Abstract
Genomic abnormalities, such as deletions in 11q22 or 17p13, are associated with poorer prognosis in patients with chronic lymphocytic leukemia (CLL). We hypothesized that unknown regions of copy number variation (CNV) affect clinical outcome and can be detected by array-based single-nucleotide polymorphism (SNP) genotyping. We compared SNP genotypes from 168 untreated patients with CLL with genotypes from 73 white HapMap controls. We identified 322 regions of recurrent CNV, 82 of which occurred significantly more often in CLL than in HapMap (CLL-specific CNV), including regions typically aberrant in CLL: deletions in 6q21, 11q22, 13q14, and 17p13 and trisomy 12. In univariate analyses, 35 of total and 11 of CLL-specific CNVs were associated with unfavorable time-to-event outcomes, including gains or losses in chromosomes 2p, 4p, 4q, 6p, 6q, 7q, 11p, 11q, and 17p. In multivariate analyses, six CNVs (ie, CLL-specific variations in 11p15.1-15.4 or 6q27) predicted time-to-treatment or overall survival independently of established markers of prognosis. Moreover, genotypic complexity (ie, the number of independent CNVs per patient) significantly predicted prognosis, with a median time-to-treatment of 64 months versus 23 months in patients with zero to one versus two or more CNVs, respectively (P = 3.3 × 10−8). In summary, a comparison of SNP genotypes from patients with CLL with HapMap controls allowed us to identify known and unknown recurrent CNVs and to determine regions and rates of CNV that predict poorer prognosis in patients with CLL.
Several recurrent genomic abnormalities have been identified in chronic lymphocytic leukemia (CLL) using traditional cytogenic methods and had prognostic importance.1,2 For example, deletions in chromosome 11q22 (10% to 20% of CLL cases), which includes the ataxia-telangiectasia mutated (ATM) gene, or 17p13 (3% to 5% of CLL cases), which includes the tumor protein p53 (TP53) gene, have been associated with rapid disease progression, treatment resistance, and inferior survival in patients.1–5 Thus, fluorescence in situ hybridization analysis of interphase nuclei using a panel of probes is commonly part of the clinical workup of CLL cases at diagnosis of CLL.1,6 In addition to the common recurrent chromosomal gains and losses, the presence of balanced or unbalanced translocations and cytogenetic complexity (three or more abnormalities) have been associated with shorter treatment-free and overall survival (OS).2,7–9 Furthermore, the acquisition of new and high-risk abnormalities during the clinical course of disease can render CLL B cells resistant to therapy.10,11 These data indicate that genomic instability is common in CLL, and that genetic abnormalities evolve over time, with a subset of changes that correlate with therapeutic resistance and/or prognosis.
Modern array-based genomic technologies, such as array comparative genomic hybridization and single-nucleotide polymorphism (SNP) genotyping, allow rapid and sensitive identification of small DNA copy number changes that are beyond the resolution of conventional cytogenetic techniques. For example, we and other researchers have demonstrated that bacterial artificial chromosome and oligonucleotide-based array comparative genomic hybridization allow the identification of novel recurrent genomic imbalances in CLL involving chromosomes 5q, 2p, 8, 19, and 22.12,13 Although the average resolution of bacterial artificial chromosome arrays is approximately 1 Mb, commercially available SNP arrays offer the possibility of genome-wide DNA genotyping at a mean resolution of <1000 bp by assessing thousands or millions of SNP loci per patient. Several groups have evaluated such SNP arrays as a screening tool for copy number abnormalities in various types of cancers. SNP arrays applied to CLL have identified acquired chromosomal imbalances in 66% to 82% of investigated cases.14–18 However, the interpretation of results from high-resolution DNA microarrays, especially without comparison of tumor to matched nontumor DNA, can be challenging. In particular, SNP arrays detect constitutional copy number variants and allelic polymorphisms, which tend to be smaller (median, approximately 150 kb) than acquired genomic aberrations in tumors.19 It is conceivable that both acquired and constitutional copy number changes, both included in the term copy number variation (CNV) herein, provide a genetic advantage or disadvantage to leukemic cells, and contribute to disease stage and clinical outcome. Considerable evidence from family and case-control studies suggests a nonexclusive, but presumably polygenic, predisposition for CLL by the presence of DNA variants in distinct susceptibility loci, such as 2q37.3, 8q24.21, 15q21.3, and 16q24.1.20–22
In the current study, we assessed total CNV by genome-wide SNP genotyping in 168 previously untreated patients with CLL using the Illumina610Quadv1 BeadChips (Illumina Inc., San Diego, CA). We developed analytical methods that allow us to determine acquired and constitutional CNVs associated with CLL, and to use the genome-wide copy number data for time-to-event outcome prognostication, without the availability of matched nontumor DNA. We hypothesized that CNVs acquired by CLL cells or inherently involved in CLL biological characteristics, are detected more frequently in SNP genotypes from patients with CLL than in SNP genotypes from healthy HapMap individuals who had been assessed on the same array platform. Furthermore, we hypothesized that any CNVs of biological interest in CLL should be associated with clinical time-to-event outcomes. We calculated CLL-specific CNVs as segments that were identified with statistical significance more frequently in patients with CLL than in HapMap controls. These segments included known recurrent abnormalities in chromosomes 2, 6, 11, 12, 13, and 17, as well as previously unknown CNVs in CLL. By using either all or CLL-specific CNV, we designed multivariate models that determined individual CNVs and increasing levels of genotypic complexity to predict time-to-event outcomes independent of established markers of prognosis.
Materials and Methods
Sample Collection, Purification, and Clinicopathologic Characterization
Between August 17, 2000, and December 15, 2008, with informed consent, we collected peripheral blood samples from 176 previously untreated patients with CLL at The University of Texas M. D. Anderson Cancer Center, Houston. The study was approved by the Institutional Review Board and conducted according to the principles expressed in the Declaration of Helsinki. During sample workup and data analysis, we removed eight cases because of false diagnosis (n = 1), false treatment status (previously treated; n = 3), double collection (n = 1), or poor genotype data quality (n = 3). All remaining 168 CLL cases had morphological features of CLL and met the diagnostic criteria established by the International Workshop on CLL.6 Clinical and routine laboratory data were obtained by review of the medical records. The somatic mutation status of immunoglobulin heavy chain variable region (IGHV) genes and ZAP70 expression, measured by either flow cytometry or immunohistochemistry, were assessed on blood or bone marrow samples, according to established protocols.23–25 Fluorescence in situ hybridization analysis for common abnormalities associated with CLL was performed on interphase nuclei obtained from cultured bone marrow cells using a probe panel designed to detect deletions of 11q22.3 (ATM), 13q14.3 (D13S319), 17p13.1 (TP53), and trisomy 12 (12p11.1-q11), according to the manufacturer’s instructions (Abbott Molecular, Abbott Park, IL). The CLL immunophenotypes were scored as either typical or atypical, as previously described.26,27 The CLL cells were enriched by negative selection and processed as previously described.28 Enriched cell preparations contained ≥95% CD5+/CD19+ copositive CLL cells, assessed by flow cytometry.
SNP Genotyping Analysis
Genomic DNA was extracted from enriched CLL cells using DNEasy spin columns (Qiagen, Valencia, CA). We performed SNP genotyping using Illumina Infinium high-density DNA Analysis BeadChips (HUMAN610-QUADv1; Illumina Inc., San Diego, CA), according to the manufacturer’s instructions. These arrays provide whole genome-wide coverage by 620,901 tag and nontag SNPs and additional CNV-targeted probes, with a median spacing of 2.7 kb (mean, 4.7 kb). Briefly, for each CLL case, 200 ng of DNA was denatured and isothermally amplified at 37°C for 24 hours. The amplified product was enzymatically fragmented, precipitated, resuspended, and hybridized onto the chip overnight. After allele-specific, single-base extension of bead- and sample-bound primers, extension products were fluorescently stained and assessed using the Illumina BeadArray Reader (Illumina Inc.).
SNP Copy Number Data Analysis
BeadChip readings derived from 225 HapMap controls assessed on Human610-Quadv1 BeadChips were downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number 17205, 73 CEU samples; accession number17206, 75 CH + JP; accession number 17207, 77 YRI). Raw BeadChip data from 168 patients with CLL and 225 HapMap controls were preprocessed to decode SNP/probe positions and generate genotype calls, log R ratio, and B-allele frequency (BAF) estimates using Illumina GenomeStudio, version 2010.2 (Illumina Inc.). Intensity data for each chromosome per patient were renormalized and segmented in R by applying circular binary segmentation.29 Each segment was assigned one of four calls based on the number of components in the BAF data: double loss (one component), homozygosity (two components), balanced (three components), or unbalanced (four components) heterozygosity. To determine the best fit among these four possible models, BAF data on each segment were clustered using k-means, with the quality of the clusters measured by the silhouette width.30 Adjacent segments were merged if they exhibited statistically equivalent copy number and identical BAF calls. Final call assignment was performed by pooling segments of log R ratio and BAF across patients. Gene alignments were performed using the University of California, Santa Cruz, Genome gateway (http://genome.ucsc.edu; hg18/March 2006/build 36.1 coordinates, last accessed March 17, 2011) and miRBase for noncoding RNA (http://mirnablog.com/the-mirbase-sequence-database-release-130, version 13.0/March 2009, last accessed March 17, 2011).
Quality Control, Statistical Analysis, and Interactive Web Tool
All clinical and SNP genotyping data underwent visual inspection. To assess the quality of SNP data after segmentation, we computed the median absolute deviation about the segment medians. For most samples, the median absolute deviation ranged between 0.1 and 0.2. As previously noted, three samples with a median absolute deviation of >0.3 were removed for poor genotype data quality. χ2 Tests were used to identify CLL-specific CNV by comparing the frequency of the abnormality in CLL cases with the frequency in HapMap control samples. The correlation of genomic CNV with clinical or biological covariates was assessed using a Student’s t-test for continuous variables and Pearson’s χ2 and Fisher’s exact tests for categorical variables. Outcome parameters (time-to-treatment and OS) were assessed using the Kaplan-Meier method to estimate survival functions and the log-rank test. Median follow-up was estimated using the reverse Kaplan-Meier method, which uses Kaplan-Meier estimation for patients alive as the event and deaths as censored observations to define the time interval between the origin and last follow-up.31
To construct multivariate models to predict time-to-event outcomes, we used a two-step process. First, we performed univariate analyses by applying log-rank testing and Cox proportional hazards models. Second, after filtering variables using a lenient criterion on the univariate P values, we applied a rigorous criterion to determine the final model by using the Akaike Information Criterion (AIC)32 to stepwise add or subtract variables. To construct models that only used existing clinical predictors, we applied AIC to all clinical predictors with P ≤ 0.15 in the univariate analyses. To construct models that only used CNV predictors, we applied AIC to all CNVs with P ≤ 0.10 in univariate models. We then generated a summary statistic by counting the number of CNVs in the final AIC model that were present in each patient, and used the log-rank test to determine whether this count was an independent predictor of the time-to-event outcome.
All computations were performed using the survival package (version 2.36-10) in the R statistical programming environment (version 2.14.0). The complete set of computer scripts used to perform the analyses is available at our website (http://bioinformatics.mdanderson.org/CLL-SNP). This website also contains browsable versions of all SNP data used herein.
Results
Patient Characteristics and Long-Term Follow-Up
The clinical and laboratory characteristics and follow-up for 168 previously untreated patients with CLL (110 men and 58 women; median age, 58 years at diagnosis) are presented in Table 1. When the samples were obtained, most patients had low- or intermediate-stage disease [132 (78.6%) Rai stage ≤2 and 144 (85.7%) Binet stage A/B].33,34 However, most patients [116 (69.0%)] had at least one unfavorable prognostic feature (ie, unmutated IGHV somatic mutation status, ZAP70 positivity, high CD38 expression, or elevated serum β-2-microglobulin (B2M) levels). At final follow-up, 144 patients (85.7%) had received frontline treatment using a variety of regimens, predominantly combined chemotherapy or chemoimmunotherapy [88 patients (61.1%)] (Supplemental Table S1). Forty-two patients (25%) had died. Based on Kaplan-Meier estimates, the 5-year OS from diagnosis was 90.2% (95% CI, 85.7% to 94.9%).
Table 1.
Clinical and Laboratory Characteristics of 168 Subjects
| Characteristics | Absolute value | % |
|---|---|---|
| Age at sample (years)∗ | 58 (27-83) | |
| Ethnic origin | ||
| White American | 153 | 91.1 |
| African American | 8 | 4.8 |
| Hispanic | 6 | 3.6 |
| Asian | 1 | 0.6 |
| Sex | ||
| Male | 110 | 65.5 |
| Female | 58 | 34.5 |
| Rai stage | ||
| ≤2 | 132 | 78.6 |
| ≥3 | 36 | 21.4 |
| WBC at sample date (109/L)∗ | 76.2 (7.9-372) | |
| IGHV somatic mutation status | ||
| Mutated | 90 | 53.6 |
| Unmutated | 76 | 45.2 |
| Not available | 2 | 1.2 |
| B2M (mg/L) | ||
| ≤4 | 122 | 72.6 |
| >4 | 46 | 27.4 |
| Median (range) | 3.3 (1.3-7.8) | |
| ZAP70 expression† | ||
| Positive | 73 | 43.5 |
| Negative | 73 | 43.5 |
| Not available | 22 | 13.1 |
| CD38 expression (%)‡ | ||
| <30 | 117 | 69.6 |
| ≥30 | 45 | 26.8 |
| Not available | 6 | 3.6 |
| Light chain use | ||
| κ | 104 | 61.9 |
| λ | 58 | 34.5 |
| Not available | 6 | 3.6 |
| FISH positivity (hierarchical model) | ||
| Del13q as sole abnormality | 35 | 20.8 |
| Trisomy 12 | 21 | 12.5 |
| Del11q | 17 | 10.1 |
| Del17p | 6 | 3.6 |
| Normal | 32 | 19.1 |
| Not available | 57 | 33.9 |
| Follow-up (months)∗ | ||
| Diagnosis to sample (N = 168) | 30.1 (0.0-211.5) | |
| Diagnosis to first treatment (N = 144)§ | 34.0 (0.7-211.5) | |
| Diagnosis to final follow-up (N = 168)¶ | 138.0 (0.7-271.3) | |
| Sample to final follow-up (N = 168)¶ | 72.6 (0.7-271.3) | |
FISH, fluorescence in situ hybridization; WBC, white blood cell.
Data are given as median (range).
The expression of ZAP70 was analyzed by either immunohistochemistry (96 patients) or flow cytometry (50 patients). By flow cytometry, patients were designated ZAP70 positive if expression was ≥20% in CD19-positive cells.
Immunophenotypic analysis of CD38 expression was assessed on bone marrow for 145 patients and on peripheral blood for 17 patients for whom bone marrow was unavailable.
The time from diagnosis to first treatment was assessed only for the cohort of 144 patients who have received treatment as of the last follow-up. The median was computed using Kaplan-Meier estimates.
Follow-up times are estimated using the reverse Kaplan-Meier method.
SNP Genotyping Identifies Regions of CNV
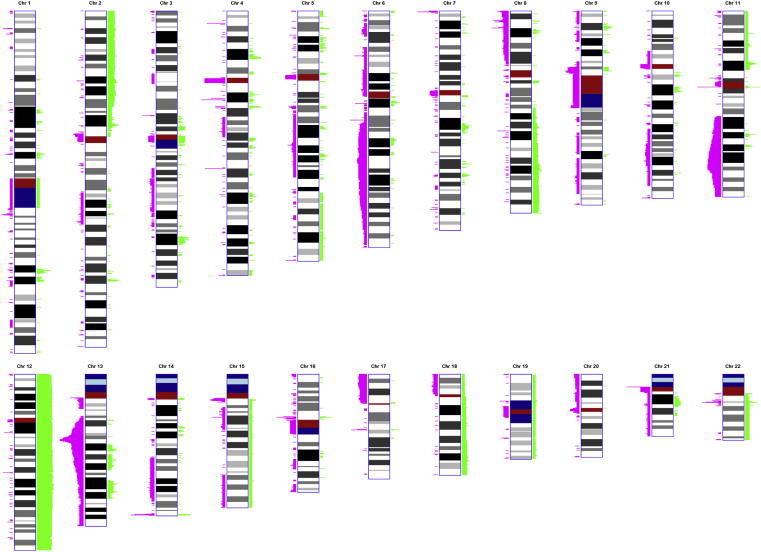
Overall, the computational segmentation of genotype data in 168 patients with CLL identified 1064 autosomal segments called as a gain or deletion (del) (Figure 1). After correcting for oversegmentation by merging adjacent segments that represented the same large event (gain or loss) in the same set of patients, we identified 322 segments of recurrent CNV, defined as a minimally deleted/gained region (MR) in a minimum of five cases (approximately 3%), for further analysis. Visual review of copy number (log R and BAF) plots revealed that the largest aberrations were detected in regions known to be aberrant in CLL. The single largest deletion was identified in chromosome 6 (90 Mb; 6q14.1-q27), followed by deletions in chromosomes 13 (78 Mb; 13q13.3-q34), 11 (46 Mb; 11q14.1-q24.1), and 17 (22 Mb; 17p11.1-p13.3).1,2 The largest chromosomal gain, other than trisomy, was identified in chromosome 2p (48.9 Mb; 2p16.3-p25.3), followed by gains in 8q (29.1 Mb; 8q23.3-q24.3) and 18q (15.7 Mb; 18q12.3-q21.32). As expected, the most common trisomy was of chromosome 12. We also identified trisomies of chromosomes 15 (one case), 18 (one case), 19 (one case), and 22 (two cases). There was one case of monosomy for chromosome 21.
Figure 1.
Cumulative SNP genotype in 168 patients with CLL. Loci of copy number abnormalities identified as autosomal gains are depicted in green; losses are depicted in pink. Chromosome G-banded ideograms are demonstrated at an approximately 700-band level of resolution. The heights of gains and losses along the horizontal axis are scaled proportionally to the square root of the number of cases with the corresponding abnormality. Centromeres, repeated sequences, and constitutive heterochromatin are indicated in red, light blue, and dark blue, respectively.
SNP Genotypes of Patients with CLL Differ from Those of HapMap Individuals
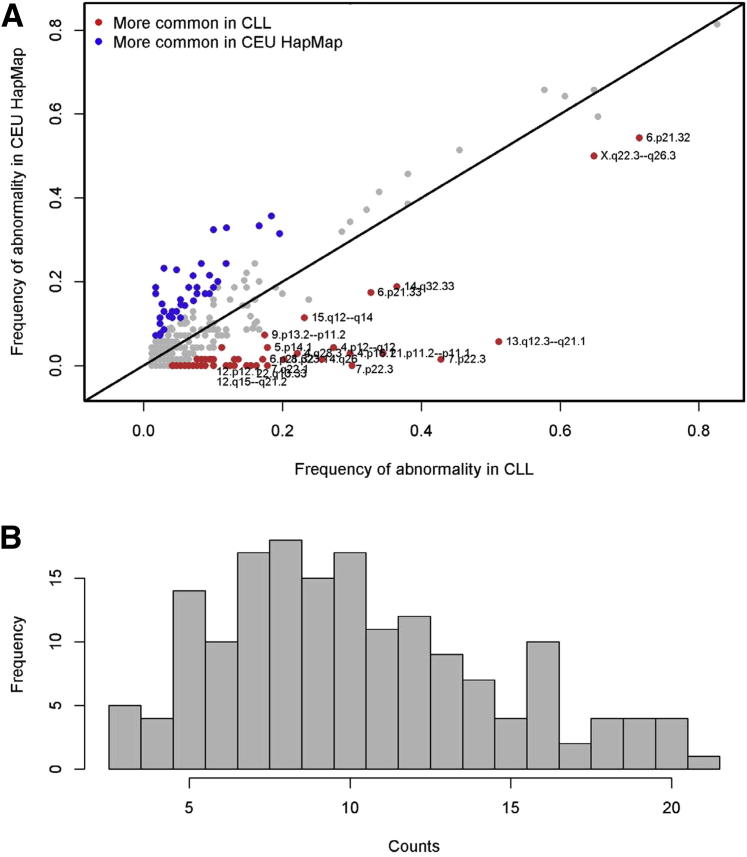
We compared the genotypes obtained from 168 CLL cases with the genotypes of 73 HapMap individuals of white descent, the predominant racial category in our CLL data set. We calculated the relative frequencies of CNVs designated as segments of gain or deletion and their MR in CLL versus all HapMap, and in CLL versus white HapMap samples only (Figure 2A). Abnormal segments were designated CLL specific if they were detected significantly more frequently in patients with CLL than in the white HapMap population (χ2 two-sided P < 0.05, corresponding to a false-discovery rate of 12%). Of 322 nonredundant CNVs, 82 segments were identified as CLL specific (37 gains and 45 deletions, P = 2.17 × 10−11 to 0.0441) (Supplemental Table S2). These CLL-specific segments included regions of CNV on each chromosome, clearly identified known common acquired abnormalities in CLL at the expected rates, and were consistent with data obtained by fluorescence in situ hybridization in bone marrow from 111 of 168 samples (Supplemental Table S3): del6q16.3-q21, 7.7% [loss (LT) 0211, 13 patients, P = 0.0083]; del11q22.3-q23.3, 13.1% (LT0374, 22 patients, P = 0.0007); del13q12.3-q21.1, 51.2% (LT0405, 86 patients, P = 5.8 × 10−14); del17p13.1-13.2, 6.0% (LT0477, 10 patients, P = 0.0185); and gains (GT) of chromosome 12, 17.3% (GT0299, 28 patients, including 25 with complete trisomy, P = 0.0002). CLL-specific segments were detected in a minimum of 7 patients (4.2%) and spanned ≥10 SNPs. The median number of CLL-specific CNVs per patient was 10 (range, 3 to 21) (Figure 2B). There was no patient without any CNV. We identified no single CNV shared by all CLL samples. In addition, this analysis identified 31 regions that were more common in the HapMap population than in CLL cases.
Figure 2.
A: Relative frequencies of recurrent segments of CNV in 168 patients with CLL versus 73 white HapMap individuals, based on their minimally deleted/gained region by SNP genotyping. Eighty-two autosomal CNVs identified as significantly more frequent in patients with CLL than in HapMap individuals are indicated in red (χ2 test). Segments of CNV identified as significantly more frequent in HapMap individuals than in patients with CLL are indicated in blue. Segments identified as equally frequent in both groups are indicated in gray. B: Frequency of recurrent CLL-specific CNVs per patient. CEU, standard HapMap designation of Utah residents with Northern and Western ancestry (ie, white).
CNVs in Chromosomes 2, 4, 6, 7, 11, and 17 Are Associated with Clinical Prognostic Parameters and Time-to-Event Outcome
Univariate analyses demonstrated that previously described prognostic factors, such as IGHV somatic mutation status, ZAP70 expression, elevated serum B2M or lactate dehydrogenase level, age, and unfavorable genetics (eg, del11q22/del17p13 or del6q21/del17p13 by SNP), are associated with time-to-event outcomes in our data set (log-rank test) (Supplemental Table S4 and Supplemental Figure S1).1,34–39 In multivariate analyses, the best model for time-to-treatment (TTT; P = 1.5 × 10−6) retained IGHV somatic mutation status [hazard ratio (HR), 2.3; 95% CI, 1.6 to 3.3], del11q22 or del17p13 (unfavorable cytogenetic abnormalities) by SNP (HR, 1.5; 95% CI, 1.04 to 2.2), and λ light chain use (HR, 1.42; 95% CI, 1.06 to 2.0) as independent predictive variables. The best model for OS (P = 9.4 × 10−6) retained IGHV somatic mutation status (HR, 2.2; 95% CI, 1.1 to 4.5), serum B2M level (categorical; cutoff, 4 mg/L; HR, 2.4; 95% CI, 1.3 to 4.6), and del6q21 or del17p13 by SNP (HR, 2.7; 95% CI, 1.3 to 5.6).
Next, we determined which of the 322 recurrent CNVs were associated with the strongest prognostic parameters in our data set (ZAP70 positivity, unmutated IGHV status, high serum B2M, and λ light chain use), with high CD38 expression or Rai stage, or with time-to-event outcomes. In univariate analyses (log-rank test), 71 of 322 nonredundant segments were associated with at least one prognostic marker, 22 correlated with TTT, and 14 correlated with OS (P < 0.05) (Supplemental Table S5); 20, 5, and 6 of these segments, respectively, were CLL specific. The CLL-specific segments associated with time-to-event parameters (Table 2) included losses in 4p12-q12 (46 patients; key genes, CWH43 and DCUN1D4), 6p21.32 (120 patients; HLA-DR region), 6q27 (13 patients; key gene, DLL1), 11q22.3-q23.2 (22 patients; MR, ATM and other genes), and 17p13.2-p13.1 (10 patients; MR, TP53 and other genes). CLL-specific gains associated with TTT or OS were detected in 2p11.1-p11.2 (seven patients; MR, immunoglobulin κ light chain locus), 4p15.1 (17 patients; MR, intergenic), 7q21.11 (12 patients; MR, SEMA3E), 7q31.1 (10 patients; MR, IMMP2L), 11q22.1 (eight patients; MR, intergenic), and 11p15.1-p15.4 (nine patients; MR, SOX6).
Table 2.
CLL-Specific Genomic Segments of CNV, Associated with Time-to-Event Outcomes in Univariate or Multivariate Analyses
| ID | Chr | Band | Start-end | No. of SNPs | No. of Pat sharing MR | Start-end MR | P value for CLL vs HapMap | P value for TTT univ | HR TTT univ | P value for TTT multiv | HR TTT multiv | P value for OS univ | Log HR OS univ | P value for OS multiv | Log HR OS multiv | Potential key gene affected in or near MR | Structural variation in DGV (MR) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GT0042 | 2 | p11.2-p11.1 | 87872343-91021213 | 265 | 7 | 89757456-89873958 | 0.0397 | 0.0810 | 0.48 | 0.2313 | 0.59 | 0.0113 | −18.17 | 0.0073 | −18.33 | IGK@ | Yes |
| LT0118 | 4 | p12-q12 | 48119281-52514330 | 131 | 46 | 49345777-49345777 | 0.0000 | 0.0467 | 1.46 | 0.2080 | 1.28 | 0.3826 | 0.31 | 0.9217 | 0.04 | DCUN1D4 | Yes |
| GT0081 | 4 | p15.1 | 31083440-34471356 | 504 | 17 | 33813772-34027766 | 0.0029 | 0.0139 | 0.48 | 0.0363 | 0.53 | 0.8769 | 0.08 | 0.9329 | 0.05 | Intergenic | Yes |
| LT0193 | 6 | p21.32 | 32158177-32608988 | 635 | 120 | 32574137-32574137 | 0.0054 | 0.0429 | 0.68 | 0.6149 | 0.91 | 0.3254 | −0.33 | 0.9168 | 0.04 | HLA,DRA, DRB1, and DRB5 | Yes |
| LT0221 | 6 | q27 | 169980638-170557777 | 138 | 13 | 170224848-170224848 | 0.0333 | 0.2640 | 1.45 | 0.1069 | 1.74 | 0.0310 | 1.09 | 0.0449 | 1.01 | LOC15449 and DLL1 | Yes |
| GT0195 | 7 | q21.11 | 82689044-83177270 | 151 | 12 | 83131205-83133610 | 0.0109 | 0.8467 | 1.07 | 0.5658 | 1.24 | 0.0492 | −17.08 | 0.0653 | −17.01 | SEMA3E | Yes |
| GT0202 | 7 | q31.1 | 110249344-111251527 | 267 | 10 | 110718711-110764580 | 0.0185 | 0.3858 | 0.72 | 0.9682 | 0.98 | 0.0272 | −17.10 | 0.0671 | −16.82 | IMMP2L and LRRN3 | Yes |
| GT0267 | 11 | p15.4-p15.1 | 3513788-16565390 | 3597 | 9 | 16167919-16285585 | 0.0242 | 0.0375 | 2.25 | 0.0489 | 2.16 | 0.5502 | −0.55 | 0.7026 | −0.37 | SOX6 | Yes |
| GT0281 | 11 | q22.1 | 97861584-98426127 | 151 | 8 | 98132117-98212355 | 0.0316 | 0.2866 | 1.51 | 0.3624 | 1.43 | 0.0418 | −17.09 | 0.0401 | −17.19 | CNTN5 | Yes |
| LT0374 | 11 | q22.3-q23.2 | 107197896-113285988 | 1094 | 22 | 107520009-113107765 | 0.0007 | 0.0400 | 1.68 | 0.4967 | 1.29 | 0.5310 | −0.36 | 0.1561 | −0.78 | ATM | No |
| LT0477 | 17 | p13.2-p13.1 | 6164128-9233788 | 767 | 10 | 6164128-9233788 | 0.0185 | 0.5450 | 1.23 | 0.3525 | 0.70 | 0.0182 | 1.21 | 0.8683 | 0.12 | TP53 | No |
Segments that reflect the recurrent abnormalities del11q22 and del17p13 are underlined. P < 0.05 values are boldfaced.
Chr, chromosome; DGV, Database of Genomic Variants (http://projects.tcag.ca/variation, last accessed January 1, 2012); multiv, in multivariate analysis; Pat, patients; univ, in univariate analysis.
Individual CNVs and CNV Rates per Patient Independently Predict Time-to-Event Outcome
We determined whether recurrent CNV, which associated with time-to-event outcomes by univariate analysis, would do so after adjustment for other known CNV and outcome predictors in a multivariate analysis. Of 39 recurrent overall CNVs that showed at least a trend to associate with TTT (P < 0.10), 25 were associated with poorer outcome (HR, >1). Two of these regions of CNV were biased through co-occurrence with unfavorable abnormalities in CLL (del11q22/del17p13) or with trisomy 12 by χ2 testing. For the remaining 23 regions, we used AIC to optimize a multivariate Cox proportional hazards model that prioritized 14 segments as independent predictors of TTT (Table 3).
Table 3.
Genomic Segments of Total CNVs That Were Independent Predictors of TTT
| ID | Chr | Band | Start-end | No. of SNPs | No. of Pat sharing MR | Start-end MR | P value for CLL vs HapMap | P value for TTT univ | HR TTT univ | P value for TTT multiv | HR TTT multiv | P value for OS univ | Log HR OS univ | P value for OS multiv | Log HR OS multiv | Potential key gene affected in or near MR | Structural variation in DGV (MR) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LT0069 | 2 | q37.1 | 232274785-232811993 | 191 | 12 | 232413576-232413900 | 0.0109 | 0.0691 | 1.87 | 0.0917 | 1.78 | 0.9761 | −0.02 | 0.9999 | 0.00 | COPS7B | Yes |
| LT0138 | 4 | q28.3 | 134301954-134519508 | 39 | 138 | 134352228-134352498 | 0.4121 | 0.0765 | 1.52 | 0.3805 | 1.25 | 0.9453 | −0.03 | 0.7228 | −0.15 | None | Yes |
| GT0117 | 4 | q34.3 | 179314101-180642935 | 481 | 5 | 180185537-180252886 | 0.0723 | 0.0971 | 2.38 | 0.0208 | 3.62 | 0.7178 | 0.39 | 0.2993 | 1.29 | None | Yes |
| GT0140 | 5 | q14.1 | 80707506-80931112 | 68 | 5 | 80759214-80849681 | 0.0723 | 0.0337 | 3.14 | 0.1987 | 1.93 | 0.1804 | 1.15 | 0.2857 | 0.93 | SSBP2 | Yes |
| LT0190 | 6 | p21.33 | 31353098-31408775 | 111 | 55 | 31400561-31400561 | 0.0083 | 0.0618 | 1.40 | 0.0869 | 1.37 | 0.9952 | 0.00 | 0.9799 | 0.01 | HLA-B | Yes |
| GT0159 | 6 | p21.32 | 32518405-32619844 | 113 | 12 | 32616865-32616865 | 0.9754 | 0.0339 | 2.19 | 0.3749 | 1.38 | 0.0738 | 0.97 | 0.0597 | 1.09 | HLA,DRA, DRB1, and DRB5 | Yes |
| LT0205 | 6 | q12 | 66565309-67421357 | 212 | 17 | 67079427-67104015 | 0.3564 | 0.0116 | 2.15 | 0.4902 | 1.24 | 0.4691 | 0.36 | 0.7703 | −0.14 | None | Yes |
| GT0182 | 6 | q27 | 167778489-168519816 | 296 | 5 | 168131062-168334983 | 0.8435 | 0.0293 | 3.28 | 0.0220 | 3.58 | 0.9838 | 0.02 | 0.8373 | 0.22 | KIF25 and FRMD1 | Yes |
| LT0253 | 7 | q35 | 143445383-143740335 | 49 | 7 | 143547020-143639837 | 0.1428 | 0.0156 | 3.04 | 0.0290 | 2.70 | 0.7718 | −0.28 | 0.8375 | −0.20 | CTAGE4 | Yes |
| LT0293 | 8 | q24.23 | 137747099-137955330 | 117 | 7 | 137897035-137900849 | 0.6978 | 0.0473 | 2.41 | 0.0099 | 3.33 | 0.1668 | 0.95 | 0.0493 | 1.45 | None | Yes |
| GT0267 | 11 | p15.4-p15.1 | 3513788-16565390 | 3597 | 9 | 16167919-16285585 | 0.0242 | 0.0375 | 2.25 | 0.0489 | 2.16 | 0.5502 | −0.55 | 0.7026 | −0.37 | SOX6 | Yes |
| LT0419 | 14 | q31.3 | 85454369-85743205 | 103 | 5 | 85454369-85743205 | 0.4802 | 0.0840 | 2.46 | 0.0648 | 2.67 | 0.9711 | 0.04 | 0.6324 | 0.53 | None | Yes |
| LT0503 | 18 | q22.1 | 61833162-61974746 | 48 | 12 | 61881775-61881930 | 0.6480 | 0.0005 | 3.58 | 0.0011 | 3.28 | 0.4746 | 0.46 | 0.2973 | 0.71 | None | Yes |
| GT0398 | 22 | q11.23 | 21820663-24259550 | 623 | 9 | 22717669-24259550 | 0.3653 | 0.0550 | 2.34 | 0.3668 | 1.48 | 0.7092 | 0.28 | 0.7672 | 0.23 | CABIN1 and ADORA2A | Yes |
P < 0.05 values are boldfaced.
Chr, chromosome; DGV, Database of Genomic Variants (http://projects.tcag.ca/variation, last accessed January 1, 2012); multiv, in multivariate analysis; Pat, patients; univ, in univariate analysis.
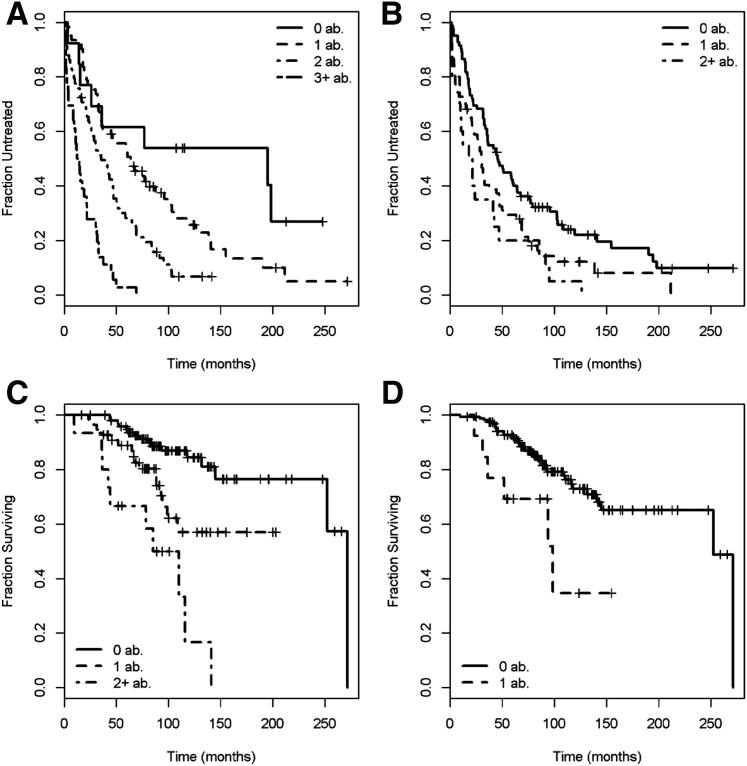
Six of these CNVs (ie, losses in 7q35, 8q24.23, and 18q22.1; and gains in 4q34.3, 6p21.32, and 11p15.1-15.4) (CLL specific) added significant predictive value as individual variables to our established clinical model (P ≤ 0.05), after adjusting for IGHV somatic mutation status, light chain use, and presence of del11q22/del17p13 by SNP. Moreover, the number of CNVs per patient (either as a continuous or a categorical variable) in the 14 independent segments was identified as a significant predictor of TTT in multivariate analyses beyond the statistical effect of the clinical model, including unfavorable genetics, IGHV somatic mutation status, and light chain use (P ≤ 1.68 × 10−7) (Table 4). Each additional count of a gain or loss in one of the segments increased the HR for the patient to require treatment by 2.1-fold (95% CI, 1.7 to 2.5; P = 7.8 × 10−16). Patients with three or more CNVs had an 8.1 times higher risk (95% CI, 3.48 to 18.9) to be treated than patients with no abnormalities (P = 2.0 × 10−9) (Figure 3A). The median TTT for patients with zero to one abnormality was 64 months (95% CI, 44.7 to 103.2), compared with 23 months for patients with two or more abnormalities (95% CI, 17.1 to 33.1) (P = 3.3 × 10−8).
Table 4.
Cox proportional hazards analyses testing the count of CNV as a predictive variable for TTT
| Independent segments (n=14) |
Independent CLL-specific segments (n=3) |
|||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P variable | P overall model (Likelihood ratio test) | HR | 95% CI | P variable | P overall model (Likelihood ratio test) | |
| Count of CNV as a continuous variable | 2.1 | 1.7–2.7 | 6.1 × 10−12 | 2.1 × 10−12 | 1.5 | 1.2–1.9 | 0.0005 | 0.0006 |
| Count of CNV as a categorical variable | 5.9 × 10−13 | 0.0023 | ||||||
| 1 CNV | 1.9 | 0.9–4.2 | 0.0907 | 1.6 | 1.1–2.2 | 0.0131 | ||
| 2 CNV | 3.6 | 1.6–7.9 | 0.0014 | 2.2 | 1.3–3.7 | 0.0022 | ||
| 3 CNV | 9.5 | 4.2–21.7 | 9.1 × 10−8 | NA | NA | NA | ||
| Count of CNV as a continuous variable compared to unfavorable cytogenetics | 2.3 × -10−11∗ | 1.1 × 10−12 | 0.0014∗ | 2.6x10−5 | ||||
| Count of CNV | 2.1 | 1.67–2.59 | 6.7 × 10−11 | 1.5 | 1.2–1.9 | 0.0010 | ||
| Del11q22 and/or del17p13 | 1.6 | 1.04–2.40 | 0.0323 | 1.9 | 1.2–2.8 | 0.0032 | ||
| Count of CNV as a categorical variable compared to known prognostic variables | 1.7 × 10−7∗ | 5.4 × 10−14 | 0.0709∗ | 1.3 × 10−6 | ||||
| 1 CNV | 1.8 | 0.8–3.9 | 0.1478 | 1.3 | 0.9–1.9 | 0.1424 | ||
| 2 CNV | 2.9 | 1.3–6.6 | 0.0103 | 1.8 | 1.1–3.0 | 0.0262 | ||
| ≥3 CNV | 7.5 | 3.1–17.8 | 5.8 × 10−6 | NA | NA | NA | ||
| Unmutated IGHV status | 1.9 | 1.3–2.7 | 0.0013 | 2.2 | 1.5–3.2 | 2.0 × 10−5 | ||
| Del11q22 and/or dell7p13 | 1.2 | 0.8–1.8 | 0.3899 | 1.5 | 1.0–2.2 | 0.0820 | ||
| Lambda light chain use | 1.1 | 0.78–1.6 | 0.5507 | 1.4 | 1.0–2.0 | 0.0526 | ||
CI, confidence interval; HR, hazard ratio; n, number of patients; NA, not applicable; p, p-value.
P-values for the significance of addition of the CNV count to clinical variables was tested using an analysis of deviance and is based on the chi- square statistic.
Figure 3.
Kaplan-Meier curves of time-to-event outcomes. A: Kaplan-Meier curves describing the time-to-treatment function in 36 patients with three or more CNVs versus 57 patients with two CNVs versus 60 patients with one CNV versus 13 patients with 0 CNVs. Considered in this analysis were 14 segments of CNV, independently associated with TTT. B: Kaplan-Meier curves describing the time-to-treatment function in 20 patients with two or more CNVs versus 66 patients with one CNV versus 82 patients without any CNV. Included in this analysis were three CLL-specific segments of CNV, independently associated with TTT. C: Kaplan-Meier curves describing the survival function in 15 patients with two or more CNVs versus 55 patients with one CNV versus 98 patients without any CNV. Considered in this analysis were nine segments of CNV that independently associated with OS. D: Kaplan-Meier curves describing the survival function in 16 patients with one or more CNVs versus 150 patients without any CNV. Considered in this analysis was only one CLL-specific segment of CNV (del6q27) that independently predicted OS. ab., abnormality.
We repeated this analysis beginning only with eight CLL-specific segments that demonstrated at least a trend to associate with decreased TTT in univariate analyses (P < 0.10). After accounting for bias by known common abnormalities or mutual dependence, we found an optimal set of three segments of CNV in 4p12-q12, 6p21.33, and 11p15 that predicted TTT in a multivariate model. The per-patient count of CNV in these three segments again performed as a significant predictor of TTT as either a continuous (P = 2.7 × 10−4) or ordinal (P = 0.0029) variable (Figure 3B) and was independent of the known genetic markers (P = 0.0014). Included in the clinical model, which incorporated unfavorable genetics, IGHV somatic mutation status, and light chain use, the count still demonstrated a statistical trend for an independent predictive value (P = 0.0709).
We repeated the same type of multivariate model building for OS starting with 39 recurrent CNVs associated with OS in univariate testing (P < 0.10), 14 of which were associated with a poor outcome. We identified nine CNVs as independent predictors of adverse survival (Table 5), four of which (losses in 8p22, 8q24.3, and 11p15.4, and gain in 20p11.1) added significant value as individual variables to our clinical model beyond IGHV somatic mutation status, serum B2M level, and unfavorable genetics (del6q21/del17p13) (P < 0.0336). The count of CNV in these segments also significantly predicted OS, both as a continuous and a categorical integer, independent of the same clinical factors (P ≤ 3.5 × 10−5) (Table 6 and Figure 3C). None of the predictive segments were CLL specific. Starting the analysis with only CLL-specific CNV associated with OS, only deletions in 6q27 (LT0221) (Supplemental Table S2) performed as a significant predictor of OS (P = 0.0093) (Figure 3D). As a predictor of OS, this segment was independent of unfavorable genetics (P = 0.01734), as well as independent of the clinical model that incorporated unfavorable genetics, IGHV mutation status, and B2M (P = 0.0449).
Table 5.
Genomic Segments of Total CNVs That Were Independent Predictors of OS
| ID | Chr | Band | Start-end | No. of SNPs | No. of Pat sharing MR | Start MR | P value for CLL vs HapMap | P value for TTT univ | HR TTT univ | P value for TTT multiv | HR TTT multiv | P value for OS univ | Log HR OS univ | P value for OS multiv | Log HR OS multiv | Potential key gene in MR or overall segment | Structural variation in DGV (MR) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LT0001 | 1 | p36.21 | 12767427-13614996 | 102 | 10 | 12821696-12836483 | 0.3031 | 0.6416 | 1.20 | 0.5141 | 1.30 | 0.0309 | −17.09 | 0.0914 | −16.94 | HNRNPCL1 | Yes |
| GT0137 | 5 | q13.2 | 68862825-69475807 | 105 | 11 | 69328189-69379245 | 0.5663 | 0.0607 | 1.91 | 0.0366 | 2.09 | 0.0305 | 1.09 | 0.0504 | 0.99 | SMA4 and SERF1A/B | Yes |
| GT0159 | 6 | p21.32 | 32518405-32619844 | 113 | 12 | 32616865-32616865 | 0.9754 | 0.0339 | 2.19 | 0.3749 | 1.38 | 0.0738 | 0.97 | 0.0597 | 1.09 | HLA,DRA, DRB1, and DRB5 | Yes |
| LT0267 | 8 | p22 | 13620726-13654984 | 26 | 5 | 13620726-13654984 | 0.0723 | 0.7886 | 1.15 | 0.8109 | 1.13 | 0.0916 | 1.19 | 0.0336 | 1.59 | None | Yes |
| LT0294 | 8 | q24.3 | 141086472-141321409 | 152 | 10 | 141189636-141189852 | 0.0699 | 0.4621 | 1.32 | 0.2940 | 1.51 | 0.0695 | 1.12 | 0.0428 | 1.28 | TRAPPC9 | Yes |
| LT0354 | 11 | p15.4 | 3383178-3888105 | 110 | 5 | 3588683-3686252 | 0.0723 | 0.1159 | 2.52 | 0.2153 | 2.05 | 0.0117 | 1.96 | 0.0211 | 1.78 | ART1/5 and NUP98 | Yes |
| GT0311 | 13 | q13.2--q13.3 | 33937199-35109143 | 303 | 5 | 34542952-34877387 | 0.0723 | 0.6773 | 1.22 | 0.5043 | 1.38 | 0.0950 | 1.18 | 0.1028 | 1.16 | NBEA | Yes |
| GT0380 | 20 | p11.1 | 25710186-25971373 | 64 | 5 | 25894996-25897274 | 0.4773 | 0.3422 | 1.59 | 0.6163 | 1.27 | 0.0050 | 1.87 | 0.0273 | 1.44 | None | Yes |
| LT0551 | 22 | q11.23 | 22448589-23491733 | 260 | 25 | 22688572-22715105 | 0.1200 | 0.9409 | 0.98 | 0.4202 | 0.82 | 0.0226 | 0.88 | 0.0936 | 0.67 | GSTT1 | Yes |
P < 0.05 values are boldfaced.
Chr, chromosome; DGV, Database of Genomic Variants (http://projects.tcag.ca/variation, last accessed January 1, 2012); multiv, in multivariate analysis; Pat, patients; univ, in univariate analysis.
Table 6.
Cox proportional hazards analyses testing the count of CNV as a predictive variable for OS
| Independent segments (n=9) |
Independent CLL-specific segments (n=1) |
|||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P variable | P overall model (Likelihood ratio test) | HR | 95% CI | P variable | P overall model (Likelihood ratio test) | |
| Count of CNV as a continuous variable | 2.7 | 1.8–4.1 | 1.5 × 10−6 | 3.5 × 10−7 | 3.0 | 1.3–7.1 | 0.0133 | 0.0093 |
| Count of CNV as a categorical variable | 3.8 × 10−7 | NA | NA | NA | ||||
| 1 CNV | 2.6 | 1.3–5.4 | 0.0083 | |||||
| 2 CNV | 7.6 | 3.3–17.2 | 1.4 × 10−6 | |||||
| Count of CNV as a continuous variable compared to unfavorable cytogenetics | 6.0 × 10−6 | 7.6 × 10−7 | 0.0173∗ | 0.0058 | ||||
| Count of CNV | 2.7 | 1.8–4.1 | 2.2 × 10−6 | 3.4 | 1.4–8.4 | 0.0066 | ||
| Del6q21 and/or del17p13 | 1.7 | 0.8–3.6 | 0.1630 | 2.1 | 1.0–4.5 | 0.0549 | ||
| Count of CNV as a categorical variable compared to known prognostic variables | 3.9 × 10−6 | 7.3 × 10−11 | 0.0449∗ | 1.2 × 10−6 | ||||
| 1 CNV | 3.0 | 1.4–6.4 | 0.0036 | 2.7 | 1.1–6.7 | 0.0250 | ||
| 2 CNV | 7.6 | 3.1–18.4 | 7.4 × 10−6 | NA | NA | NA | ||
| Unmutated/GHV status | 1.7 | 0.8–3.5 | 0.1537 | 2.2 | 1.1–4.5 | 0.0278 | ||
| High B2M | 2.1 | 1.1–4.2 | 0.0246 | 2.2 | 1.2–4.3 | 0.0147 | ||
| Del6q21 and/or del17p13 | 4.0 | 1.8–9.0 | 0.0005 | 2.7 | 1.3–5.7 | 0.0104 | ||
CI, confidence interval; HR, hazard ratio; n, number of patients; NA, not applicable; p, p-value.
P-values for the significance of addition of the CNV count to clinical variables was tested using an analysis of deviance and is based on the χ2 statistic.
Discussion
We report one of the largest data sets of CNV in a total of 168 previously untreated patients with CLL, combined with extensive clinical and laboratory data. Several earlier studies have provided data on global DNA copy number profiling in CLL cases using SNP-based microarrays. However, unlike the current study, many of these older data sets were significantly smaller than the current study, had a shorter follow-up or limited clinical data, were based on mixed patient cohorts with respect to treatment status, or were performed using unenriched blood and/or bone marrow samples, which decreases the sensitivity to detect abnormalities in small subclones.14–17,35
For stored tumor samples, one limiting factor in identifying regions of acquired CNV has been the lack of corresponding nontumor DNA for comparison. This is particularly true for tumors collected and processed before the advent of cost-effective, high-throughput genomic technologies, as were many of the samples in the current study, or for small biopsy specimens that lack surrounding normal tissue. Alternative approaches are needed to analyze genomic data from tumors in the absence of patient-matched benign tissue. Two studies of CNV in CLL by SNP genotyping have compared purified CLL DNA with nontumor DNA prepared from either purified T cells or buccal swabs.14,18 Other studies have identified abnormalities by either visual inspection of the plots or selection of only those abnormalities that demonstrated <50% overlap with copy number events in the Toronto Database of Genomic Variants (http://projects.tcag.ca/variation, last accessed January 1, 2012).16,36 In this study, we developed a statistical approach that addresses the limitation of the lack of nontumor DNA by comparing genotypes from patients with CLL with the genotypes from healthy white HapMap individuals whose DNA had been analyzed on the same SNP genotyping platform. Furthermore, we focused our study on identifying regions of CNV that were associated with time-to-event outcomes, rather than cataloguing regions of acquired CNV. This approach should filter out array noise (eg, detection errors, repetitive sequences, or population-specific genomic polymorphisms), but it still allows us to identify new genomic regions of potential clinical or biological relevance in CLL.
We identified 322 nonoverlapping genomic segments of CNV in a subset of at least five patients (3%). We chose 3% as a reasonable minimum patient subset of clinical interest (similar to the incidence of TP53 deletion) in which we could expect statistically significant and clinically relevant associations in our data set. The 322 segments included 111 gains (34.5%) and 211 losses (65.5%). Consistent with data provided in studies previously reported, chromosomal losses were more common than gains.14–18 In a recent study using Affymetrix 6.0 SNP arrays (>900,000 SNP and copy number probes), Ouillette et al18 identified 584 different somatic changes in 255 patients with CLL. Approximately 20% of patients presented without any somatic abnormality. Recurrent somatic abnormalities other than the common abnormalities were relatively infrequent, occurring in subsets of 6 to 10 patients (2.4% to 3.9%), and included losses on chromosomes 8p, 10q, 14q, and 18q and gains on 8q, 17q, and 18p. Abnormalities in these regions often showed variability in the start and end sites, leading to more than one minimally deleted or gained region (MR).18 By comparing the 322 genomic segments in CLL to HapMap samples, we identified three groups of CNV: CNV equally present in CLL and in HapMap, CNV significantly more frequent in CLL than in HapMap (CLL specific), and CNV significantly more frequent in HapMap than in CLL. Among the 82 CLL-specific segments, we detected well-established recurrent abnormalities, such as deletions in 6q, 11q, 13q, and 17p and trisomy 12, in the expected frequencies. Our finding that some CNVs occur more frequently in normal HapMap individuals compared with patients with CLL raises the possibility that some polymorphisms may be associated with a decreased risk of developing CLL. Larger population-based studies are needed to test this hypothesis.
Visual inspection of the copy number plots on our website (http://bioinformatics.mdanderson.org/CLL-SNP) indicated that gains in 2p and 8q are the most commonly acquired CNVs after del6q, 11q, 13q, 17p, and trisomy 12. By using conventional or high-throughput technologies, researchers have described gains in 2p as a frequent genomic event in advanced-stage CLL, and this abnormality has potentially unfavorable prognostic significance. REL and BCL11A (2p15-p16.1) or MYCN (2p24.3) have been reported as the genes most frequently gained.14,16,36–38 We identified a segment in this region (GT0038, 2p12-16.3) as a CLL-specific event in eight patients (P = 0.0316) (Supplemental Table S2), with an MR in 2p15-p14. Of these eight patients, seven presented with an overlap in 2p16.1-p15 that included REL and BCL11A; six patients shared >6 Mb of gained chromosomal material that included MYCN and >350 other genes. Furthermore, four patients also demonstrated losses in ATM/11q22.3, indicating a potential biological relationship between gains in 2p and ATM losses. However, we found no association between gains in GT0038 or gains solely in BCL11A/REL or MYCN and clinical outcome. Gains in 8q24.21, which lead to overexpression of MYC, have previously been found in approximately 3% of CLL cases, and may be associated with an unfavorable prognosis.39 We identified similar gains in this region in five patients (CLL-specific segment GT0232) (Supplemental Table S2). These gains were relatively large, with a minimum overlap of 19.1 Mb, and contained >90 genes. However, we found no association between gains in 8q and time-to-event outcomes.
Algorithms that attempt to identify MRs using SNP genotyping face major statistical challenges. These challenges begin with the segment boundaries found when applying segmentation algorithms one sample at a time. In our data, the SD of the SNP log ratios is between one-half and two-thirds of the distance between the expected levels for a single copy gain or loss. As a consequence, there is inherent uncertainty in the segment boundaries for individual patients, on the order of two to three SNPs or 10 to 15 KB. This uncertainty is compounded when intersecting segments from multiple patients are considered. Statistically, finding the intersection requires estimating the extreme values of a distribution (the left end point is a maximum; the right end point, a minimum), which is intrinsically more variable than an estimate of the mean. Estimates of extreme values are not robust because they can be heavily influenced by a single outlier. For example, the MR determined algorithmically for the 8q24.21 gain failed to include MYC because one subsegment for one patient (CL104) had a median log ratio that was just short of the cutoff needed to call a gain. The MR determined algorithmically for the 2p15-16.1 gain failed to include REL or BCL11A because of the inclusion of one patient (CL048) for whom the evidence for the segment boundary is weak. The Genomic Identification of Significant Targets in Cancer (GISTIC) method improves on the simple intersection of segments, but this method still relies on the (possibly inaccurate) results of the individual segmentation algorithms.40 There is substantial room for improvement in statistical algorithms that combine the raw data with the segments to identify MR across sets of patients. Meanwhile, visual inspection of copy number plots is still needed to confirm relevant minimally gained or deleted regions. Our website (http://bioinformatics.mdanderson.org/CLL-SNP) provides multiple images of both the raw and segmented data to facilitate visual inspection.
Visual inspection revealed additional regions of CNV shared by fewer than our cutoff of five cases. These regions included 78 events of ≥1 Mb in 42 patients (25%), and often appeared to target potential tumor suppressor genes or oncogenes (eg, PTEN in patient sample CLL213 or hsa-mir-181a-1/b-1 in sample CLL137) (http://bioinformatics.mdanderson.org/CLL-SNP), most of them as a single-case event. Patients with such rare events also tended to have higher overall numbers of total or CLL-specific abnormalities, and carried a deletion in 17p13 significantly more frequently (P ≤ 0.0324) (data not shown), indicating a higher level of genomic instability in those cases. Thus, our SNP array data demonstrate that CLL cases exhibit substantial genomic heterogeneity. The finding is also consistent with results obtained using next-generation sequencing technologies, which have shown many small subsets of patients with CLL who contain mutations in a wide variety of genes.41–43 It is conceivable that these rare recurrent abnormalities affect the disease outcome in individual patients and might need therapeutic consideration in the future, but their clinical relevance is difficult to characterize.
We identified previously unknown regions of CNV that were associated with time-to-event outcomes, and that were retained in multivariate models of prognosis, independent of established prognostic markers in CLL. For most of these CNVs, structural variants in the minimally deleted or gained region have been described in the Database of Genomic Variants (http://projects.tcag.ca/variation, last accessed January 1, 2012). Interestingly, variations in the HLA region (LT0193 and GT0159; MR, HLA-DR region) demonstrated significant associations with time-to-treatment. The HLA gene locus comprises a highly polymorphic genomic region prone to structural changes and recombination, resulting in a characteristically diverse SNP log R ratio/BAF profile obtained by array genotyping. In a recent report, Shah et al44 demonstrated that homozygosity for distinct HLA-A, HLA-B, HLA-C, or HLA-DR haplotypes, including HLA-DR5, occurs more frequently in patients with CLL than in healthy control individuals and is associated with progression-free-survival. Although we cannot distinguish single HLA alleles by SNP genotyping, the variation in the genomic segments we found in this region may reflect this phenotypic bias in HLA haplotypes that characterize CLL, and may potentially influence tumor immunogenicity and immunosurveillance. In particular, a biased presentation of antigens in patients with CLL carrying distinct HLA haplotypes might contribute to the clonogenic activation of CLL cells in the B-cell compartment. Thus, a detailed analysis of HLA haplotypes in patients with CLL of larger data sets is desirable to assess this hypothesis.
Because we lacked matched normal DNA samples, we were unable to distinguish between acquired somatic and nonsomatic CNV. Nevertheless, we were able to build multivariate models of predictive value using total or CLL-specific CNV detected by the SNP platform. These models were statistically independent of clinical variables when we used all CNVs, but were not independent when we restricted to CNVs that were CLL specific based on a comparison with white HapMap samples. The statistical significance also decreased if we included only segments that contained at least one protein-coding gene or miRNA. Most previous studies of CLL using SNP genotyping have focused on CNVs within coding regions. By using recent builds of Entrez Gene, the University of California, Santa Cruz, Genome Browser, and miRBase, we found that 93 (28.9%) of the 322 nonredundant abnormalities contained no protein-coding or miRNA genes in the overall segment, and 165 (51.2%) had no known genes within the MR. Of the 165 segments, 29 were associated with outcome in univariate analysis, and 10 in multivariate models. CNV in these segments might include nonsomatic variation in susceptibility regions within segmentation-dependent boundaries in our study. Large genome-wide association studies have demonstrated CLL-associated SNPs of unknown function in noncoding regions, such as variations in 2q13 (rs17483466), 11q24.1 (rs735665), and 15q23 (rs7176508).21,45 These regions may contain regulatory elements involved in transcriptional activation, silencing, epigenetic modification, chromatin organization, or unidentified noncoding RNA genes. Further validation and biological studies are necessary to elucidate the significance of somatic and nonsomatic variations in noncoding genomic regions in CLL.
In summary, SNP genotyping of 168 untreated patients with CLL identified regions of CNV in segments of chromosomes previously known to be gained or lost in CLL, as well as new segments of CNV, including loci of suspected DNA structural variants, associated with clinical and biological covariates and time-to-event outcomes. By using a segmentation approach and a focused analysis of outcome-related CNV, we constructed predictive models that allowed us to use this information to predict inferior outcome based on regions of CNV. We found that some of the genomic loci associated with regions of CNV found in the normal population are highly related with clinical outcome in our data set of previously untreated patients with CLL.
Genome-wide population studies using high-throughput whole genome sequencing methods have unveiled millions of SNPs and copy number changes, including short insertions/deletions present in the general population (The 1000 Genomes Project Consortium).46 Family and epidemiological case-control studies have provided evidence for the role of inherited genetic susceptibility in CLL. Our study indicates that acquired and nonacquired genomic variants, including CNV in distinct chromosomal segments, might affect long-term outcome in patients with CLL. Validation of our findings in larger studies, using higher-resolution technologies (eg, next-generation sequencing), and/or meta-analyses of genome-wide profiling data from several CLL groups are needed to comprehensively address the clinical relevance of acquired and common genomic variation in patients with CLL.
Footnotes
Supported by a European Hematology Association and American Society of Hematology grant (C.D.S.), CLL Global Research Foundation grant (C.D.S. and L.V.A.), Commonwealth Foundation for Cancer Research and Mr. and Mrs. William H. Goodwin, Jr., grants (L.V.A.), and National Cancer Institute grant R01CA123252 (L.V.A.).
Supplemental Data
Kaplan-Meier survival functions for variables independently predicting either time from diagnosis (Dx) to treatment (Rx) or time from diagnosis (Dx) to the date of last documented survival (OS) in multivariate models [log lactate dehydrogenase (LDH) and age are not shown because they were used as continuous variables]. BadCyto, cases with del11q22 or del17p13 (time to Rx) or with del6q21 or del17p13 (OS from Dx); CatB2M, categorization according to β-2-microglobulin serum level (high, >4 mg/L; low, ≤4 mg/L); CYTO2, categorization according to cytogenetic hierarchy; GoodCyto, without del11q22 or del17p13 (time to Rx) or without del6q21 or del17p13 (OSl from Dx); M, mutated; NEG, negative; POS, positive; U, unmutated.
References
- 1.Dohner H., Stilgenbauer S., Benner A., Leupolt E., Krober A., Bullinger L., Dohner K., Bentz M., Lichter P. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 2.Juliusson G., Oscier D.G., Fitchett M., Ross F.M., Stockdill G., Mackie M.J., Parker A.C., Castoldi G.L., Guneo A., Knuutila S., Elonen E., Gahrton G. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323:720–724. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 3.Dohner H., Stilgenbauer S., Dohner K., Bentz M., Lichter P. Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis. J Mol Med. 1999;77:266–281. doi: 10.1007/s001090050350. [DOI] [PubMed] [Google Scholar]
- 4.Zenz T., Eichhorst B., Busch R., Denzel T., Habe S., Winkler D., Buhler A., Edelmann J., Bergmann M., Hopfinger G., Hensel M., Hallek M., Dohner H., Stilgenbauer S. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;24:4473–4479. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]
- 5.Austen B., Powell J.E., Alvi A., Edwards I., Hooper L., Starczynski J., Taylor A.M., Fegan C., Moss P., Stankovic T. Mutations in the ATM gene lead to impaired overall and treatment-free survival that is independent of IGVH mutation status in patients with B-CLL. Blood. 2005;106:3175–3182. doi: 10.1182/blood-2004-11-4516. [DOI] [PubMed] [Google Scholar]
- 6.Hallek M., Cheson B.D., Catovsky D., Caligaris-Cappio F., Dighiero G., Dohner H., Hillmen P., Keating M.J., Montserrat E., Rai K.R., Kipps T.J. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huh Y.O., Abruzzo L.V., Rassidakis G.Z., Parry-Jones N., Schlette E., Brito-Bapabulle V., Matutes E., Wotherspoon A., Keating M.J., Medeiros L.J., Catovsky D. The t(14;19)(q32;q13)-positive small B-cell leukaemia: a clinicopathologic and cytogenetic study of seven cases. Br J Haematol. 2007;136:220–228. doi: 10.1111/j.1365-2141.2006.06416.x. [DOI] [PubMed] [Google Scholar]
- 8.Mayr C., Speicher M.R., Kofler D.M., Buhmann R., Strehl J., Busch R., Hallek M., Wendtner C.M. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2006;107:742–751. doi: 10.1182/blood-2005-05-2093. [DOI] [PubMed] [Google Scholar]
- 9.Van Den Neste E., Robin V., Francart J., Hagemeijer A., Stul M., Vandenberghe P., Delannoy A., Sonet A., Deneys V., Costantini S., Ferrant A., Robert A., Michaux L. Chromosomal translocations independently predict treatment failure, treatment-free survival and overall survival in B-cell chronic lymphocytic leukemia patients treated with cladribine. Leukemia. 2007;21:1715–1722. doi: 10.1038/sj.leu.2404764. [DOI] [PubMed] [Google Scholar]
- 10.Stilgenbauer S., Sander S., Bullinger L., Benner A., Leupolt E., Winkler D., Krober A., Kienle D., Lichter P., Dohner H. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–1245. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- 11.Grubor V., Krasnitz A., Troge J.E., Meth J.L., Lakshmi B., Kendall J.T., Yamrom B., Alex G., Pai D., Navin N., Hufnagel L.A., Lee Y.H., Cook K., Allen S.L., Rai K.R., Damle R.N., Calissano C., Chiorazzi N., Wigler M., Esposito D. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA) Blood. 2009;113:1294–1303. doi: 10.1182/blood-2008-05-158865. [DOI] [PubMed] [Google Scholar]
- 12.Gunn S.R., Bolla A.R., Barron L.L., Gorre M.E., Mohammed M.S., Bahler D.W., Mellink C.H., van Oers M.H., Keating M.J., Ferrajoli A., Coombes K.R., Abruzzo L.V., Robetorye R.S. Array CGH analysis of chronic lymphocytic leukemia reveals frequent cryptic monoallelic and biallelic deletions of chromosome 22q11 that include the PRAME gene. Leuk Res. 2009;33:1276–1281. doi: 10.1016/j.leukres.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Gunn S.R., Mohammed M.S., Gorre M.E., Cotter P.D., Kim J., Bahler D.W., Preobrazhensky S.N., Higgins R.A., Bolla A.R., Ismail S.H., de Jong D., Eldering E., van Oers M.H., Mellink C.H., Keating M.J., Schlette E.J., Abruzzo L.V., Robetorye R.S. Whole-genome scanning by array comparative genomic hybridization as a clinical tool for risk assessment in chronic lymphocytic leukemia. J Mol Diagn. 2008;10:442–451. doi: 10.2353/jmoldx.2008.080033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeifer D., Pantic M., Skatulla I., Rawluk J., Kreutz C., Martens U.M., Fisch P., Timmer J., Veelken H. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202–1210. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 15.Ouillette P., Erba H., Kujawski L., Kaminski M., Shedden K., Malek S.N. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008;68:1012–1021. doi: 10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 16.Gunnarsson R., Isaksson A., Mansouri M., Goransson H., Jansson M., Cahill N., Rasmussen M., Staaf J., Lundin J., Norin S., Buhl A.M., Smedby K.E., Hjalgrim H., Karlsson K., Jurlander J., Juliusson G., Rosenquist R. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia. 2009;24:211–215. doi: 10.1038/leu.2009.187. [DOI] [PubMed] [Google Scholar]
- 17.Kujawski L., Ouillette P., Erba H., Saddler C., Jakubowiak A., Kaminski M., Shedden K., Malek S.N. Genomic complexity identifies patients with aggressive chronic lymphocytic leukemia. Blood. 2008;112:1993–2003. doi: 10.1182/blood-2007-07-099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouillette P., Collins R., Shakhan S., Li J., Peres E., Kujawski L., Talpaz M., Kaminski M., Li C., Shedden K., Malek S.N. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118:3051–3061. doi: 10.1182/blood-2010-12-327858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crowther-Swanepoel D., Broderick P., Di Bernardo M.C., Dobbins S.E., Torres M., Mansouri M., Ruiz-Ponte C., Enjuanes A., Rosenquist R., Carracedo A., Jurlander J., Campo E., Juliusson G., Montserrat E., Smedby K.E., Dyer M.J., Matutes E., Dearden C., Sunter N.J., Hall A.G., Mainou-Fowler T., Jackson G.H., Summerfield G., Harris R.J., Pettitt A.R., Allsup D.J., Bailey J.R., Pratt G., Pepper C., Fegan C., Parker A., Oscier D., Allan J.M., Catovsky D., Houlston R.S. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat Genet. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Bernardo M.C., Crowther-Swanepoel D., Broderick P., Webb E., Sellick G., Wild R., Sullivan K., Vijayakrishnan J., Wang Y., Pittman A.M., Sunter N.J., Hall A.G., Dyer M.J., Matutes E., Dearden C., Mainou-Fowler T., Jackson G.H., Summerfield G., Harris R.J., Pettitt A.R., Hillmen P., Allsup D.J., Bailey J.R., Pratt G., Pepper C., Fegan C., Allan J.M., Catovsky D., Houlston R.S. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 22.Slager S.L., Rabe K.G., Achenbach S.J., Vachon C.M., Goldin L.R., Strom S.S., Lanasa M.C., Spector L.G., Rassenti L.Z., Leis J.F., Camp N.J., Glenn M., Kay N.E., Cunningham J.M., Hanson C.A., Marti G.E., Weinberg J.B., Morrison V.A., Link B.K., Call T.G., Caporaso N.E., Cerhan J.R. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood. 2011;117:1911–1916. doi: 10.1182/blood-2010-09-308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Admirand J.H., Knoblock R.J., Coombes K.R., Tam C., Schlette E.J., Wierda W.G., Ferrajoli A., O’Brien S., Keating M.J., Luthra R., Medeiros L.J., Abruzzo L.V. Immunohistochemical detection of ZAP70 in chronic lymphocytic leukemia predicts immunoglobulin heavy chain gene mutation status and time to progression. Mod Pathol. 2010;23:1518–1523. doi: 10.1038/modpathol.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rassenti L.Z., Huynh L., Toy T.L., Chen L., Keating M.J., Gribben J.G., Neuberg D.S., Flinn I.W., Rai K.R., Byrd J.C., Kay N.E., Greaves A., Weiss A., Kipps T.J. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 25.Schweighofer C.D., Huh Y.O., Luthra R., Sargent R.L., Ketterling R.P., Knudson R.A., Barron L.L., Jeffrey Medeiros L., Keating M.J., Abruzzo L.V. The B cell antigen receptor in atypical chronic lymphocytic leukemia with t(14;19)(q32;q13) demonstrates remarkable stereotypy. Int J Cancer. 2011;128:2759–2764. doi: 10.1002/ijc.25605. [DOI] [PubMed] [Google Scholar]
- 26.Delgado J., Matutes E., Morilla A.M., Morilla R.M., Owusu-Ankomah K.A., Rafiq-Mohammed F., del Giudice I., Catovsky D. Diagnostic significance of CD20 and FMC7 expression in B-cell disorders. Am J Clin Pathol. 2003;120:754–759. doi: 10.1309/FNGC-YEMJ-E3MA-E5L2. [DOI] [PubMed] [Google Scholar]
- 27.Matutes E., Owusu-Ankomah K., Morilla R., Garcia Marco J., Houlihan A., Que T.H., Catovsky D. The immunological profile of B-cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia. 1994;8:1640–1645. [PubMed] [Google Scholar]
- 28.McCarthy H., Wierda W.G., Barron L.L., Cromwell C.C., Wang J., Coombes K.R., Rangel R., Elenitoba-Johnson K.S., Keating M.J., Abruzzo L.V. High expression of activation-induced cytidine deaminase (AID) and splice variants is a distinctive feature of poor-prognosis chronic lymphocytic leukemia. Blood. 2003;101:4903–4908. doi: 10.1182/blood-2002-09-2906. [DOI] [PubMed] [Google Scholar]
- 29.Olshen A.B., Venkatraman E.S., Lucito R., Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman L, Rousseeuw P: Finding Groups in Data: An Introduction to Cluster Analysis. Edited by Wiley Series in Probability and Statistics. New York, Wiley & Sons, 1990
- 31.Shuster J.J. Median follow-up in clinical trials. J Clin Oncol. 1991;9:191–192. doi: 10.1200/JCO.1991.9.1.191. [DOI] [PubMed] [Google Scholar]
- 32.Burnham K.P., Anderson D.R. ed 2. Springer Verlag; New York: 2002. Model Selection and Multi-Model Inference: A Practical Information-Theoretic Approach. [Google Scholar]
- 33.Binet J.L., Lepoprier M., Dighiero G., Charron D., D’Athis P., Vaugier G., Beral H.M., Natali J.C., Raphael M., Nizet B., Follezou J.Y. A clinical staging system for chronic lymphocytic leukemia: prognostic significance. Cancer. 1977;40:855–864. doi: 10.1002/1097-0142(197708)40:2<855::aid-cncr2820400239>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 34.Rai K.R., Sawitsky A., Cronkite E.P., Chanana A.D., Levy R.N., Pasternack B.S. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–234. [PubMed] [Google Scholar]
- 35.Gunnarsson R., Staaf J., Jansson M., Ottesen A.M., Goransson H., Liljedahl U., Ralfkiaer U., Mansouri M., Buhl A.M., Smedby K.E., Hjalgrim H., Syvanen A.C., Borg A., Isaksson A., Jurlander J., Juliusson G., Rosenquist R. Screening for copy-number alterations and loss of heterozygosity in chronic lymphocytic leukemia: a comparative study of four differently designed, high resolution microarray platforms. Genes Chromosomes Cancer. 2008;47:697–711. doi: 10.1002/gcc.20575. [DOI] [PubMed] [Google Scholar]
- 36.Forconi F., Rinaldi A., Kwee I., Sozzi E., Raspadori D., Rancoita P.M., Scandurra M., Rossi D., Deambrogi C., Capello D., Zucca E., Marconi D., Bomben R., Gattei V., Lauria F., Gaidano G., Bertoni F. Genome-wide DNA analysis identifies recurrent imbalances predicting outcome in chronic lymphocytic leukaemia with 17p deletion. Br J Haematol. 2008;143:532–536. doi: 10.1111/j.1365-2141.2008.07373.x. [DOI] [PubMed] [Google Scholar]
- 37.Ma D., Chen Z., Patel K.P., Mishra B.M., Yao H., Abruzzo L.V., Medeiros L.J., Wierda W., Keating M., Sargent R., Luthra R. Array comparative genomic hybridization analysis identifies recurrent gain of chromosome 2p25.3 involving the ACP1 and MYCN genes in chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2011;11(Suppl 1):S17–S24. doi: 10.1016/j.clml.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapiro E., Leporrier N., Radford-Weiss I., Bastard C., Mossafa H., Leroux D., Tigaud I., De Braekeleer M., Terre C., Brizard F., Callet-Bauchu E., Struski S., Veronese L., Fert-Ferrer S., Taviaux S., Lesty C., Davi F., Merle-Beral H., Bernard O.A., Sutton L., Raynaud S.D., Nguyen-Khac F. Gain of the short arm of chromosome 2 (2p) is a frequent recurring chromosome aberration in untreated chronic lymphocytic leukemia (CLL) at advanced stages. Leuk Res. 2010;34:63–68. doi: 10.1016/j.leukres.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 39.Rinaldi A., Mian M., Kwee I., Rossi D., Deambrogi C., Mensah A.A., Forconi F., Spina V., Cencini E., Drandi D., Ladetto M., Santachiara R., Marasca R., Gattei V., Cavalli F., Zucca E., Gaidano G., Bertoni F. Genome-wide DNA profiling better defines the prognosis of chronic lymphocytic leukaemia. Br J Haematol. 2011;154:590–599. doi: 10.1111/j.1365-2141.2011.08789.x. [DOI] [PubMed] [Google Scholar]
- 40.Beroukhim R., Getz G., Nghiemphu L., Barretina J., Hsueh T., Linhart D., Vivanco I., Lee J.C., Huang J.H., Alexander S., Du J., Kau T., Thomas R.K., Shah K., Soto H., Perner S., Prensner J., Debiasi R.M., Demichelis F., Hatton C., Rubin M.A., Garraway L.A., Nelson S.F., Liau L., Mischel P.S., Cloughesy T.F., Meyerson M., Golub T.A., Lander E.S., Mellinghoff I.K., Sellers W.R. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puente X.S., Pinyol M., Quesada V., Conde L., Ordonez G.R., Villamor N. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quesada V., Conde L., Villamor N., Ordonez G.R., Jares P., Bassaganyas L. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 43.Wang L., Lawrence M.S., Wan Y., Stojanov P., Sougnez C., Stevenson K., Werner L., Sivachenko A., DeLuca D.S., Zhang L., Zhang W., Vartanov A.R., Fernandes S.M., Goldstein N.R., Folco E.G., Cibulskis K., Tesar B., Sievers Q.L., Shefler E., Gabriel S., Hacohen N., Reed R., Meyerson M., Golub T.R., Lander E.S., Neuberg D., Brown J.R., Getz G., Wu C.J. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah N., Decker W.K., Lapushin R., Xing D., Robinson S.N., Yang H., Parmar S., Tung S.S., O’Brien S., Fernandez-Vina M., Shpall E.J., Wierda W.G. HLA homozygosity and haplotype bias among patients with chronic lymphocytic leukemia: implications for disease control by physiological immune surveillance. Leukemia. 2011;25:1036–1039. doi: 10.1038/leu.2011.30. [DOI] [PubMed] [Google Scholar]
- 45.Crowther-Swanepoel D., Mansouri M., Enjuanes A., Vega A., Smedby K.E., Ruiz-Ponte C., Jurlander J., Juliusson G., Montserrat E., Catovsky D., Campo E., Carracedo A., Rosenquist R., Houlston R.S. Verification that common variation at 2q37.1, 6p25.3, 11q24.1, 15q23, and 19q13.32 influences chronic lymphocytic leukaemia risk. Br J Haematol. 2010;150:473–479. doi: 10.1111/j.1365-2141.2010.08270.x. [DOI] [PubMed] [Google Scholar]
- 46.Pennisi E. Genomics: 1000 Genomes Project gives new map of genetic diversity. Science. 2010;330:574–575. doi: 10.1126/science.330.6004.574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kaplan-Meier survival functions for variables independently predicting either time from diagnosis (Dx) to treatment (Rx) or time from diagnosis (Dx) to the date of last documented survival (OS) in multivariate models [log lactate dehydrogenase (LDH) and age are not shown because they were used as continuous variables]. BadCyto, cases with del11q22 or del17p13 (time to Rx) or with del6q21 or del17p13 (OS from Dx); CatB2M, categorization according to β-2-microglobulin serum level (high, >4 mg/L; low, ≤4 mg/L); CYTO2, categorization according to cytogenetic hierarchy; GoodCyto, without del11q22 or del17p13 (time to Rx) or without del6q21 or del17p13 (OSl from Dx); M, mutated; NEG, negative; POS, positive; U, unmutated.