Abstract
G protein-coupled receptors (GPCRs) can modulate diverse signaling pathways, often in a ligand-specific manner. The full range of functionally relevant GPCR conformations is poorly understood. Here we use NMR spectroscopy to characterize the conformational dynamics of the transmembrane core of the β2-adrenergic receptor (β2AR), a prototypical GPCR. We labeled β2AR with 13CH3ε-methionine and obtained HSQC spectra of unliganded receptor as well as receptor bound to an inverse agonist, an agonist, and a G protein-mimetic nanobody. These studies provide evidence for conformational states not observed in crystal structures, as well as substantial conformational heterogeneity in agonist- and inverse-agonist-bound preparations. They also show that for β2AR, unlike rhodopsin, an agonist alone does not stabilize a fully active conformation, suggesting that the conformational link between the agonist-binding pocket and the G-protein-coupling surface is not rigid. The observed heterogeneity may be important for β2AR’s ability to engage multiple signaling and regulatory proteins.
Classification: Biological sciences, Biophysics and Computational biology, Pharmacology
Introduction
When first characterized in the 1970s, G protein-coupled receptors (GPCRs) were often viewed as binary signaling proteins. Simple two-state models, with inactive (R) and active (R*) states in equilibrium, could describe most of their known behaviors. However, functional and pharmacological studies over the subsequent three decades have revealed many GPCRs to be very versatile signaling proteins that can modulate the activity of more than one second messenger system, often in a ligand-specific manner. These observations suggest that GPCRs are dynamic proteins that assume multiple distinct conformations depending on the bound ligand, the associated signaling proteins, and the membrane environment (Deupi and Kobilka, 2010; Yao et al., 2009). Subsequent biophysical and biochemical studies provided some direct evidence for multiple conformational states for specific GPCRs (Altenbach et al., 2008; Liu et al., 2012; Yao et al., 2006), and more recent progress in GPCR structural biology has provided the first high-resolution pictures of GPCRs in both inactive and active states (Rasmussen et al., 2011a; Rasmussen et al., 2011b; Rosenbaum et al., 2007). These crystal structures represent single snapshots of just a few conformational states, however, and relatively little is known about what other conformational states these receptors can adopt, how dynamic each conformational state is, or how different conformational states are stabilized by binding of ligands and associated signaling proteins. This uncertainty poses an obstacle to structure-based drug design for GPCR targets.
The β2AR is an ideal model system for investigating the role of GPCR protein dynamics in signaling. The β2AR activates more than one G protein, and signals through at least one G protein-independent pathway, arrestin (Rajagopal et al., 2005) (Fig. 1A). There is a rich diversity of available ligands for the β2AR. These ligands are often characterized as inverse agonists that suppress basal activity, full agonists that maximally activate the receptor, partial agonists that produce submaximal activity even at saturating concentrations, and neutral antagonists that occupy the orthosteric binding site but do not affect basal activity (Rosenbaum et al., 2009) (Fig. 1B). To complicate matters further, the efficacy of a ligand may depend on the down-stream signaling pathway used to quantify activity. For example, carvedilol is an inverse agonist for the β2AR activation of Gs, but a partial agonist for β2AR activation of arrestin (Wisler et al., 2007). Finally, the β2AR is the only GPCR that has been crystallized in both inactive and G protein-coupled conformations (Rasmussen et al., 2011b; Rosenbaum et al., 2007).
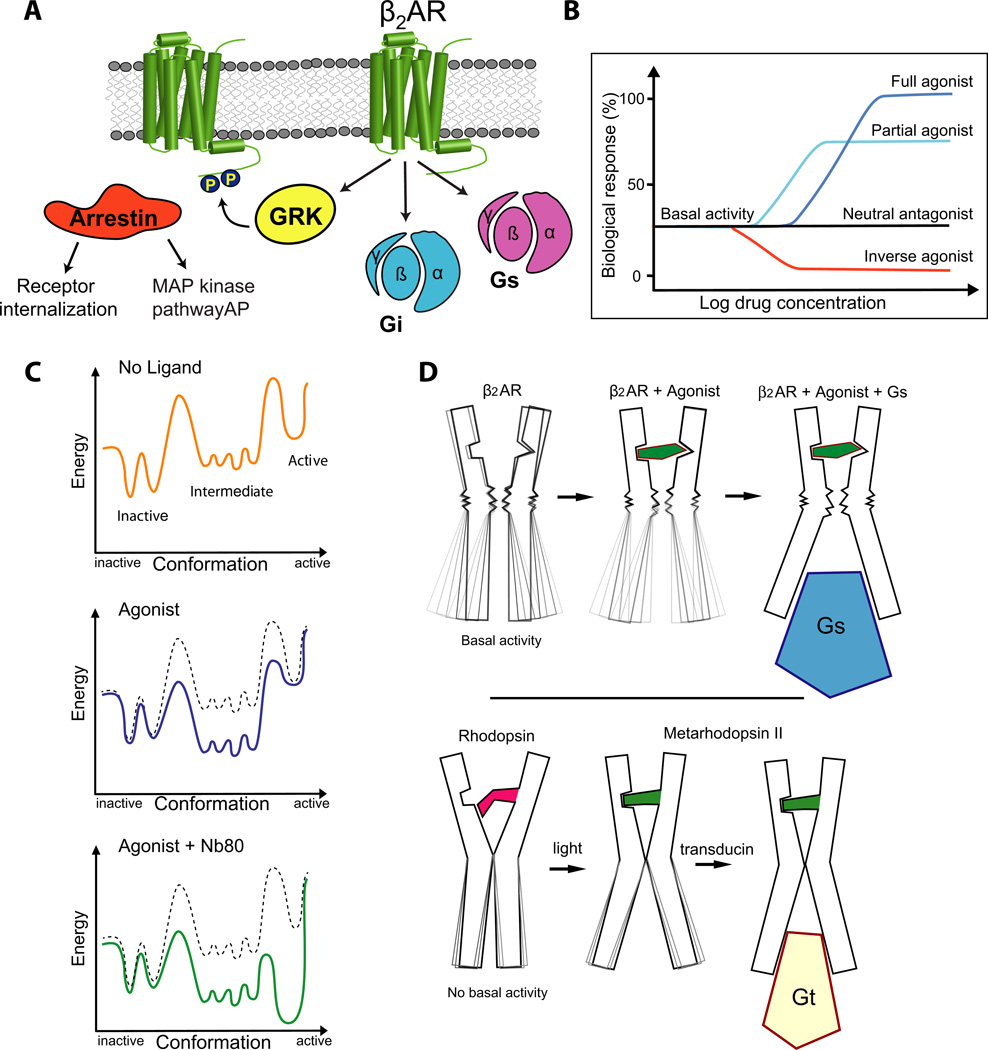
Figure 1. The β2AR is a versatile signaling protein.
A) The β2AR interacts with several signaling and regulatory proteins in an agonist-dependent manner. The β2AR can activate the heterotrimeric G proteins Gs and Gi. G-protein coupled receptor kinases (GRKs) phosphorylate the agonist-bound receptor, which can subsequently bind to arrestin and be either internalized or signal through the MAP kinase and other pathways.
B) The β2AR exhibits basal agonist-independent activation of Gs. Drugs that can suppress basal activity are called inverse agonists (for example carazolol). Neutral antagonists (for example alprenolol) can block binding of other ligands, but don’t impose any biological response. Agonists can be divided into two categories: full agonists (for example BI-167107) and partial agonists. Full agonists produce the full biological response, whereas partial agonists can only produce a partial biological response even at saturating concentrations. These properties are independent of ligand affinity.
C) Schematic free energy landscapes illustrating the energy of the receptor along the activation pathway. The top, middle, and bottom panels show the energy landscape with no ligand bound, with agonist bound, and with both agonist and nanobody 80 (Nb80) bound, respectively. The middle and bottom panels also show the unliganded landscape as a dashed line for comparison.
D) The NMR experiments show that agonist binding to the β2AR does not fully stabilize the active conformation suggesting a relatively weak conformational link between the agonist binding pocket and the G protein coupling surface.
Results from crystal structures of Metarhodopsin II and double electron-electron resonance spectroscopy suggest that covalently bound trans-retinal can stabilize the active state of the G protein coupling surface.
The dynamic behavior of proteins is well recognized, but often difficult to characterize. Proteins exhibit small-scale movements at the level of amino acid side-chains and larger-scale movements between domains, on timescales ranging from picoseconds to seconds (Henzler-Wildman and Kern, 2007). As a result, proteins exist in many conformations, each with a distinct energy resulting from intramolecular chemical bonds and non-covalent interactions. Overall, the thermodynamically most favorable conformations predominate. The most stable conformations correspond to those structures likely to be captured in X-ray crystallography. Thus, our view of protein structure is biased towards these stable conformations. However, the less stable (i.e. less populated) conformations may also be important for function (Hansen et al., 2008).
Protein dynamics are often described in terms of a free energy landscape (Fig. 1C), where the energy is plotted as a function of conformation. The depths of the energy wells determine the relative populations of the various conformational states at equilibrium, and the heights of the barriers determine the rates of transitions between states (Deupi and Kobilka, 2010). By employing nuclear magnetic resonance (NMR) spectroscopy to study dynamic properties of the β2AR, we can begin to understand its energy landscape. In this paper, we present evidence for several conformational states that have not been observed crystallographically. In particular, our studies reveal an alternative inactive conformational state, which exchanges slowly with the crystallographically observed inactive state in the presence of inverse agonists or in the absence of a ligand. Our studies also demonstrate that a receptor bound to an agonist alone is structurally heterogeneous and that the most populated conformation differs from both the inactive conformational states that dominate in inverse-agonist-bound and unliganded receptors and the active states that dominate when the receptor is bound to both an agonist and a G protein mimetic nanobody (Fig. 1D). We use molecular dynamics simulations to provide a structural framework in which to interpret these results. For the purpose of illustration and discussion, we will describe the conformational ensembles induced by different ligands using the simple energy landscapes shown in Fig. 1C.
Results and Discussion
Application of NMR spectroscopy and MD simulations to characterize conformational dynamics in the β2AR
NMR spectroscopy has been used to study the structure and dynamics of a range of proteins. Specific labeling of methyl groups, such as the ε-methyl of methionine (13CH3ε-Met), has been shown to be an effective method for studying large molecules by NMR (Beatty et al., 1996; Bose-Basu et al., 2004; DellaVecchia et al., 2007). 13CH3ε-Mets are ideal NMR probes for high molecular weight proteins because the length and flexibility of the methionine side-chain compensates for the slow tumbling of large proteins, improving the resolution and sensitivity of the spectra (Tugarinov et al., 2003).
13CH3ε-Mets are also excellent NMR probes for studying conformational changes in the β2AR. The β2AR has 9 methionine residues (not counting the N-terminal methionine, which is removed by signal peptidase) dispersed throughout its primary sequence, many of which are found in the transmembrane region of the receptor (Fig. 2A and Fig. S1A). This region has not been accessible to study using fluorescence spectroscopy or fluorine NMR, which require the addition of small molecule probes to surface-exposed cysteines (Liu et al., 2012; Yao et al., 2006).
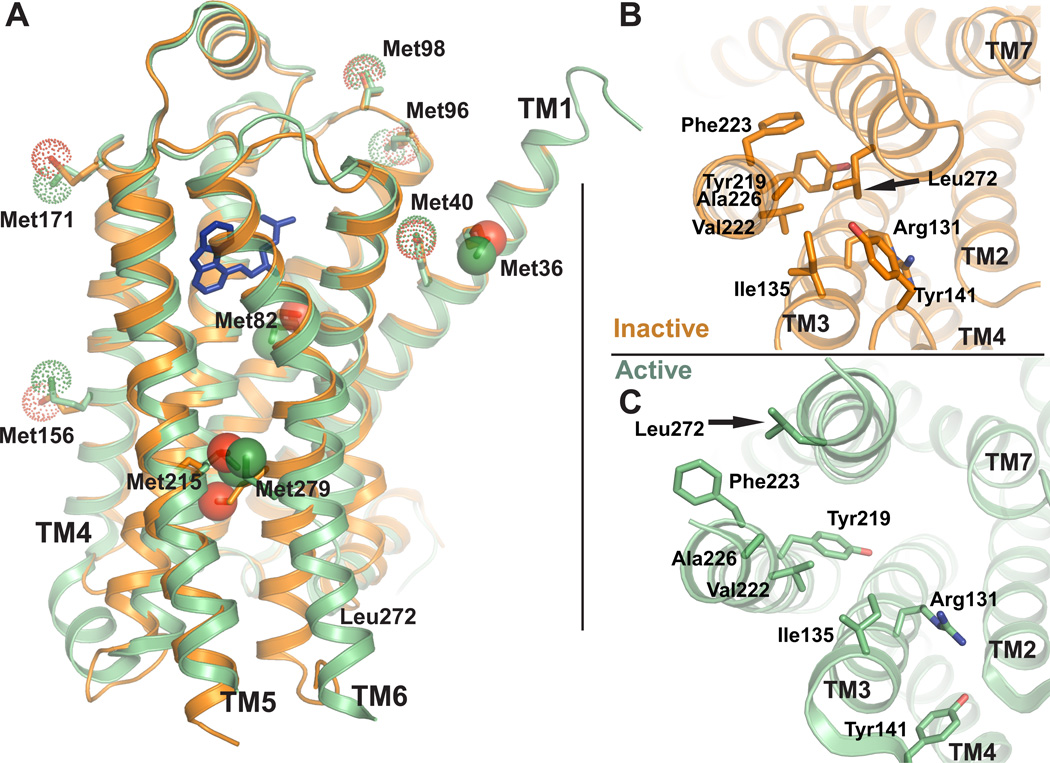
Figure 2. Positions of methionines in active and inactive β2AR structures.
A) Methionine residues shown as sticks in the active and inactive β2AR crystal structures. Solid spheres represent the methionine methyl carbons left in β2AR-Δ5, whereas dotted spheres represent methyls of methionines mutated to other residues in β2AR-Δ5. The functional properties of β2AR-Δ5 are similar to those of wild-type β2AR (See also Fig. S1). B) Structure of β2AR in the crystallographic inactive conformation (2RH1) seen from the intracellular side. Leu2726.34 and surrounding hydrophobic residues are shown as sticks. C) Structure of β2AR in the crystallographic active conformation (3P0G) seen from the intracellular side.
The location of a peak (also known as the chemical shift or resonance) representing a 13C-methyl group in an NMR spectrum is dependent on the microenvironment of the methyl in the protein. In proteins, the protons and carbon of 13C-methyls experience a wide variety of chemical environments depending on local and global protein structure and on solvent exposure (Butterfoss et al., 2010; Wishart, 2011). As a result, the position of an NMR peak is very sensitive to changes in protein conformation or protein-protein interactions. NMR is also sensitive to molecular motion; if a methyl group is in conformational exchange between two structurally distinct states, the appearance of the peak is dependent on the timescale at which this exchange occurs (Mittermaier and Kay, 2009). If the exchange between the two states is slow (several milliseconds or more), one observes two distinct peaks representing the two different environments, whereas if the exchange is fast (nanosecond–microsecond) one observes a peak at the weighted average of the two chemical shifts. For methyl groups that exchange conformations in the intermediate time regime (microseconds–milliseconds), the corresponding peaks will be very weak or not visible (Mittermaier and Kay, 2009). Therefore NMR spectra can provide information about changes in protein structure as well as the timescales over which the changes occur.
To provide a structural framework for interpreting the results of our NMR experiments, we utilized atomic-level long timescale MD simulations of the β2AR. We analyzed an extensive set of simulations—including some reported previously (Dror et al., 2011a; Dror et al., 2011b) and others performed especially for this purpose—with a focus on specific receptor regions and dynamic properties probed by the NMR experiments. These simulations were performed on Anton (Shaw et al., 2009), a special-purpose computer designed to accelerate classical MD simulations by orders of magnitude, allowing them to describe conformational changes on much longer timescales than previously possible.
Modified β2ARs for NMR experiments
To study specific regions of the β2AR we prepared a modified β2AR (β2AR-Δ5M) by making the following mutations: M40L, M96T, M98T, M156L, M171L. These methionines, which face the exterior of the receptor (Fig 2A), were mutated to simplify the spectrum and eliminate signals from 13CH3ε-Mets that do not undergo structural changes when comparing active and inactive structures. Antagonist and agonist binding affinities were determined for β2AR-Δ5M and other modified receptors described below based on saturation and competition binding experiments (See Fig. S1B, C, Table S1). We also determined that β2AR-Δ5M couples efficiently to the G protein Gs as determined by a GTPγS binding assay (Fig. S1D). We retained Met361.35 at the extracellular end of TM1 as a reference since it is facing outward in a region of the protein that does not undergo conformational changes upon activation. The other three methionines that we retained—Met822.53, Met2155.54 and Met2796.41—are especially interesting with regards to ligand binding and signal transduction (Fig. 2A and Fig. S1A). Met822.53 is found just below the ligand-binding pocket in TM2 (Fig. 2A), and is within 4 Å of several amino acids that directly interact with both agonists and antagonists. Met822.53 is therefore sensitive to changes in the chemical environment around the ligand-binding pocket. Met2155.54 and Met2796.41 are located between the ligand-binding pocket and the cytoplasmic ends of TM5 and TM6 respectively (Fig. 2A). The cytoplasmic ends of TM5 and TM6 undergo relatively large structural changes to accommodate binding to Gs and to Nb80 (Fig 2A).
To enable characterization of the dynamic features at the cytoplasmic end of TM6, we also created a L272M mutation in β2AR-Δ5M (β2AR-Δ5M-L272M). In the inactive-state crystal structure of the β2AR, Leu2726.34 packs into a pocket formed by residues in TM5 and TM3 (Fig. 2B), whereas in active-state structures, Leu2726.34 is solvent exposed (Fig. 2C) and does not interact with either Nb80 or Gs. The L272A mutation is known to lead to constitutive activity (Samama et al., 1993) and structural instability (Gether et al., 1997), but we expected that Met would be a better structural substitute for Leu, causing little constitutive activity. Indeed the L272M mutation in β2AR-Δ5M resulted in only a modest (4 fold) increase in agonist binding affinity (Fig. S1B, Table S1), while the L272A mutation in wild type β2AR leads to an approximately 20 fold increase in binding affinity (Samama et al., 1993). The agonist binding affinity for β2AR-Δ5M-L272M is indistinguishable from the unmodified β2AR, and exhibits none of the biochemical instability associated with the constitutively active mutant L272A (Gether et al., 1997).
β2AR-Δ5M and β2AR-Δ5M-L272M were expressed in methionine-deficient media supplemented with 13CH3ε-methyl labeled methionine (13CH3ε-Met) and purified to homogeneity (see Experimental Procedures). The 13CH3ε-Met resonances from β2AR-Δ5M and β2AR-Δ5M-L272M appear in a region of the cHSQC spectrum that is free of signals from buffer, detergent or unlabeled β2AR protein (Fig. S2). We assigned methionines 36, 82, 215 and 279 in β2AR-Δ5M by mutagenesis (Fig. S3). 13CH3ε-Met NMR spectroscopy was recently used to investigate the effect of agonists and partial agonists on the structure of the β2AR, and the spectral assignment of specific 13CH3ε-Met resonances is in agreement with ours (Kofuku et al., 2012).
Ligand-specific changes in cHSQC spectra of the β2AR
Figures 3 and 4 show the assigned cHSQC spectra of 13CH3ε-Met-β2AR-Δ5M and 13CH3ε-Met-β2AR-Δ5M-L272M under the following conditions: unliganded, bound to the inverse agonist carazolol, bound to the high-affinity agonist BI-167107, and bound to both BI-167107 and the G protein mimetic nanobody Nb80. Both carazolol and BI-167107 have very long dissociation half-lives (Rasmussen et al., 2011a; Rasmussen et al., 2007). For NMR experiments, these ligands were added in a 10-fold stoichiometric excess over receptor at concentrations that far exceed their Kd/Ki values. In studies done with purified β2AR-Δ5M under these conditions, the agonist BI-167107 has a dissociation halflife of 403 min and an association half-life of less than 4.4 min (Fig. S1D). We therefore do not expect to observe conformational heterogeneity due to ligand dissociation and re-association. All spectra were obtained at room temperature. We used Nb80 as a substitute for the G protein, because the substantially smaller size of Nb80 leads to higher resolution in the resulting NMR spectra (large complexes tumble more slowly, leading to line broadening). Recent crystal structures have shown that the receptor conformations stabilized by Nb80 and the G protein (Gs) are very similar (Rasmussen et al., 2011a; Rasmussen et al., 2011b).
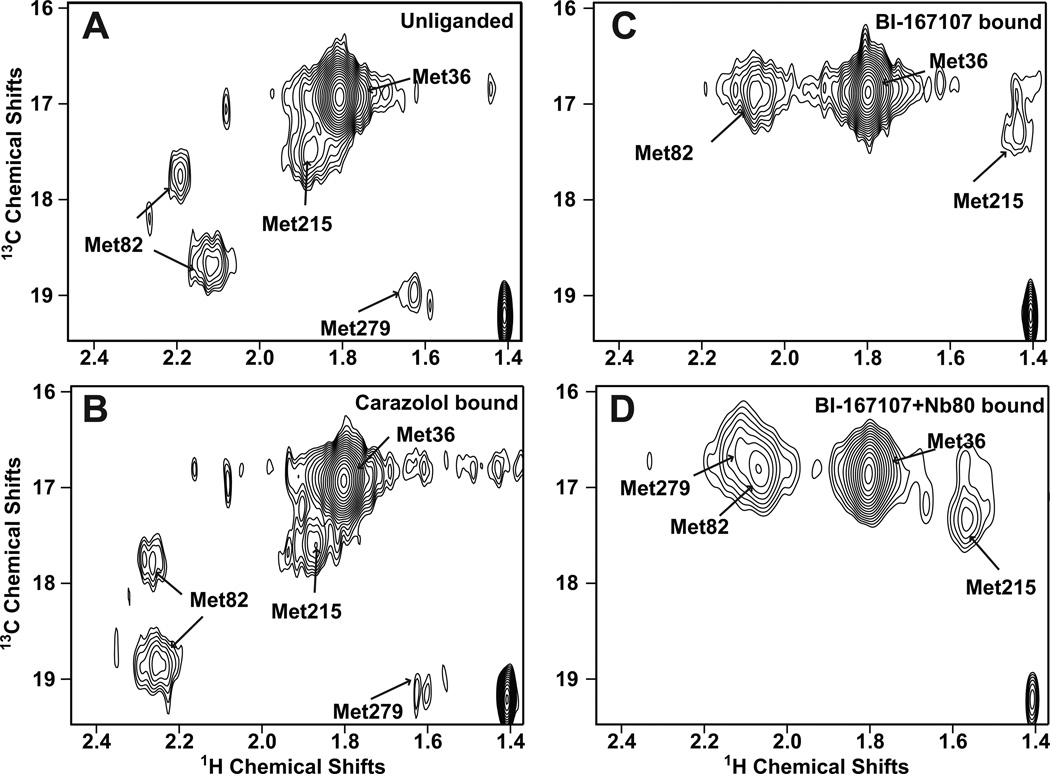
Figure 3. Ligand-specific effects on the HSQC spectrum of 13C-Met-β2AR-Δ5.
Carbon-HSQC of spectra of β2AR-Δ5 were obtained under the following conditions: A) unliganded, B) bound to the inverse agonist carazolol, C) bound to the agonist BI-167107, and D) bound to BI-167107 and the G protein mimetic Nb80. In contrast to the complex spectra of unmodified β2AR containing nine methionines (See Fig. S2), resonances for individual methionines are clearly distinguishable in the spectra for β2AR-Δ5. Assignments of 13C-Met resonances in β2AR-Δ5 were made by obtaining spectra of β2AR-Δ5+M36L, β2AR-Δ5+M82V and β2AR-Δ5+M215I (See Fig. S3). Spectra A–C were recorded at room temperature on a 900 MHz Bruker spectrometer. Spectrum D was recorded at room temperature on an 800 MHz Varian spectrometer. See Table S2 for details about acquisition of NMR spectra. In all spectra, a peak at 1.4 ppm [1H] and 19.2 ppm [13C] is observed, we expect this peak represents non-methionine methyl groups in the receptor that are observed because of the natural abundance 13C in the sample.
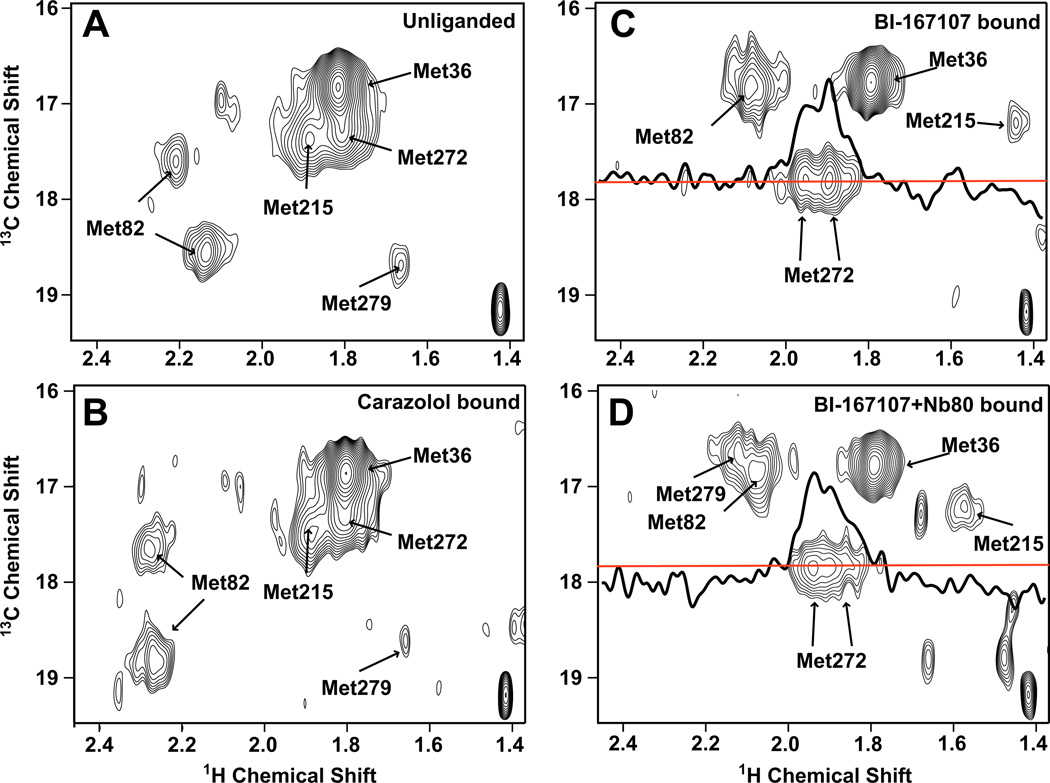
Figure 4. Ligand-specific effects on the HSQC spectrum of 13C-Met-β2AR-Δ5-L272M.
Carbon-HSQC spectra of β2AR-Δ5-L272M were obtained under the following conditions: A) unliganded, B) bound to the inverse agonist carazolol, C) bound to the agonist BI-167107, and D) bound to BI-167107 and the G protein mimetic Nb80. Spectra were recorded at room temperature on an 800 MHz Varian spectrometer (see Table S2 for details about acquisition of NMR spectra). For the spectra with BI-167107 and BI-167107 plus Nb80 bound a 1D slice illustrates the splitting of the Met2726.34. The red line represents the carbon chemical shift where this 1D slice was taken.
The peaks observed in our 2-D spectra are relatively broad and irregularly shaped. This is due in part to the large size of β2AR-Δ5M together with its detergent micelle, but may also reflect inherent conformational heterogeneity.
The Met361.35 peak is the most intense peak in all spectra (Fig. 3 and Fig. S3A). This was expected, as Met361.35 is the most solvent-exposed methionine in β2AR-Δ5M and β2AR-Δ5M-L272M. Inactive- and active-state β2AR crystal structures show that TM1 does not undergo a major conformational change during activation, so it is not surprising that we observe no ligand-specific change in the chemical shift for Met361.35. Met361.35 therefore serves as a reference for the remaining 13CH3-Met resonances.
Unliganded and inverse-agonist-bound β2AR are conformationally heterogeneous
With the exception of Met822.53, each of the methionines present in β2AR-Δ5M corresponds to a single peak in the spectra of the unliganded receptor (Fig. 3 and Fig. S3). The positions of these peaks do not change upon binding to the inverse agonist carazolol, and they thus likely correspond to the inactive state of the receptor (Fig. 3).
In contrast, Met822.53, which is located just below the ligand-binding pocket in TM2 (Fig. 2A), is represented by two peaks of moderate intensity (and possibly a third weak peak) in the unliganded and inverse-agonist-bound samples (Fig. 3 and Fig. S3B). The observation of two peaks from Met822.53 suggests that its 13CH3ε-methyl is detecting at least two conformations that exchange on a millisecond or longer timescale. Molecular dynamics simulations indicate that although the Met822.53 side-chain transitions between different rotameric states, it does so very rapidly, on a nanosecond timescale, which is far faster than the millisecond timescales that would lead to multiple peaks in NMR spectra (Fig. S4). The two peaks thus probably reflect larger structural changes in the receptor.
Several factors might contribute to the presence of dual Met822.53 peaks, including changes to the rotameric state of Trp2866.48 or the protonation state of Asp792.50. Long-timescale molecular dynamics simulations offer an alternative explanation. In certain simulations of the crystallographic inactive state (both with the inverse agonist carazolol bound and with no ligand bound), we observed a transition to an alternative conformation in which the intracellular half of TM7 (Asn3187.45–Cys3277.54, including the conserved NPxxY motif) rotates ~40° clockwise relative to the crystal structure (viewed from the intracellular side) and shifts toward the center of the helical bundle by ~3 Å (Fig. 5A, B). This conformational change would allow the ε-methyl Met822.53 to move into a more hydrophobic environment and increases its distance to the aromatic ring of Trp2866.48 from ~4.5 Å to ~7 Å (Fig. 5C), which would be expected to alter its chemical shift. The conformational change does not alter the chemical environments of the other methionines present in β2AR-Δ5M and β2AR-Δ5M-L272M. Exchange between these two inactive conformations appears to be slow. In roughly 400 µs of unbiased simulation (split roughly evenly between the crystallographic inactive and alternative conformational states), we have observed three transitions from the crystallographic conformation to the alternative conformation, and a single reverse transition. Millisecond-timescale exchange between these two conformations in the NMR experiments seems quite plausible, given that rates estimated by MD simulations often differ from experimentally measured ones by several fold (Dror et al., 2010), and that the receptor is surrounded by lipids in the simulations and detergent in the experiments. To further probe these long-timescale events, we performed temperature-accelerated MD simulations of the carazolol-bound receptor. These simulations exhibited repeated transitions between the crystallographic inactive and alternative conformations, with the crystallographic conformation present in a slightly higher population (Fig. S4D). It should be noted that while the timescale of transitions between inactive states observed in MD simulations is consistent with our NMR results, we cannot be certain that either state is responsible for the observed chemical shifts for M82.
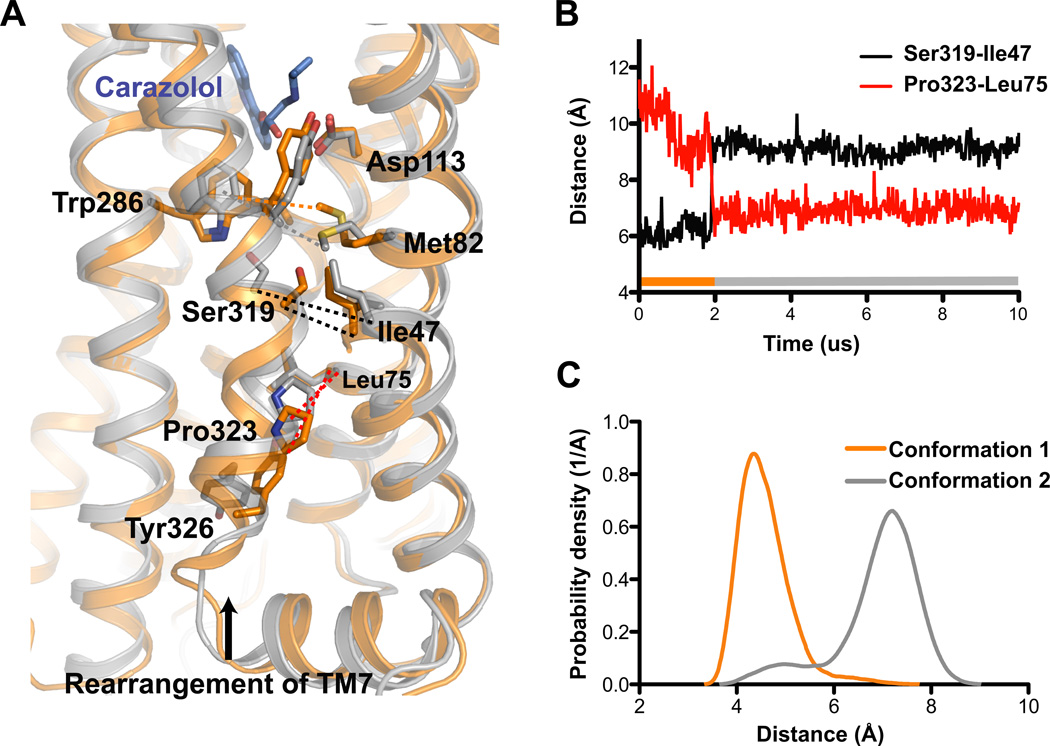
Figure 5. Alternative inactive state observed in unliganded and inverse-agonist-bound MD simulations may explain dual Met822.53 peaks.
A) The transition from the crystallographic inactive conformation (orange, 2RH1) to the alternative inactive conformation (gray, snapshot from MD simulation) involves a rotation of the intracellular part of TM7. Pro3237.50 moves ~4 Å towards TM2 into the space between Asp792.50 and Asn511.50, displacing a conserved water molecule linking these two residues (Pardo et al., 2007), and Tyr3267.53 adopts the trans χ1 rotamer, pointing its phenol hydroxyl down towards the ionic lock. Ser3197.46 shifts ~2 Å towards Trp2866.48 and forms a hydrogen bond, and its displacement opens a small hydrophobic cavity at the interface of TM1, TM2, and TM7 into which Met822.53’s ε-methyl group docks. B) Distances between the alpha-carbon atoms of Ser3197.46 and Ile471.46 (black) and Pro3237.50 and Leu757.46 (red) over the course of a simulation of β2AR which transitions from the crystallographic inactive conformation to the alternative inactive conformation after 1.93 µs. The bar at the bottom of the plot illustrates when we see the crystallographic inactive conformation (orange) and the alternative inactive conformation 2 (gray). C) Distributions of the distance from Cε of Met822.53 to the closest non-hydrogen atom in the aromatic ring of Trp2866.48, plotted for the crystallographic inactive and alternative inactive conformation. See Fig. S4 for additional analysis these two inactive conformations, and Table S3 for additional details about molecular dynamics experiments.
In Fig. 1C, we illustrate the conformational sub-states observed with NMR as a pair of local minima, corresponding to the crystallographic and alternative inactive states, within the larger well representing inactive conformations. The local minima are separated by a relatively large energy barrier that is responsible for the slow exchange observed in NMR experiments. These inactive sub-states are themselves dynamic; within each, for example, our simulations exhibit fast exchange between conformations with and without the TM3–TM6 ionic lock formed (Dror et al., 2009).
Although carazolol binding does not change the relative intensities of the two main Met822.53 peaks, it does induce a small shift in their position relative to the unliganded condition (Fig. 3). The aromatic component of carazolol is relatively far away (7–9 Å) from 13CH3ε-Met822.53 and would thus not be expected to influence its chemical shift (Fig. S4A). The shift of the Met822.53 peaks upon carazolol binding might reflect subtle changes around the ligand-binding pocket propagated to Met822.53 through Trp2866.48 and Tyr3167.43 (Fig. S4C).
Agonists do not fully stabilize an active conformation of the β2AR
Not surprisingly, all peaks apart from the one corresponding to Met361.35 change substantially upon receptor activation, as evidenced by the spectra of receptor bound to both a strong agonist and the nanobody (BI-167107+Nb80) (Fig. 3 and Fig. 4). Under this condition, each methionine has one corresponding peak. The peak representing Met2796.41 is best seen in Fig. S3B, where the overlapping Met822.53 has been mutated, or in Fig. S5, which shows a 1D slice. The peaks are broad and irregular suggesting that when in complex with Nb80 the β2AR exists in predominantly one conformation with substantial dynamic behavior, as indicated by a single broad energy well in Fig. 1C).
When only the agonist BI-167107 is bound (Figs. 3 and 4), the spectra indicate the presence of a receptor conformation distinct from those which dominate either when both BI-167107 and Nb80 are bound, or when an inverse agonist or no ligand are bound. In the presence of BI-167107 alone, the peaks corresponding to Met822.53, just below the binding pocket, and Met2726.34, at the cytoplasmic end of TM6, display only subtle changes from those observed in the BI-167107+Nb80 condition. In contrast, the peaks corresponding to the residues between the binding pocket and G protein coupling regions change substantially: the Met2155.54 peak becomes weaker and shifts upfield, while the Met2796.41 peak disappears from spectra of β2AR-Δ5M. We cannot exclude the possibility that a peak or peaks representing Met2796.41 in the presence of agonist alone may overlap with that for Met822.53. Very weak peaks are observed at 2.07 [1H] and 17.1 ppm [13C] and 2.1 [1H] and 16.5 ppm [13C], in the spectrum of β2AR-Δ5M-M82V (Fig. S3B).
These results indicate that, with only a strong agonist bound, the receptor does not simply populate a mixture of the crystallographically observed active and inactive conformations. Instead, it populates a set of conformations in which the chemical environment of Met822.53 and Met2726.34 is similar to the active conformation of the BI-167107+Nb80 condition, but that of Met2155.54 and possibly Met2796.41 is different. Not only do we observe a different set of conformations around Met2155.54 and Met2796.41 when comparing agonist alone with no ligand and agonist+Nb80, we observed differences in peak intensities (Fig. 6A). These observations can be interpreted using an energy landscape (Fig. 1C). Our results suggest that in the presence of agonist alone, the receptor transitions between several sub-states on an intermediate time scale, thus the weaker signals. Nb80 appears to stabilize a more uniform conformation as suggested by the strength of the NMR signals for Met822.53, Met2155.54 and Met2726.34 (Fig. 6A).
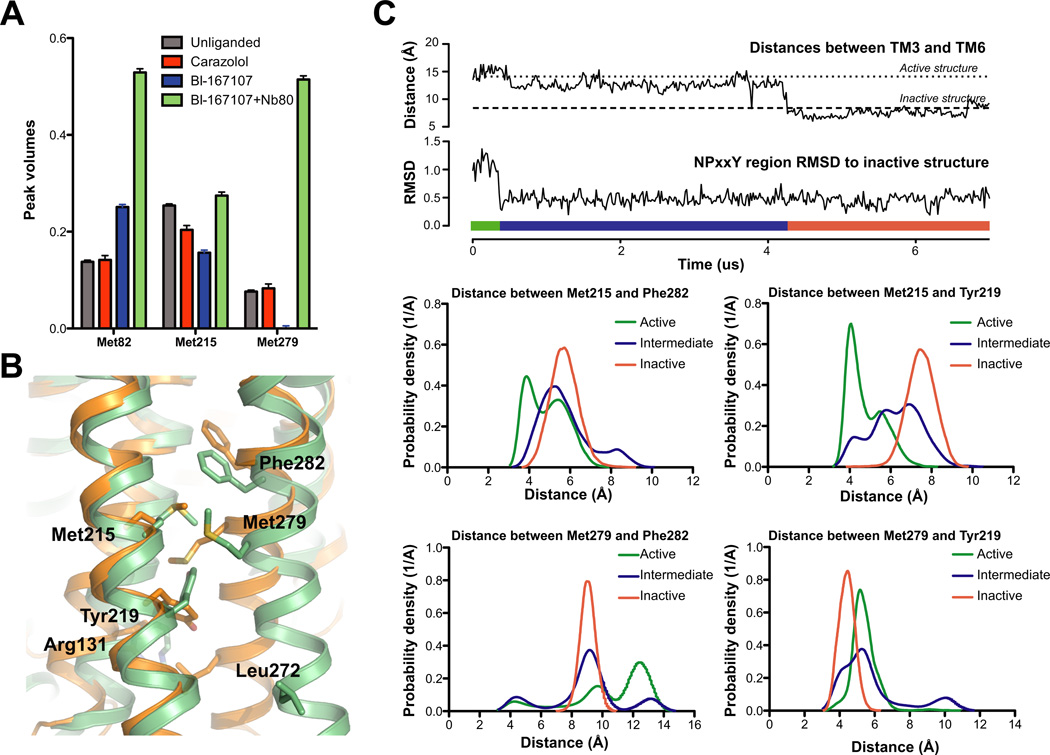
Figure 6. Agonist binding promotes conformational heterogeneity around Met2155.54 and Met2796.41.
A) Peak volumes for individual methionines. Automated 2D line-shape fitting was performed on the spectra using NMRpipe (Delaglio et al., 1995). From the line-shape analysis volumes of the different peaks were extracted. The peak volumes were normalized to the peak volume of Met36. After extracting peak volumes we estimated their uncertainties by evaluating the RMS from the volumes of 10 peaks chosen from regions of the spectrum where no signal could be detected in the 2D spectrum. For Met82 the peak volumes in the carazolol and the unliganded state are averages of peak intensities of the two most intense Met82 peaks. Although NB80 would be expected to reduce tumbling of the receptor in solution and thereby weaken signals, we see signals intensify suggesting a stabilization of the receptor and a more uniform distribution of conformations compared to the agonist bound form (see also Fig. S5).
B) Region around Met2155.54 and Met2796.41 shown for the active (green, 3P0G) and inactive (orange, 2RH1) structures of β2AR. Met2155.54 and Met2796.41 and aromatic residues in the vicinity of Met2155.54 and Met2796.41 are shown as sticks. Arg1313.50 and Leu2726.34 are also shown as sticks.
C) Simulations of β2AR starting in the active conformation with Nb80 removed spontaneously transition back to the inactive conformation. Top two plots illustrate a transition from the active conformation to the inactive conformation during an MD simulation (adapted from Fig. 2 of Dror et al. (2011a)). The transition starts with the re-arrangement of TM7 into its inactive conformation, as illustrated by the plot of TM7’s RMSD to the inactive crystal structure; subsequently TM6 moves inward towards TM3, as illustrated by the plot of distances between TM3 (Cα of Arg1313.50) and TM6 (Cα of Leu2726.34). We call the state between the re-arrangement of TM7 and the inward movement of TM6 the intermediate state. The bar in the bottom of the plot illustrates what part of the simulation is considered the active (green), intermediate (blue) and inactive (orange) state.
The bottom four plots illustrate the distance distributions during the simulation between two NMR probes (Met2155.54 or Met2796.41) and two nearby aromatic residues (Tyr2195.58 or Phe2826.44). The distance distribution of the active conformation is based on simulations of β2AR with BI-167107 and Nb80 bound; the inactive state is based on simulations of the carazolol bound receptor, and the intermediate state is based on simulations with only BI-167107 bound, as illustrated in the two top plots. See Table S3 for additional details about molecular dynamics experiments.
Again, long-timescale simulations can provide a possible structural explanation consistent with this data. As described previously (Dror et al., 2011a), β2AR with BI-167107 bound but without an intracellular binding partner transitions spontaneously from the crystallographic active state to the crystallographic inactive state in simulation. En route, it pauses in an intermediate state in which TM7 adopts a conformation similar to that seen in inactive structures, but TM5 and TM6 do not (Figs. 6 and 7). In this intermediate conformational state, as in the crystallographic state, the intracellular end of TM6 maintains its separation from TM3, but the intracellular ends of TM5 and TM6 display substantial mobility.
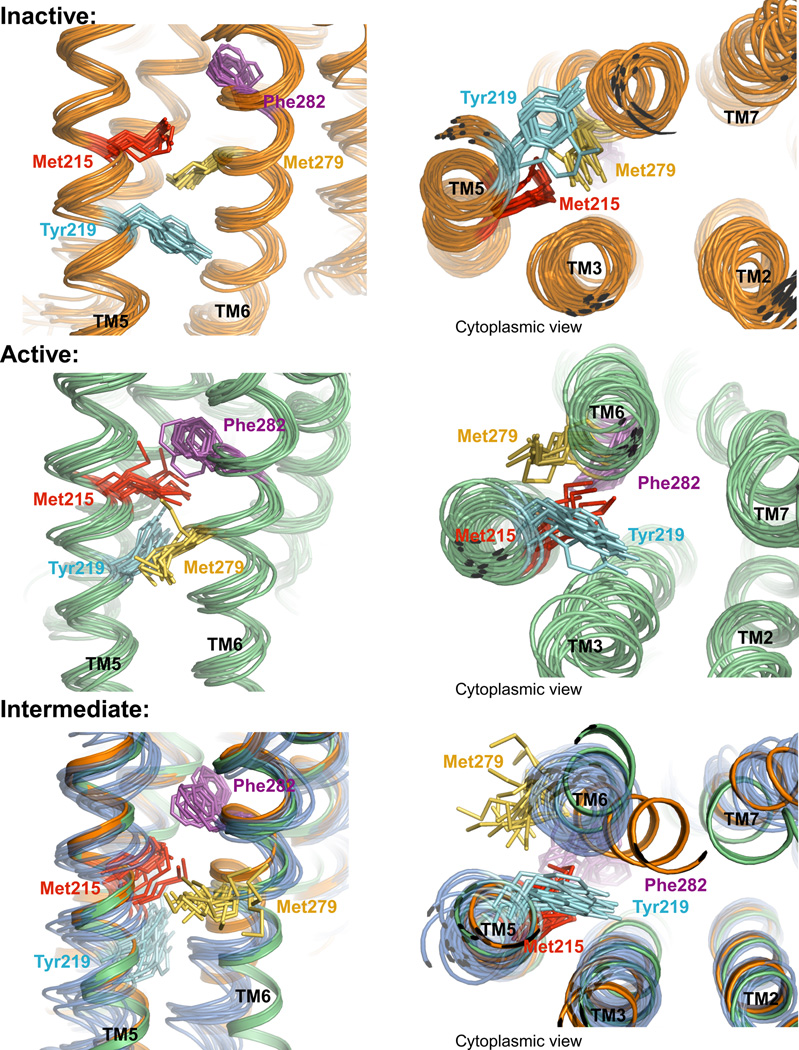
Figure 7. Conformational diversity in inactive, active and intermediate states.
Snapshots from MD simulations are shown every 18 ns. Met2155.54, Met2796.41, Tyr2195.58 and Phe2826.44 are shown as sticks. The top panel shows snapshots from a crystallographic- inactive-state simulation with carazolol bound. The middle panel shows snapshots from an active-state simulation with BI-167107 and Nb80 bound. The bottom panel shows frames from the intermediate state (blue), with only BI-167107 bound. In the bottom panel we also show the backbone of crystallographic inactive (orange) and active (green) states.
If we assume that the receptor preferentially populates the intermediate state when bound only to a strong agonist, the mobility observed in the intermediate state (Fig. 7) explains the weakness of the Met2155.54 peak and particularly the Met2796.41 peak. Met2796.41, whose side-chain points toward TM3 in the inactive crystal structure and toward TM5 in the active crystal structure, displays the greatest mobility in simulations of the intermediate state; its side-chain not only transitions between its active and inactive conformations, but frequently points out of the helix bundle into the lipids (Fig. 7). Tyrosine Tyr2195.58, just below Met2155.54, also adopts two distinct side-chain rotamers.
The region of the receptor just above residues Met2796.41 and Met2155.54, and just below the binding pocket, also displays substantial mobility in the intermediate state. This connector region (Dror et al., 2011a), particularly residues Phe2826.44 and Ile1213.40, displays a substantially different conformation in active and inactive crystal structures (Rasmussen et al., 2011b)(Fig. 2). In the intermediate state of the receptor, this region toggles between its two crystallographically observed conformations on timescales of a few microseconds in simulation; by contrast, it is locked in its inactive-like conformation in inactive-state receptor simulations, and it adopts a predominantly active-like conformation in active-state simulations. The observed fluctuations of the Phe2826.44/Ile1213.40 connector region in the intermediate state would also be expected to weaken the Met2796.41 and Met2155.54 NMR peaks; in particular, these intermediate-timescale fluctuations cause variations in the distance from both Met2796.41 and Met2155.54 to the aromatic ring of Phe2826.44 (Fig. 6 B, C, Fig. 7).
While the intracellular end of TM6 adopts a broader range of conformations in the intermediate state than in the active state in simulation, the chemical environment of Leu2726.34 changes little; it consistently points out into the lipid bilayer. This is consistent with the NMR spectra, where the Met2726.34 peaks are similar in the BI-167107- and BI-167107+Nb80-bound conditions. It also explains the strength of the Met2726.34 peak under these conditions.
The close similarity of the Met822.53 peak in the BI-167107-bound and BI-167107+NB80-bound NMR spectra suggests that, in the presence of a strong agonist, the chemical environment around Met822.53 near the binding pocket is very similar to that in the fully active-like conformation. This suggests that the conformational coupling between the binding pocket and the cytoplasmic ends of TMs 5 and 6 is relatively weak, and is in agreement with simulations showing that the conformation of the ligand-binding pocket is only loosely coupled to the conformation of the remainder of the receptor (Dror et al., 2011a). However, it should be noted that the absence of a chemical shift for Met822.53 does not exclude the possibility of a conformational changes in the binding pocket that does not alter the chemical environment of Met822.53.
We note that while both our NMR data and the simulations suggest that an intermediate state is more stable than the active state when only the strong agonist BI-167107 is bound, the simulations—unlike the NMR data—suggest that the inactive state is even more stable under these conditions (Dror et al., 2011a). These simulation results are consistent with a β2AR crystal structure with only an agonist bound (Rosenbaum et al., 2011), which closely matches the inactive structure. These differences suggest that the relative populations of the various states may be highly sensitive to environmental conditions, including lipids/detergent, temperature, etc. Indeed, minor changes in the energetics of the different conformational states (e.g., 1–2 kcal/mol) can have substantial effects on their relative populations.
Conclusion
Crystal structures provide snapshots of the β2AR at the beginning and end of the activation process. The full range of functionally relevant conformations, however, is difficult or perhaps even impossible to characterize by crystallography. In an effort to investigate the process of receptor activation, we used NMR spectroscopy to monitor the chemical environment of native methionines strategically located between the ligand-binding pocket and the G protein coupling interface. Our studies show that the conformational link between the ligand-binding pocket and the G protein coupling interface of the β2AR is not rigid. The results are compatible with a model (Fig. 1D) whereby agonist binding destabilizes the inactive state, but does not stabilize the fully active conformation observed in the β2AR-Gs complex. Instead, binding of a high affinity, high efficacy agonist is associated with conformational heterogeneity that may be important for allowing the β2AR to engage several alternative signaling or regulatory proteins depending on the physiological context (Gs, Gi, kinases and arrestins, Fig. 1A). This is in agreement with recent biophysical studies that suggest biased agonists that preferentially activate the arrestin pathway may have a more limited effect on receptor structure (Kahsai et al., 2011; Liu et al., 2012; Rahmeh et al., 2012).
Our studies highlight the complex role of protein dynamics in signal transduction and raise new questions about the universality of the structural changes underlying GPCR signal transduction. While many aspects of GPCR structure appear to be highly conserved, the dynamic properties we observe may reflect our choice of receptor, its environment, or the specific ligands we employed. Rhodopsin has been the most extensively studied GPCR model system and there is a wealth of structural and biophysical data that characterize the process of activation (Farrens, 2010; Hofmann et al., 2009; Hubbell et al., 2003). Several lines of evidence suggest that rhodopsin is somewhat less dynamic than other GPCRs (Fig. 1D). Covalently bound trans retinal, for example, appears to be effective at stabilizing the active state of rhodopsin even in the absence of a G protein (Fig. 1D), as shown by recent crystal structures of Metarhodopsin II and double electron electron resonance (DEER) spectroscopy studies (Choe et al., 2011; Knierim et al., 2008). This together with the lack of constitutive activity reflects its highly specialized role in detection of light. For GPCRs activated by peptide ligands, it is possible that a more extensive interface between GPCR and agonist might reduce structural heterogeneity. The effect of the lipid environment and associated signaling proteins (including other GPCRs) will likely also contribute to protein dynamics. Further investigation will be necessary to elucidate such variations and their importance in both cellular signal transduction and GPCR drug design.
Materials and Methods
NMR spectroscopy of 13C-ε-methyl-Met-β2AR
Between 15–20 L of Sf9 cells were infected by baculovirus encoding one of the β2AR constructs described (see more details in suppl. material) and grown in methionine deficient media with 13C methyl labeled methionine added into the media at 250 mg/L concentration. Subsequently, the cells were lysed and the receptor was purified by antibody affinity chromatography followed by ligand affinity chromatography and then a final antibody chromatography step.
The receptor was dialyzed twice against a buffer containing 20 mM HEPES pH 7.5, 100 mM NaCl, 0.1 % DDM and prepared in 98% D2O and concentrated to a final volume of 270 µL.
NMR
The HSQC pulse sequence used is a modified version of the basic HC-HSQC experiment as described in Bokoch et al (Bokoch et al., 2010). Data were acquired on the 800 MHz Varian INOVA spectrometer at Stanford Magnetic Resonance Laboratory (SMRL) and on the 900 MHz Bruker Avance 2 at the Central California 900 MHz NMR Facility.
For all the samples, except the β2AR-365N-construct samples, 128 complex t1 points were collected, for the β2AR-365N sample only 64 complex t1 points were collected (See Table S2). To allow temperature equilibration 64 steady state scans preceded data acquisition and a relaxation delay of 1.5 s were inserted to allow spin to relax back to equilibrium. The spectra were processed and visualized using NMRPipe/NMRDraw (Delaglio et al., 1995) software and NMRViewJ software, respectively. To eliminate horizontal noise from the very intense detergent methyl peak at 0.85 ppm [1H] and 16.8 ppm [13C] we used WET suppression of this methyl peak in some of the experiments (See Table S2). A common threshold, based upon a natural abundance peak at 1.4 ppm [1H] and 19.25 ppm [13C], was chosen for all spectra.
Molecular Dynamics Simulation Methods
We analyzed MD simulations of β2AR totaling 588 µs, including 15 new simulations (Table S3) as well as previously published simulations (Dror et al., 2011a; Dror et al., 2011b).
We performed all-atom simulations of β2AR under the four primary conditions of our NMR experiments: unliganded, bound to carazolol, bound to BI-167107, and bound to both BI-167107 and Nb80. The initial coordinates of unliganded and carazolol-bound simulations were based on a carazolol-bound β2AR crystal structure (PDB entry 2RH1). The initial coordinates of BI-167107-bound simulations, with and without Nb80, were based on the crystal structure of β2AR bound to BI-167107 and Nb80 (PDB entry 3P0G).
All simulations included an explicitly represented lipid bilayer, water, and salt ions. We used the CHARMM27 parameter set (MacKerell et al., 1998; Mackerell et al., 2004), with a modified lipid force field (Klauda et al., 2010), a modification to Asp, Glu, and Arg side-chains (Piana et al., 2011), and previously designed parameters for carazolol, palmitoyl-cysteine, and BI-167107 (Dror et al., 2009; Rosenbaum et al., 2011). Simulations were unbiased, apart from two that used temperature-accelerated MD (Maragliano and Vanden-Eijnden, 2006) to accelerate sampling of the distance between the centers of mass of the backbone atoms of Leu752.46 and Pro3237.50.
For more details on methods and materials used for this work see Supplementary Material.
Supplementary Material
Highlights.
-
-
NMR spectra of β2 adrenergic receptor (β2AR) labeled with 13CH3–ε-Met were obtained
-
-
NMR spectra reveal conformational states not observed in β2AR crystal structures
-
-
Conformational heterogeneity is observed in unliganded and antagonist-bound β2AR
-
-
Agonist alone does not fully stabilize the active conformation of the β2AR
Acknowledgements
We acknowledge support from National Institutes of Health Grant NS028471(B.K.K.) and the Mathers Foundation (B.K.K.). We thank Prof. Joseph Puglisi for encouragement and advice. We thank Dr. Søren Rasmussen for help with protein preparation. We acknowledge support from the Lundbeck Foundation (R.N.). We thank Paul Maragakis and Kresten Lindorff-Larsen for helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altenbach C, Kusnetzow AJ, Ernst O, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of the helix movement due to activation. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty EJ, Cox MC, Frenkiel TA, Tam BM, Mason AB, MacGillivray RT, Sadler PJ, Woodworth RC. Interlobe communication in 13C-methionine-labeled human transferrin. Biochemistry. 1996;35:7635–7642. doi: 10.1021/bi960684g. [DOI] [PubMed] [Google Scholar]
- Bose-Basu B, DeRose EF, Kirby TW, Mueller GA, Beard WA, Wilson SH, London RE. Dynamic characterization of a DNA repair enzyme: NMR studies of [methyl-13C]methionine-labeled DNA polymerase beta. Biochemistry. 2004;43:8911–8922. doi: 10.1021/bi049641n. [DOI] [PubMed] [Google Scholar]
- Butterfoss GL, DeRose EF, Gabel SA, Perera L, Krahn JM, Mueller GA, Zheng X, London RE. Conformational dependence of 13C shielding and coupling constants for methionine methyl groups. Journal of biomolecular NMR. 2010;48:31–47. doi: 10.1007/s10858-010-9436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DellaVecchia MJ, Merritt WK, Peng Y, Kirby TW, DeRose EF, Mueller GA, Van HB, London RE. NMR analysis of [methyl-13C]methionine UvrB from Bacillus caldotenax reveals UvrB-domain 4 heterodimer formation in solution. J Mol Biol. 2007;373:282–295. doi: 10.1016/j.jmb.2007.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deupi X, Kobilka BK. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (Bethesda) 2010;25:293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Arlow DH, Borhani DW, Jensen MO, Piana S, Shaw DE. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc Natl Acad Sci U S A. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, Borhani DW, Shaw DE. Activation mechanism of the beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2011a;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Jensen MO, Borhani DW, Shaw DE. Exploring atomic resolution physiology on a femtosecond to millisecond timescale using molecular dynamics simulations. The Journal of general physiology. 2010;135:555–562. doi: 10.1085/jgp.200910373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror RO, Pan AC, Arlow DH, Borhani DW, Maragakis P, Shan Y, Xu H, Shaw DE. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci U S A. 2011b;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrens DL. What site-directed labeling studies tell us about the mechanism of rhodopsin activation and G-protein binding. Photochemical & photobiological sciences : Official journal of the European Photochemistry Association and the European Society for Photobiology. 2010;9:1466–1474. doi: 10.1039/c0pp00283f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether U, Ballesteros JA, Seifert R, Sanders-Bush E, Weinstein H, Kobilka BK. Structural instability of a constitutively active G protein-coupled receptor. Agonist-independent activation due to conformational flexibility. J Biol Chem. 1997;272:2587–2590. doi: 10.1074/jbc.272.5.2587. [DOI] [PubMed] [Google Scholar]
- Hansen DF, Vallurupalli P, Kay LE. Using relaxation dispersion NMR spectroscopy to determine structures of excited, invisible protein states. Journal of biomolecular NMR. 2008;41:113–120. doi: 10.1007/s10858-008-9251-5. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, Heck M, Ernst OP. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- Kahsai AW, Xiao K, Rajagopal S, Ahn S, Shukla AK, Sun J, Oas TG, Lefkowitz RJ. Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nature chemical biology. 2011;7:692–700. doi: 10.1038/nchembio.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. The journal of physical chemistry B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knierim B, Hofmann KP, Gartner W, Hubbell WL, Ernst OP. Rhodopsin and 9-demethyl-retinal analog: effect of a partial agonist on displacement of transmembrane helix 6 in class A G protein-coupled receptors. J Biol Chem. 2008;283:4967–4974. doi: 10.1074/jbc.M703059200. [DOI] [PubMed] [Google Scholar]
- Kofuku Y, Ueda T, Okude J, Shiraishi Y, Kondo K, Maeda M, Tsujishita H, Shimada I. Efficacy of the beta(2)-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nature communications. 2012;3:1045. doi: 10.1038/ncomms2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins†. The Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. Journal of computational chemistry. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Maragliano L, Vanden-Eijnden E. A temperature accelerated method for sampling free energy and determining reaction pathways in rare events simulations. Chem Phys Lett. 2006;426:168–175. [Google Scholar]
- Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Pardo L, Deupi X, Dolker N, Lopez-Rodriguez ML, Campillo M. The role of internal water molecules in the structure and function of the rhodopsin family of G protein-coupled receptors. Chem bio chem. 2007;8:19–24. doi: 10.1002/cbic.200600429. [DOI] [PubMed] [Google Scholar]
- Piana S, Lindorff-Larsen K, Shaw DE. How robust are protein folding simulations with respect to force field parameterization? Biophysical journal. 2011;100:L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh R, Damian M, Cottet M, Orcel H, Mendre C, Durroux T, Sharma KS, Durand G, Pucci B, Trinquet E, et al. Structural insights into biased G protein-coupled receptor signaling revealed by fluorescence spectroscopy. Proc Natl Acad Sci U S A. 2012;109:6733–6738. doi: 10.1073/pnas.1201093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal K, Lefkowitz RJ, Rockman HA. When 7 transmembrane receptors are not G protein-coupled receptors. The Journal of clinical investigation. 2005;115:2971–2974. doi: 10.1172/JCI26950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011a;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Devree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. Crystal structure of the beta(2) adrenergic receptor-Gs protein complex. Nature. 2011b doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samama P, Cotecchia S, Costa T, Lefkowitz RJ. A mutation-induced activated state of the beta 2-adrenergic receptor. Extending the ternary complex model. J Biol Chem. 1993;268:4625–4636. [PubMed] [Google Scholar]
- Shaw DE, Dror RO, Salmon JK, Grossman JP, Mackenzie KM, Bank JA, Young C, Deneroff MM, Batson B, Bowers KJ, et al. Millisecond-scale molecular dynamics simulations on Anton. Proceedings of the Conference on High Performance Computing Networking, Storage and Analysis; ACM; Portland, Oregon. 2009. pp. 1–11. [Google Scholar]
- Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H[bond]13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- Wishart DS. Interpreting protein chemical shift data. Prog Nucl Magn Reson Spectrosc. 2011;58:62–87. doi: 10.1016/j.pnmrs.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Wisler JW, DeWire SM, Whalen EJ, Violin JD, Drake MT, Ahn S, Shenoy SK, Lefkowitz RJ. A unique mechanism of beta-blocker action: carvedilol stimulates beta-arrestin signaling. Proc Natl Acad Sci U S A. 2007;104:16657–16662. doi: 10.1073/pnas.0707936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Parnot C, Deupi X, Ratnala VR, Swaminath G, Farrens D, Kobilka B. Coupling ligand structure to specific conformational switches in the beta2-adrenoceptor. Nat Chem Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]
- Yao XJ, Velez RG, Whorton MR, Rasmussen SG, DeVree BT, Deupi X, Sunahara RK, Kobilka B. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.