point mutation analysis. Site-directed mutagenesis was used to generate a construct with a 5′ end point at nucleotide −457 and with a structure similar to that described in A. This construct, and the control wild-type construct, were analysed as in A but only in A×2 cells.
A construct with a 5′ end point at −619 was strongly expressed in the prespore zone of parental cells but deletion to −457 reduced expression (Fig. 8A). Comparison of the signals for parental and cudA-null cells shows that expression of both these constructs is strongly dependent upon CudA. The next deletion end point, just 16 nucleotides downstream from −457 at −441, showed a greatly reduced level of expression. Again, the low level of expression that was observed with the −441 construct was decreased even further in the cudA-null strain. This deletion, to −441, removes the distal half of the dyad, but there is another half site, very near the 3′ end of oligonucleotide B, that could be interacting with CudA (Fig. 8A). When all three half sites were removed, by deletion to −416, there was no detectable level of expression.
The effect of less drastic manipulations was determined by introducing point mutations into all three potential half sites within the context of the −457 construct (Fig. 8B). The effect of the mutations was to greatly reduce the level of prespore expression. The fact that the point mutant construct retained more activity than the deletion construct could have a number of explanations, including: a low level residual activity of the point mutated sites; other, cryptic binding sites within the 41-mer; or ‘junctional inhibition’ brought about when the plasmid sequences upstream of the 5′ end points of the −457 and −441 constructs inhibit the activity of cryptic CudA binding sites downstream of −441.
A domain that has sequence similarity with the CudA core is present in proteins from other amoebozoans and several plants
Having ascribed a function to CudA, we performed a bioinformatic analysis. The Dictyostelium genome sequence is essentially complete (Eichinger et al., 2005) and it is possible to identify within it six genes that share a region of very high similarity with cudA. This ‘core’ region is, in different homologues, between 120 and 180 aa in length. Outside the core region there is very little sequence similarity between the seven Dictyostelium genes (data not shown). We extended the search to other databases. Fig. 9A shows an alignment of CudA with 14 representative sequences found after iterative searching with PSI-BLAST and iSCANPS (G.J.B., unpublished) of the NCBI and UniRef databases. The 14 sequences include two from Arabidopsis and one from Oryza. The plant proteins were found with E-values close to 1.0 in the third iteration of PSI-BLAST against the NCBI nr protein database. A dendrogram for the 14 sequences is shown in Fig. 9B. The plant proteins cluster with the CudA family of sequences with a Z-score of over 7σ, strongly suggesting that the proteins share a similar three-dimensional structure. The searches also revealed a further two fragment sequences, one from Hartmanella and one from Acanthamoeba, but these are omitted for clarity. When the reciprocal search was performed, using the domain from Oryza as the search sequence, the same group of proteins was detected.
The sequences show similarity over their entire length, but there are three regions of strong conservation in all but the AtSHA sequence, which is missing the N-terminus. Positions 6-21 include a hydrophobic VVV/I motif suggestive of a β-strand. Positions 85-109 include a conserved LSS motif, and 174-189 contain a basic [R/K]IVSK motif. Quantitative comparisons derived from the BLAST scores, pairwise comparisons and clustering, indicate that the similarity between CudA and the best-conserved sequences from other species is higher than with most of the other Dictyostelium sequences (Fig. 9B). This is consistent with a family of cudA-like genes that pre-dates the divergence of the amoebozoa. The notion of a relatively ancient domain is further supported by the existence of the plant proteins. All three plant proteins contain the CudA core domain similarity in the same, approximately central position. All three also contain an SH2 domain, with an associated linker region, located near their C-termini (Gao et al., 2004; Williams and Zvelebil, 2004), i.e. the presumptive DNA-binding, linker and SH2 domains are in the same relative positions as in the STAT proteins (Fig. 2B).
The conserved core in ECudA is necessary for efficient DNA binding
The fact that the core region is the only significant region of homology between CudA and ECudA, coupled with the fact that both proteins bind to similar DNA-binding sites, strongly imply that the core is the DNA-binding site. In order to test this supposition directly, we inserted clustered mutations into two of the most highly conserved regions in the core domain of ECudA (Fig. 9A). In ECudA:LSS, the conserved sequence LSS (141-143) is mutated to AAA; in ECudA:RVISK, the conserved sequence RVISK (173-177) is mutated to AAAAA. These proteins were expressed in E. coli, as His-tagged species, in parallel with unmutated His-tagged ECudA. The proteins were purified by metal resin affinity chromatography, the amount of each fusion protein was quantitated by SDS-PAGE and equivalent amounts of fusion protein were used in band shifts with the 4×14-mer (i.e. the 4×WT′) probe (Fig. 10). There was a higher level of binding to ECudA than to either mutant form: the average relative decrease was almost 60% for the LSS mutant and almost 90% for the RVISK mutant. This further strengthens the notion that the core is the DNA-binding region and also bolsters the validity of the alignments (Fig. 9A).
Fig. 10. Mutational evidence that the ECudA core region is the DNA-binding domain.
His-tagged ECudA, either wild-type (WT) or mutated in the two positions indicated in Fig. 9A (LSS, RVISK) and described in the text, was analysed by band shift using the 4-fold multimerised 14-mer element as a probe. In the bar chart, the averaged results of four experiments are quantitated and presented with the s.d.
DISCUSSION
The CudA-cotC interaction is, to our knowledge, the first example of Dictyostelium cell type-specific gene expression mediated by a form of regulation that commonly underlies pattern formation in higher organisms, whereby, the expression of a transcription factor in a particular embryonic region leads to the specific activation of its target genes. Early development in Drosophila is a paradigm for such transcription factor cascades, and here we face the same challenge of identifying first causes, i.e. in order to understand how CudA activates cotC gene expression selectively in prespore cells, we need to understand how CudA itself becomes localised in prespore nuclei. This will require identification of the activator and repressor proteins that confer localisation in the prespore cells (Fukuzawa and Williams, 2000) and determination of the mechanisms that regulate their expression.
Another important goal will be to determine how CudA is able to regulate different gene sets in two very different cell types: the prespore cells and the tip cells. In the case of prespore-specific gene expression there is a significant body of relevant information. The null mutant for CudA expresses cotC at a reduced but finite level (Fukuzawa et al., 1997); thus, CudA would appear to be an ancillary regulator of cotC gene expression. The cotC promoter (Fig. 2A) contains binding sites for GBF, a non-cell-type-specific transcriptional regulator (Schnitzler et al., 1994). These sites, the CA-rich elements (CAEs), have been proposed to act in synergy with an AT-rich element to direct prespore-specific gene expression (Powell-Coffman et al., 1994). This same configuration of sites is present in several other prespore-specific promoters (Powell-Coffman and Firtel, 1994; Powell-Coffman et al., 1994), but the protein or proteins that bind the AT-rich element are unknown. There are no CAEs mapped to oligonucleotide B, but the adjacent fragment, oligonucleotide C, contains a CAE and a TA element (Fig. 2A). Hence, there is the potential for an interaction with CudA.
The core region of CudA is, we conclude, a previously unrecognised kind of DNA-binding domain. The evidence for this derives in part from comparing the Dictyostelium and Entamoeba data. CudA is a specific DNA-binding protein: it is bound to the cotC gene in vivo, it binds in vitro to a specific region of the cotC promoter and point mutations at this site reduce prespore-specific expression of lacZ reporter constructs in vivo. CudA and ECudA are closely related functionally because they bind the same site within the cotC promoter. The fact that the only significant similarity between CudA and ECudA is in the core domain strongly implies, therefore, that the core domain is the DNA-binding domain. This was directly tested by mutagenesis of two of the most highly conserved regions within the core: both mutations reduced specific DNA binding.
Proteins containing a region of similarity to the CudA core domain are present in five amoebozoan species and there are two homologues in Arabidopsis and one in Oryza. These are also the only genes in either plant species to possess an SH2 domain. At the time of their identification, analogies were drawn with the STATs, but these were inconclusive because it was not possible to discern sequence similarity with STAT DNA-binding domains (Williams and Zvelebil, 2004; Gao et al., 2004). The presence of a homologue to a DNA-binding domain upstream of an SH2 domain in the three plant proteins suggests very strongly that they function similarly to STATs, but this does not necessarily indicate a direct evolutionary relatedness; there could be ‘mix and match’ gene evolution, with a DNA-binding domain that is unrelated to the STAT DNA-binding domain becoming juxtaposed to an SH2 domain.
Is the presumptive CudA DNA-binding domain structurally related to the STAT DNA-binding domain? Profile-profile alignment methods are currently the most sensitive method to detect weak similarity between two protein families (Ohlson and Elofsson, 2005). However, comparison of an alignment of STAT DNA-binding domain sequences with the CudA alignment shown in Fig. 9A using the program PRC (http://supfam.mrc-lmb.cam.ac.uk/PRC/) failed to show a significant match. Although there is no detectable significant sequence similarity between the CudA sequences and the STATs, the secondary structure prediction of the CudA-like domains (Fig. 9A) suggests that the domain has a predominantly β-sheet architecture. This is also true for STAT DNA-binding domains (Chen et al., 1998; Becker et al., 1998; Soler-Lopez et al., 2004). Fold recognition searches by 3D-PSSM (Kelley et al., 2000) predominantly identify all-β-domains, but did not show the STAT DNA-binding domain as a significant hit.
Although the above analyses yield no clear evidence to identify CudA as a STAT DNA-binding domain fold, functional evidence in favour of CudA and STAT DNA-binding domains being structurally related derives from the sequence of the ECudA binding site. The binding site for ECudA in oligonucleotide B has as its half site the sequence GAA. This is identical to the half site sequence of the GAS element, the dyad sequence that is bound by STAT dimers in the majority of STAT-regulated promoters. The GAS half sites are separated by between two and four nucleotides (Seidel et al., 1995) and the ECudA half sites are separated by two nucleotides. The radical difference between the GAS and the ECudA dyads is in the relative positioning of the two arms of the dyad: whereas the GAS site reads TTCnGAA, the ECudA dyad reads GAAnTTC.
Because of their shared specificity, we suggest that the STAT and CudA DNA-binding domains derive from a common ancestor, which we will term the GAA-binding protein (Fig. 11). In Dictyostelium, these two classes of descendant from the common ancestor survive and STATa, at least, retains the ability to bind GAA half sites when present as either a direct repeat or an inverted repeat (Kawata et al., 1997; Fukuzawa and Williams, 2000). We propose that the ancestral form of the GAA-binding protein was, like CudA, a constitutive dimer that bound the sequence GAAnTTC. We assume that the ancestral STAT arose from it by the recruitment of an SH2 domain and a site of tyrosine phosphorylation (Fig. 11). This allowed for regulatable dimerisation, but the transition must also have been accompanied by a shift in the relative positions of the two halves of the dyad. The fact that STATa can recognise both direct and inverted GAA repeats might be a reflection of this ancestral flexibility.
Fig. 11. A model for the evolution of STATs and their DNA binding sites.
The model proposes an ancestral, constitutively dimeric protein (GAA). It has a GAA-binding half site and binds DNA co-operatively. This latter feature favoured the evolution of tandem, dyad-binding sites of the form (GAAnTTC)n. The junctions between these sites (shown at A) constitute potential binding sites for a protein recognising the reversed order sequence TTCnGAA. STAT proteins are proposed to have arisen, by the recruitment of a site of tyrosine phosphorylation (Y) and an SH2 domain (SH2), to bind such junctional sequences.
There is an implicit ‘chicken and egg’ conundrum in this notion: how could the configuration of half sites recognised by STAT dimers evolve prior to the appearance of STAT proteins? We suggest that the answer lies in the binding behaviour of CudA dimers. Just as with STAT proteins and STAT binding sites (John et al., 1999; Vinkemeier et al., 1998), the presence of multiple copies of the CudA dyad in a target sequence increases CudA binding dramatically (Fig. 5C). This could provide an evolutionary pressure favouring multiple adjacent sites. If two CudA dyad elements with the same relative orientation became very closely apposed on a promoter then, at the interface between the two, there would exist a dyad with the present-day GAS site configuration (Fig. 11). We propose that STAT proteins evolved to utilise such sites.
Acknowledgments
We thank Dr Graham Clark for generously providing Entamoeba histolytica DNA, Scott Cameron and Bill Hunter for permission to cite their unpublished result on CudA dimerisation, and the Wellcome Trust (Programme Grant 053640/Z/98 and Project Grant 078971/Z/05/Z) for funding the work.
References
- Barton GJ. The AMPS package for multiple protein sequence alignment. Methods Mol. Biol. 1994;5:327–347. doi: 10.1385/0-89603-276-0:327. [DOI] [PubMed] [Google Scholar]
- Barton GJ, Sternberg MJ. A strategy for the rapid multiple alignment of protein sequences. Confidence levels from tertiary structure comparisons. J. Mol. Biol. 1987;198:327–337. doi: 10.1016/0022-2836(87)90316-0. [DOI] [PubMed] [Google Scholar]
- Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- Bromberg J, Darnell JE., Jr The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- Cann JR. Phenomenological theory of gel electrophoresis of protein-nucleic acid complexes. J. Biol. Chem. 1989;264:17032–17040. [PubMed] [Google Scholar]
- Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Dormann D, Abe T, Weijer CJ, Williams J. Inducible nuclear translocation of a STAT protein in Dictyostelium prespore cells: implications for morphogenesis and cell-type regulation. Development. 2001;128:1081–1088. doi: 10.1242/dev.128.7.1081. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glöckner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa M, Williams JG. Analysis of the promoter of the cudA gene reveals novel mechanisms of Dictyostelium cell type differentiation. Development. 2000;127:2705–2713. doi: 10.1242/dev.127.12.2705. [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Hopper N, Williams J. cudA: a Dictyostelium gene with pleiotropic effects on cellular differentiation and slug behaviour. Development. 1997;124:2719–2728. doi: 10.1242/dev.124.14.2719. [DOI] [PubMed] [Google Scholar]
- Gao Q, Hua J, Kimura R, Head JJ, Fu X, Chin YE. Identification of the linker-SH2 domain of STAT as the origin of the SH2 domain using two-dimensional structural alignment. Mol. Cell. Proteomics. 2004;3:704–714. doi: 10.1074/mcp.M300131-MCP200. [DOI] [PubMed] [Google Scholar]
- John S, Vinkemeier U, Soldaini E, Darnell JE, Jr, Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Mol. Cell. Biol. 1999;19:1910–1918. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata T, Early A, Williams J. Evidence that a combined activator-repressor protein regulates Dictyostelium stalk cell differentiation. EMBO J. 1996;15:3085–3092. [PMC free article] [PubMed] [Google Scholar]
- Kawata T, Shevchenko A, Fukuzawa M, Jermyn KA, Totty NF, Zhukovskaya NV, Sterling AE, Mann M, Williams JG. SH2 signaling in a lower eukaryote: A STAT protein that regulates stalk cell differentiation in Dictyostelium. Cell. 1997;89:909–916. doi: 10.1016/s0092-8674(00)80276-7. [DOI] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Kirsten JH, Xiong Y, Dunbar AJ, Rai M, Singleton CK. Ammonium transporter C of Dictyostelium discoideum is required for correct prestalk gene expression and for regulating the choice between slug migration and culmination. Dev. Biol. 2005;287:146–156. doi: 10.1016/j.ydbio.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol. Cell. 2006;22:851–868. doi: 10.1016/j.molcel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Ohlson T, Elofsson A. ProfNet, a method to derive profile-profile alignment scoring functions that improves the alignments of distantly related proteins. BMC Bioinformatics. 2005;6:253. doi: 10.1186/1471-2105-6-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell-Coffman JA, Firtel RA. Characterization of a novel Dictyostelium discoideum prespore-specific gene, PspB, reveals conserved regulatory sequences. Development. 1994;120:1601–1611. doi: 10.1242/dev.120.6.1601. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman JA, Schnitzler GR, Firtel RA. A GBF-binding site and a novel AT element define the minimal sequences sufficient to direct prespore-specific expression in Dictyostelium discoideum. Mol. Cell. Biol. 1994;14:5840–5849. doi: 10.1128/mcb.14.9.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel L, Finley JR, Brent R. DNA Cloning-Expression Systems. Oxford University Press; Oxford, UK: 1996. [Google Scholar]
- Schnitzler GR, Fischer WH, Firtel RA. Cloning and characterization of the G-box binding factor, an essential component of the developmental switch between early and late development in Dictyostelium. Genes Dev. 1994;8:502–514. doi: 10.1101/gad.8.4.502. [DOI] [PubMed] [Google Scholar]
- Seidel HM, Milocco LH, Lamb P, Darnell JE, Jr, Stein RB, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc. Natl. Acad. Sci. USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler-Lopez M, Petosa C, Fukuzawa M, Ravelli R, Williams JG, Muller CW. Structure of an activated Dictyostelium STAT in its DNA-unbound form. Mol. Cell. 2004;13:791–804. doi: 10.1016/s1097-2765(04)00130-3. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;15:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerke-van Wijk I, Fukuzawa M, Devreotes PN, Schaap P. Adenylyl cyclase A expression is tip-specific in Dictyostelium slugs and directs StatA nuclear translocation and CudA gene expression. Dev. Biol. 2001;234:151–160. doi: 10.1006/dbio.2001.0232. [DOI] [PubMed] [Google Scholar]
- Vinkemeier U, Moarefi I, Darnell JE, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- Williams JG, Zvelebil M. SH2 domains in plants imply new signalling scenarios. Trends Plant Sci. 2004;9:161–163. doi: 10.1016/j.tplants.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Zhukovskaya NV, Fukuzawa M, Yamada Y, Araki T, Williams JG. The Dictyostelium bZIP transcription factor DimB regulates prestalk-specific gene expression. Development. 2006;133:439–448. doi: 10.1242/dev.02190. [DOI] [PubMed] [Google Scholar]