Abstract
MCM7 is one of the pivotal DNA replication licensing factors in controlling DNA synthesis and cell entry into S phase. Its expression and DNA copy number are some of the most predictive factors for the growth and behavior of human malignancies. In this study, we identified that MCM7 interacts with the receptor for activated protein kinase C 1 (RACK1), a protein kinase C (PKC) adaptor, in vivo and in vitro. The RACK1 binding motif in MCM7 is located at the amino acid 221-248. Knocking down RACK1 significantly reduced MCM7 chromatin association, DNA synthesis, and cell cycle entry into S phase. Activation of PKC by 12-O-tetradecanoylphorbol-13-acetate dramatically decreased MCM7 DNA replication licensing and induced cell growth arrest. Activation of PKC induced redistribution of RACK1 from nucleus to cytoplasm and decreased RACK1-chromatin association. The MCM7 mutant that does not bind RACK1 has no DNA replication licensing or oncogenic transformation activity. As a result, this study demonstrates a novel signaling mechanism that critically controls DNA synthesis and cell cycle progression.
Miniature chromosome maintenance (MCM) proteins were initially identified from autonomously replicating sequence in Saccharomyces cerevisiae. Mutations of some of these proteins, such as MCM7 or MCM3 result in loss of the large chunk of yeast chromosomes in yeast. MCM7 cDNA encodes a 543-amino acid protein and is ubiquitously expressed in all tissues. A large body of studies indicate that MCM7 is a critical component of DNA replication licensing complex in the yeast and xenopus.1–4 Some studies suggest that MCM4, MCM6, and MCM7 complex contains DNA helicase activity.5,6 DNA replication licensing complex is multimeric and phase specific. In yeast, DNA replication licensing proteins, such as MCM2-7 and several replication origin binding proteins, such as Cdc6, germinin, and Cdt1, form DNA replication licensing complex in G1 phase to enable DNA replication and to promote cell cycle entry into S phase. Initial implication of MCM7 involvement in human malignancies came from positive immunostaining of MCM7 in several human malignancies, including endometrial carcinoma,7 melanoma,8 esophageal adenocarcinoma,9 colorectal adenocarcinoma,10 oral squamous cell carcinoma,11 glioblastoma,12 and thyroid cancer.13 The first study addressing the oncogenic role of MCM7 in prostate cancer came from genome analysis of prostate cancer by performing a genome wide copy number analysis using biotin-labeled genome DNA on Affymetrix U95av2 chip.14 The DNA copy number of MCM7 was found to increase severalfold accompanied with a concomitant increase of MCM7 mRNA level. Subsequent validation analyses suggest that either copy number and/or protein level increase of MCM7 are associated with prostate cancer relapse and metastasis. Amplification of MCM7 was also found in esophageal carcinoma.9 The magnitude of MCM7 amplification correlates with the expression of MCM7, tumor grades, and the aggressiveness of esophageal cancer.9 It is presumed that amplification of MCM7 is the driving force of MCM7 overexpression in primary human malignancies. MCM7 is probably the primary target of Rb, the tumor suppressor that controls cell entry into S phase.15 There is growing evidence that other signaling pathways also regulate MCM7 activity.
Receptor for activated protein kinase C 1 (RACK1), was initially identified as an adaptor of several protein kinase C (PKC) isoforms.16 The binding of RACK1 and PKC anchor PKC to its substrate to initiate second messenger signaling. It is suggested, according to recent studies that RACK1 interacts with a variety of other signaling molecules, including ras-GTPase activating protein,17 dynamin-1,18 src,19 integrins,20 PTPμ,21 phosphodiesterase,22 hypoxia induced factor-1,23 and so forth, that play an important role in several physiological processes, including, growth, hypoxia response, migration, adhesion, and cell differentiation. RACK1 only binds PKC activated by diacylglycerol or phorbol ester, but not quiescent PKC. In this study, we showed that RACK1 binds with MCM7 N-terminus. The MCM7/RACK1 interaction appears essential for DNA replication activity of MCM7.
Materials and Methods
Plasmid Construction
For construction of pCDNA4-TO-TO-MCM7, primers (5′-TGCATAAGCTTACGTTTCGCGCCAATTTCGGTT-3′/5′-TAGTTCTAGAGACAAAAGTGATCCGTGTCCGGGA-3′) corresponding to the sequence encompassing the full length MCM7 were used in a PCR, using template of PCR-MCM7 vector. The PCR product was restricted with HindIII and XbaI, and ligated into a similarly restricted pCDNA4-TO vector. To construct deletion mutant of MCM7, mutagenesis PCR was performed to generate upstream MCM7 fragment using primers 5′-TGCATAAGCTTACGTTTCGCGCCAATTTCGGTT-3′/5′-CCAGCACCGTGATACTACGAGGGATCATGAAAGTGGGAGACT-3′ and downstream MCM7 fragment using primers 5′-GATATGCCGATCCAGTCTCCCACTTTCATGATCCCTCGTAGTATCAC-3′/5′-TAGTTCTAGAGACAAAAGTGATCCGTGTCCGGGA-3′. These PCR products were combined and re-amplifed in PCR using primers 5′-TGCATAAGCTTACGTTTCGCGCCAATTTCGGTT-3′/5′-TAGTTCTAGAGACAAAAGTGATCCGTGTCCGGGA-3′. The PCR product was then restricted with HindIII and XbaI, and was ligated into similarly restricted pCDNA4-TO vector to produce 28 amino acid deletion (aa221-248). For construction of pET28a-RACK1 vector, a mutagenic primer set 5′-AACTGCTAGCATGACTGAGCAGATGACCCT-3′ and 5′-AACTGCGGCCGCTCTAGCGTGTGCCAATGGTCA-3′) was designed to create two restriction sites (NheI and NotI). A PCR was performed on pCMV-RACK1. The PCR product was restricted with NheI and NotI, and ligated into a similarly restricted pET28a vector to generate a Histag-RACK1 (aa2-317). To construct pCMV-RACK1, PCR product on RACK1 cDNA using primers 5′-CAGTGCCGGATCCCTCGTCGCTGCAGCGACACAC-3′/5′-TTATTTGGGTACCCTCTGCCATAAACTTCTAGCGTGTG-3′ was digested with BamH1 and KpnI. The digested product was ligated into similarly digested pCMVscript.
Construction of Inducible MCM7 and ΔMCM7 Expression in PC-3 Cell Line
The plasmid pCDNA4-TO-MCM7 or the pCDNA4-TO-ΔMCM7 was then co-transfected into PC3 cells with pcDNA6. Transfected cells were selected with 500 μg/mL Blasticidin and 1 μg/mL Zeocin (Invitrogen, Carlsbad, CA). Clones were expanded and tested for inducible MCM7 expression by exposure to 5 μg/mL tetracycline and by Western blot analysis with an antibody specific for MCM7 and β-actin. For RACK1 knockdown assays, siRNA specific for RACK1 (5′-GAGAGGUUGUGGUGCUAGUUUCUCdT-3′/5′-AGAGAAACUAGCACCACAACCUCUCCU-3′) or for PKCBii (5′-CCACAAUGACGACCUGCUUUGAUdTdT-3′/5′-AAAUCAAAGCAGGUCGUCAUUGUGGUU-3′) or scramble siRNA (5′-UAAUGUAUUGGAACGCAUAUU-3′/5′-UAUGCGUUCCAAUACAUUA-3′) was transfected into cultured cells using lipofectamine 2000 (Invitrogen). The detailed procedure followed the manufacturer’s manual.
Yeast Two-Hybrid Analysis
The yeast-competent cell preparation was previously described.24 One hundred microliters of freshly prepared competent AH109 cells were mixed with 0.25 to 0.50 μg plasmid DNA plus 0.5 μg DNA from prostate yeast two-hybrid cDNA library constructed in pACT2 in 0.5 mL of polyethylene glycol/LiAc, incubated at 30°C for 30 minutes. After this initial incubation with plasmid DNA, the cell solution was combined with 20 μL of DMSO and subjected to incubation for 15 minutes at 42°C. The cells were pelleted, resuspended in 1 mL YPD medium, and shaken at 30°C for 40 minutes. The transformed cells were then pelleted, resuspended in 0.5 mL 0.9% NaCl, and plated onto the appropriate SD agar plate. The transformants were first plated on low and medium stringency plates of SD-Leu/-Trp and SD-Leu/-Trp/-His, respectively. The grown colonies were subjected to the β-galactosidase assay, as previously described,24 and were allowed to grow further in the high stringency plate (SD-Ade/-His/-Leu/-Trp).
Immunoprecipitation
Protein extracts of PC3 cells or RWPE1 cells were incubated with MCM7 (mouse monoclonal, 1:400) or RACK1 (1:500, goat polyclonal) antibodies for 16 hours, then with protein G Sepharose for 3 hours. The complex was washed five times with radioimmunoprecipitation assay buffer, and the bound proteins were eluted with SDS-PAGE sample buffer. The bound RACK1 or MCM7 was electrophoresed in 10% SDS-PAGE and immunoblotted with anti-RACK1 antibodies or MCM7 antibodies (1:2000).
GST Fusion Proteins Pull Down to Examine RACK1/MCM7 Binding
The Escherichia coli cells harboring phospho glutathione S-transferase (pGST)-MCM7 mutants or pGST were grown in 100 mL of Luria-Bertani medium supplemented with 100 μg/mL ampicillin overnight and induced by a final concentration of 1 mmol/L isopropyl-L-thio-B-D-galactopyranoside for 3 hours. The cells were then pelleted, resuspended in 1× PBS, and sonicated for 2 minutes. The proteins were solubilized in 1% triton X-100. The supernatant was collected after centrifugation at 15,000 g for 5 minutes. The GST, GST-MCM7c, GST-MCM7m, GST-MCM7n, and other MCM7 mutant fusion proteins were purified through a Glutathione-Sepharose 4B column (Amersham Bioscience, Piscataway, NJ). The PC3 protein extracts were pre-cleared with the column for 15 minutes at 4°C. The flow-through was collected after spinning at 3000 g for 1 minute. The pre-cleared cell lysates were then incubated with GST fusion protein-packed glutathione-Sepharose 4B at 4°C for 2 hours. The column was spun at 3000 g at room temperature for 1 minute and further washed twice with PBS. The proteins were eluted from the column with 40 μL of SDS-PAGE gel sample loading dye. SDS-PAGE and Western blot analyses were subsequently conducted.
BrdU Labeling Analysis
To perform bromodeoxyuridine (BrdU) labeling analysis, 10 μL of BrdU solution (1 mmol/L BrdU in 1× PBS) was added to 1 mL of tissue culture media. The treated cells were then incubated for 3 hours at 37°C. Cells were then resuspended with 100 μL of BD Cytofix/Cytoperm Buffer per sample (BD Pharmagen, San Jose, CA) and incubated for 30 minutes at room temperature. The cells were then pelleted and washed with 1 ml of 1× BD Perm/Wash Buffer. The cells were then incubated with Cytoperm Plus Buffer for 10 minutes on ice. The permeation procedure was repeated twice. The cells were resuspended with 100 μL of diluted DNaseI (diluted to 300 μg/mL in Dulbecco's phosphate-buffered saline) per tube, (ie, 30 μg of DNaseI to each tube), and were incubated for 1 hour at 37°C. The cells were washed with 1× BD Perm/Wash Buffer and incubated with 50 μL of BD Perm/Wash Buffer containing diluted fluorescent anti-BrdU antibody (1:50) and propidium iodide for 20 minutes at room temperature. The incubation was washed with 1 mL of 1× BD Perm/Wash Buffer. The immune-reactivity of BrdU was assessed in LSC-II flowcytometer.
Immunofluorescence Staining
PC3 or RWPE1 cells were cultured on chamber slides for 24 hours. The slides were washed with PBS twice. The cells were fixed with 4% paraformaldehyde for 1 hour at room temperature as previously described.25,26 After washing the slides with PBS twice, the cells were blocked with 5% bovine serum albumin with 0.1% Triton X-100. The cells were then incubated with mouse monoclonal antibody against RACK1 (1:100, Santa Cruz Biotechnology, Santa Cruz, CA) or mouse monoclonal antibody against RACK1 and rabbit antisera against MCM7 (1:100, Santa Cruz Biotechnology) at room temperature for 1 hour. The slides were then washed with PBS twice. Secondary antibodies from donkey against mouse (fluorescein isothiocyanate conjugated, 1:200) or goat against rabbit (tetramethylrhodamine-isothiocyanate– conjugated, 1:200) were added and incubated at room temperature for 1 hour. The slides were then washed with PBS twice before addition of 4α−6-diamidino-2-phenylindole. After additional washes with PBS, slides were mounted with immunomounting buffer. Immunofluorescence staining was examined under confocal microscope.
Chromatin Association Assay
Cells were cultured to 75% confluence, synchronized in serum-free medium for 24 hours, and then treated with tetracycline for 24 or 48 hours. These cells were washed with PBS and trypsinized. These cells were resuspended in 1 mL of Buffer A [110 mmol/L KC2H3O2, 15 mmol/L NaC2H3O2, 2 mmol/L MgC2H3O2, 0.5 mmol/L EGTA, and 20 mmol/L HEPES (pH 7.3)]. The cell suspension was treated to the final concentration of 2 mmol/L DTT and 50 μg/mL digitonin. The cells were incubated at 4°C for 10 minutes in a rotator. Nuclei were pelleted by centrifugation at 1500 g for 10 minutes. They were resuspended in hypotonic buffer [Buffer B: 1 mmol/L HEPES (pH 7.5), 0.5 mmol/L EDTA supplemented with 0.5% NP40]. The nuclear suspensions were then incubated at 4°C for 15 minutes in a rotator and laid on top of a 10-mL sucrose cushion [100 mmol/L sucrose and 0.5 mmol/L Tris-HCl (pH 8.5)] and centrifuged at 3500 g for 15 minutes at 4°C. The chromatin pellets were suspended in 0.25 mmol/L EDTA (pH 8.0) and sonicated for 10 seconds twice for each sample. The chromatin suspensions were centrifuged twice at high speed for 10 minutes at 4°C and the supernatants were retained.
Colony Formation Assay
Colony formation assay was performed similarly, as previously described.27 Five thousand or 10,000 cells were cultured in 60-mm dishes. Triplicate experiments were performed for each cell clones. Medium containing 10% fetal bovine serum (Invitrogen) was changed every 4 days. On day 7, the plates were stained with 1% crystal violet and the colonies with a diameter of >2 mm were counted.
Helicase Assay
MCM7 immunoprecipitates were incubated with a PCR product of a pUC19 template generated with primers 5′-CAAGTTGGGAAGACAACCTG-3′/5′-Cy5-CCAATATGGTGAAACCCCGT-3′ using the following condition: 20 mmol/L Tris-HCl, pH 7.4, 50 mmol/L NaCl, 3 mmol/L MgCl2, 2 mmol/L ATP, 20% glycerol, 0.1% bovine serum albumin, and capture probes (Biotin-5′-CAAGTTGGGAAGACAACCTGTAGGGCCTGCGGGGT-3′) for 30 minutes at room temperature. The reaction was stopped by 170 mmol/L EDTA, pH 8.0. The probes were then captured with streptavidin-agarose beads, and washed with Tris-Tween-20 buffer three times. Cy5 intensity was then quantified with Affymetrix 426 scanner. Triplicate experiments were performed for each assay. Assays with agarose beads without streptavidin were used as background controls. WT MCM7 immune complexes from cells not treated with siRNA were used as positive controls.
Results
To investigate signaling molecules that interact and regulate MCM7 activity, we performed a yeast two-hybrid screening analysis on a prostate cDNA library using MCM7 full length protein fused with the binding domain of GAL4 gene. Using pBD-MCM7, we have identified 36 positive colonies after three rounds of metabolic screening of a prostate yeast two-hybrid cDNA library. These colonies were subsequently isolated. After several restriction enzyme digestions, several redundant clones were eliminated. Eight unique clones were identified and sequenced. One of these clones contains a cDNA encoding RACK1.
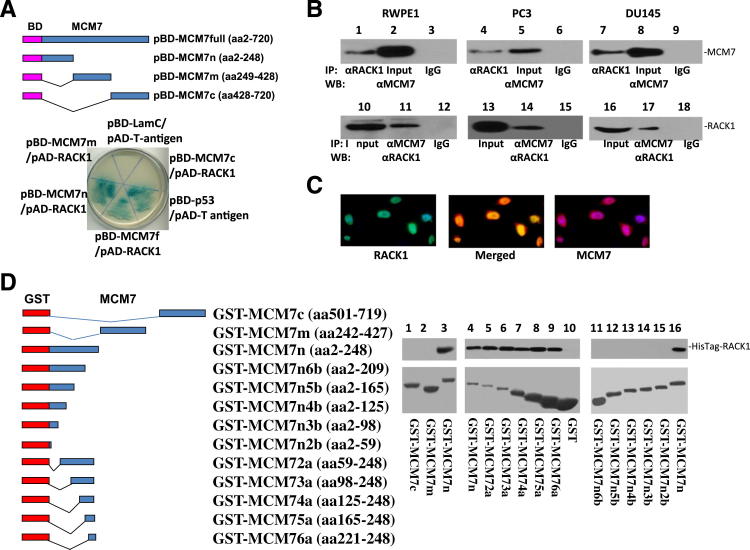
To validate the yeast two-hybrid screening results, pAD-RACK1 and pBD-MCM7 were co-transfected into Yeast AH109 cells, grown in high stringency medium, and tested for α-galactosidase activity. Both pBD-MCM7 (full length) and pBD-MCM7n (N-terminus) showed positive galactosidase activity, whereas the C-terminus or mid-segment of MCM7 was negative, suggesting that the RACK1 binding activity is mediated by a region located in the N-terminus of MCM7 (Figure 1A). Among prostate cell lines, RACK1 is abundantly expressed in RWPE-1 cells (immortalized prostate epithelial cells), PC3, and DU145 cells (prostate cancer cells). To verify the interaction, an in vivo MCM7-RACK1 binding analysis was performed in protein extracts of RWPE1, PC3, and DU145 cells. As shown in Figure 1B, co-immunoprecipitation of MCM7 and RACK1 was readily apparent. To visualize whether MCM7 and RACK1 co-localize in the nucleus, double immunofluorescence staining using antibodies against MCM7 and RACK1 were performed in RWPE1 cells. As demonstrated in Figure 1C, significant amount of RACK1 was colocalized with MCM7 in the nuclei of RWPE1 cells. Similar colocalization results were obtained with PC3 and DU145 cells (data not shown).
Figure 1.
N-terminus of MCM7 binds with RACK1. A: Constructs of full length (pBD-MCM7full), N-terminus (pBD-MCM7n), mid segment (pBD-MCM7m), and C-terminus (pBD-MCM7c) of MCM7 with bait domain (BD) in yeast two-hybrid analysis. Co-transformants of pBD-MCM7full or pBD-MCM7n, or pBD-MCM7m or pBD-MCM7c with pAD-RACK1 on SD agar plate with high stringent nutrient selection (SD-leu-Trp-His-Ade) are shown. Co-transfection of pBD-p53 and pAD-T-antigen is the positive control, whereas co-transfection of pBD-LamC and pAD-T-antigen is the negative control. B: Co-immunoprecipitation of MCM7 (lanes 1 to 9) or RACK1 (lanes 10 to 18) using antibodies specific for RACK1 or MCM7 to immunoprecipitate from RWPE1 (left panels), PC3 (middle panels), and DU145 cells (right panels). The immunoprecipitates were blotted with the indicated antibodies. C: Immunofluorescence staining of RWPE-1 cells with antibody against RACK1 bound by fluorescein isothiocyanate-conjugated secondary antibody for mouse and antibody specifically against MCM7 recognized by TRITC conjugated secondary antibody for rabbit. D: Mapping binding motif of RACK1 on MCM7. Left: Constructs of series of MCM7 deletion mutants with GST expression vectors. Right: Binding assays on GST or GST-MCM7 deletion mutants with RACK1 from PC3 cells (lanes 1 to 3) or HisTag-RACK1 (lanes 4 to 16). The bound RACK1 was blotted with anti-RACK1 antibodies. Top blots: Immunoblots with antibodies specific for RACK1. Bottom blots: Coomassie staining of proteins.
To validate the interaction between MCM7 N-terminus and RACK1 in vitro, fragments of the N-terminus (247 amino acids), mid-segment (186 amino acids), and C-terminus (219 amino acids) of MCM7 were constructed into pGEX-5T to create GST-MCM7n, GST-MCM7m, and GST-MCM7c fusion proteins, respectively. In vitro MCM7/RACK1 binding assays were performed using RACK1 extracted from PC3 cell line and these fusion MCM7 proteins. The results of the binding assays indicate that GST-MCM7n binds with RACK1 in the cell free system (Figure 1D). To rule out potential bridging proteins between RACK1 and MCM7 interaction, RACK1 cDNA was ligated into pET-28 vector to create a HisTag-RACK1 fusion protein. The binding analysis with the recombinant HisTag-RACK1 and GST-MCM7 N-terminus (2-248 amino acids) shows that the RACK1 binds with MCM7 N-terminus directly (Figure 1D). These results indicate that the interaction between MCM7 and RACK1 is direct and does not require bridge protein in their interaction. A series of deletion mutants of GST-MCM7n were constructed to identify the motifs that are required to interact with RACK1. A stretch of 28 amino acid sequence located in 221-248 of MCM7 was found crucial for MCM7 binding with RACK1, because the fusion proteins deleted of this sequence did not bind with RACK1, whereas all proteins containing this sequence bound with RACK1 (Figure 1D).
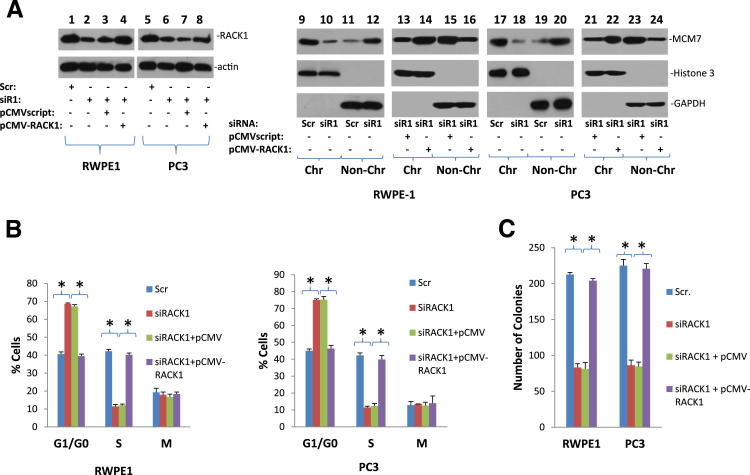
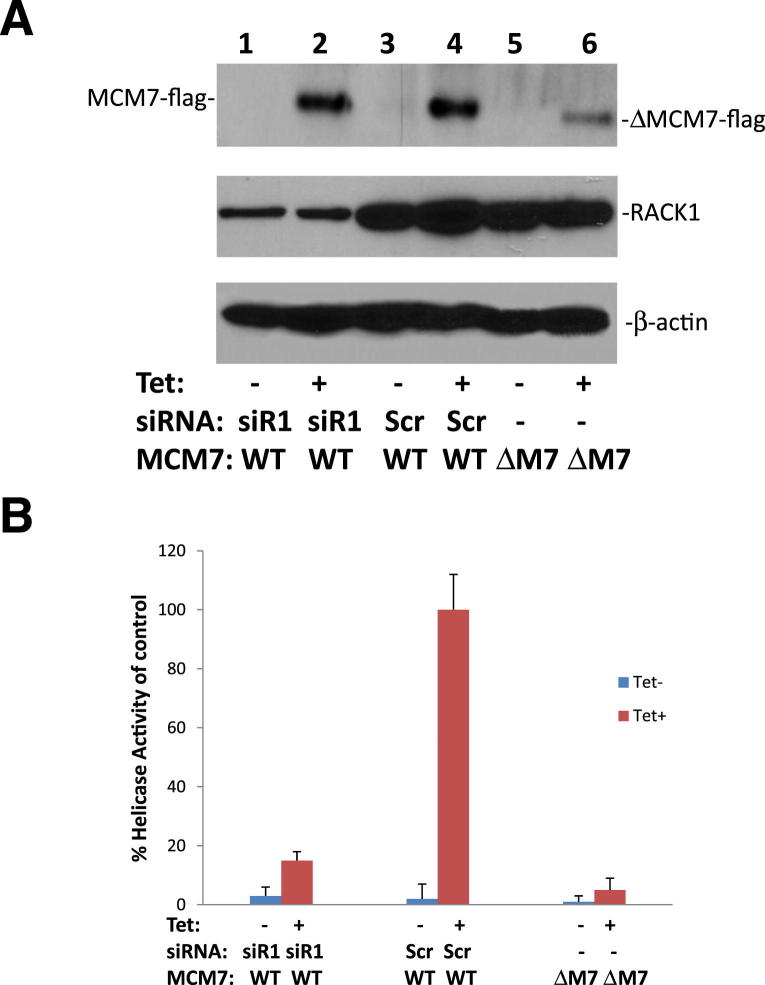
RACK1 is an adaptor of protein kinase C and several other signaling molecules. To investigate whether interaction of RACK1 and MCM7 has impact on the function of MCM7, RACK1 protein expression was knocked down using siRNA specific for RACK1 5′UTR and MCM7 chromatin association analysis was performed. As shown in Figure 2A, knocking down of RACK1 in either RWPE1 or PC3 cells significantly reduced MCM7 association with chromatin. To rule out off-target effects of siRACK1, RWPE1, and PC3 cells that were co-transfected with siRACK1 and pCMV-RACK1, which does not contain the sequence corresponding to siRACK1. The chromatin associated MCM7 was reversed on the expression of this exogenous RACK1. On knocking down of RACK1, there was a 20% reduction of BrdU labeling in RWPE1 cells and a 30% drop (P < 0.001) of cells entering S phase and concomitant 26% increase of G0/G1 phase for RWPE1 cells (P < 0.001) (Figure 2B). For PC3 cells, the decrease of S phase is 45% (P < 0.001) and the increase of G0/G1 phase is 25% (P < 0.001). Similar to chromatin association analyses, the cell growth arrest effects were reversed with pCMV-RACK1. Knocking down of RACK1 also reduced colony formation of both RWPE1 and PC3 cells by 47% (P < 0.01) and 45% (P < 0.01) (Figure 2C), respectively. Furthermore, knocking down of RACK1 had a dramatic negative impact on MCM7 helicase activity (Figure 3). Mutant MCM7 that does not interact with RACK1 had limited helicase activity. These results clearly suggest that RACK1 plays an important role in inducing MCM7 DNA replication licensing, in advancing cell cycle, and in cell growth.
Figure 2.
RACK1 is essential for MCM7 chromatin association and DNA synthesis. A: Knocking down of RACK1 reduced MCM7 chromatin association. Lanes 1 to 8 represent immunoblot analyses of RACK1 expression in RWPE1 and PC3 cells treated with siRNA specific for RACK1 (siR1) or scramble control (Scr) or pCMV-RACK1 or its vector control (pCMVscript). The chromatin (Chr) and non-chromatin (Non-Chr) fractions of these cells were purified and immunoblotted with antibodies specific for MCM7. Antibodies against histone 3 and glyceraldehydes-3-phosphate dehydrogenase were used as purity controls. Lanes 9 to 16 represent RWPE1 cells; lanes 17 to 24, PC3 cells. B: Knocking down of RACK1 reduced cell entry to S phase. RWPE1 (left panel) or PC3 (right panel) cells were synchronized at G0/G1 phase by serum starvation. These cells were treated with siR1 or Scr or pCMV-RACK1 or pCMVscript as of (A), and labeled with BrDU. Cell cycles and DNA synthesis were then quantified 4 hours after serum stimulation. *P < 0.01. C: Knocking down of RACK1 reduced colony formation of RWPE1 and PC3 cells. These cells were treated with siR1 or Scr or pCMV-RACK1 or pCMVscript as in A. The cells were grown in medium supplemented with 10% FBS for 7 days. Colonies larger than 2 mmol/L were counted. *P < 0.01.
Figure 3.
Helicase activity of MCM7 is enhanced by RACK1. A: Immunoblot analysis of MCM7 (WT), −ΔMCM7Δ221-248 (ΔM7), RACK1, and β-actin from pcDNA4-TO-MCM7/pCDNA6 (lanes 1 to 4) or pcDNA4-TO–ΔMCM7Δ221-248/pCDNA6 transformed (lanes 5 and 6) PC3 cells. These cells were transfected with siRNA specific for RACK1 (siR1) or scramble control (Scr) as indicated. B: Helicase activities of MCM7 immunocomplex. MCM7 immunocomplex was immunoprecipitated with antibodies specific for FLAG tag. Helicase assays were performed using a Cy5-labeled DNA duplex template and biotin-labeled oligo capture probe.
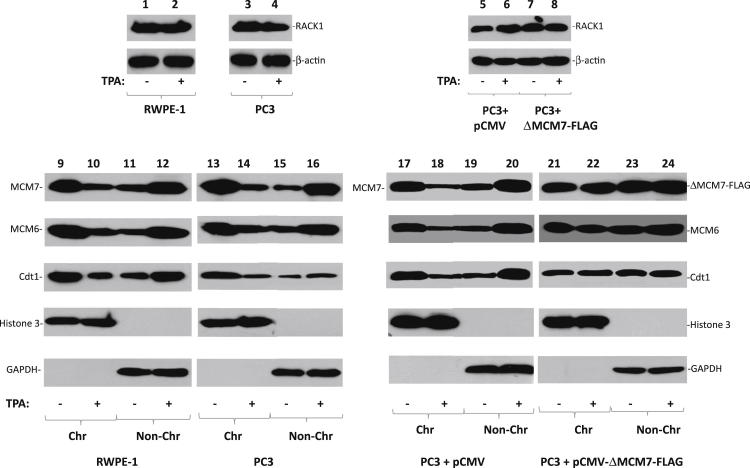
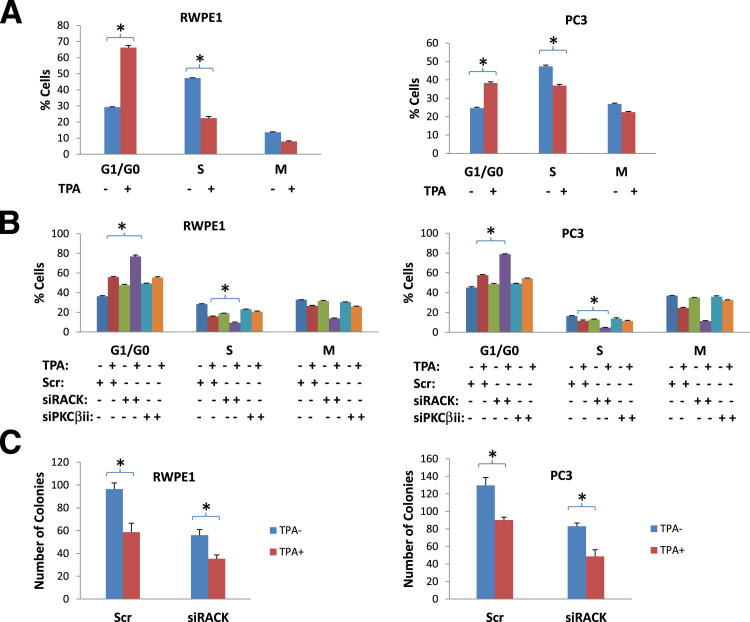
To investigate the effect of protein kinase C (PKC)-RACK1 signaling pathway on MCM7 DNA replication licensing activity, RWPE1 and PC3 cells were treated with 1 mg/mL PKC activator, 12-O-tetradecanoylphorbol-13-acetate (TPA). These cells were then examined for MCM7 replication licensing activity. As shown in Figure 4, activation of PKC is accompanied with significant dissociation of MCM7 from the chromatin fraction in both RWPE1 and PC3 cells. Interestingly, the activation of PKC not only reduced MCM7 chromatin association but also other components of DNA replication licensing complex, such as Cdt1 and MCM6 (Figure 4), suggesting a disintegration of DNA replication complex on PKC activation. Mutant MCM7 that is deleted of amino acid 221-248 and is defective of RACK1 interaction was not responsive to TPA stimulation, and appeared to inhibit other licensing factor for chromatin de-association (Figure 4). Indeed, treatment of PC3 and RWPE1 cells with TPA induced dramatic cell cycle arrest with an increase of G0/G1 phase by more than 100% in RWPE1 cells (P < 0.001) and 56% (P < 0.001) in PC3 cells, and a decrease of S phase by 49% (P < 0.001) in RWPE1 and 27% (P = 0.002) in PC3 cells (Figure 5A). Knocking down of RACK1 enhanced the TPA-induced growth arrest (G0/G1 phase) effect by an additional 50% (P < 0.001) in RWPE1 cells and 35% (P < 0.001) in PC3 cells (Figure 5B). Knocking down of PKCβII, the major RACK1-binding PKC family member, resulted in significant blunting of cell growth arrest induced by TPA (23.7% of scramble control on S phase reduction of RWPE1 cells, P < 0.05, and 38.5% for PC3 cells, P < 0.05) (Figure 5B). Colony formation analyses also showed that treatment of TPA dramatically inhibited colony formation (Figure 5C). These results indicate that PKC-RACK1 signaling serves to inactivate RACK1/MCM7 DNA replication licensing, to block cell entry into S phase, and to inhibit cell growth in the prostate cancer and immortalized prostate epithelial cells.
Figure 4.
Activation of PKC inhibits MCM7 chromatin association. RWPE1 (lanes 1 and 2) or PC3 (lanes 3 and 4) or PC3 cells transfected with pCMVscript (lanes 5 and 6) or pCMV-ΔMCM7-FLAG (lanes 7 and 8) were treated with 1 mg/mL of TPA for 24 hours. These cells were harvested. The chromatin (Chr) and non-chromatin (non-Chr) fractions were purified and immunoblotted with antibodies specific for FLAG, MCM7, MCM6, and Cdt1. Antibodies against histone 3 and glyceraldehydes-3-phosphate dehydrogenase were used as purity controls. Immunoblots using antibodies specific for β-actin were used for normalization. Lanes 9 to 12: RWPE1 cells treated with or without TPA. Lanes 13 to 16: PC3 cells treated with or without TPA. Lanes 17 to 20: PC3 transfected with pCMVscript treated with or without TPA. Lanes 21 to 24: PC3 cells transfected with pCMV-ΔMCM7-FLAG treated with or without TPA.
Figure 5.
Activation of PKC inhibits cell entry to S phase and cell growth. A: TPA inhibits cell entry to S phase. RWPE1 (left) and PC3 (right) cells were synchronized at G0/G1 phase by serum starvation. These cells were treated with or without 1 mg/mL TPA, labeled with BrdU, and stimulated with 10% fetal bovine serum. Cell cycles and DNA synthesis were then quantified 4 hours after serum stimulation. *P < 0.01. B: Knocking down of RACK1 enhances TPA-mediated cell growth arrest. RWPE1 (left) and PC3 (right) cells were synchronized at G0/G1 phase by serum starvation. These cells were silenced with siRACK, or siPKCβii, or Scr control. These cells were then treated with or without 1 mg/mL TPA, labeled with BrDU, and stimulated with 10% fetal bovine serum. Cell cycles and DNA synthesis were then quantified 4 hours after serum stimulation. *P < 0.01. C: Colony formation of RWPE1 (left) and PC3 (right) cells treated with or without TPA, in the presence of siRACK or Scr. *P < 0.05.
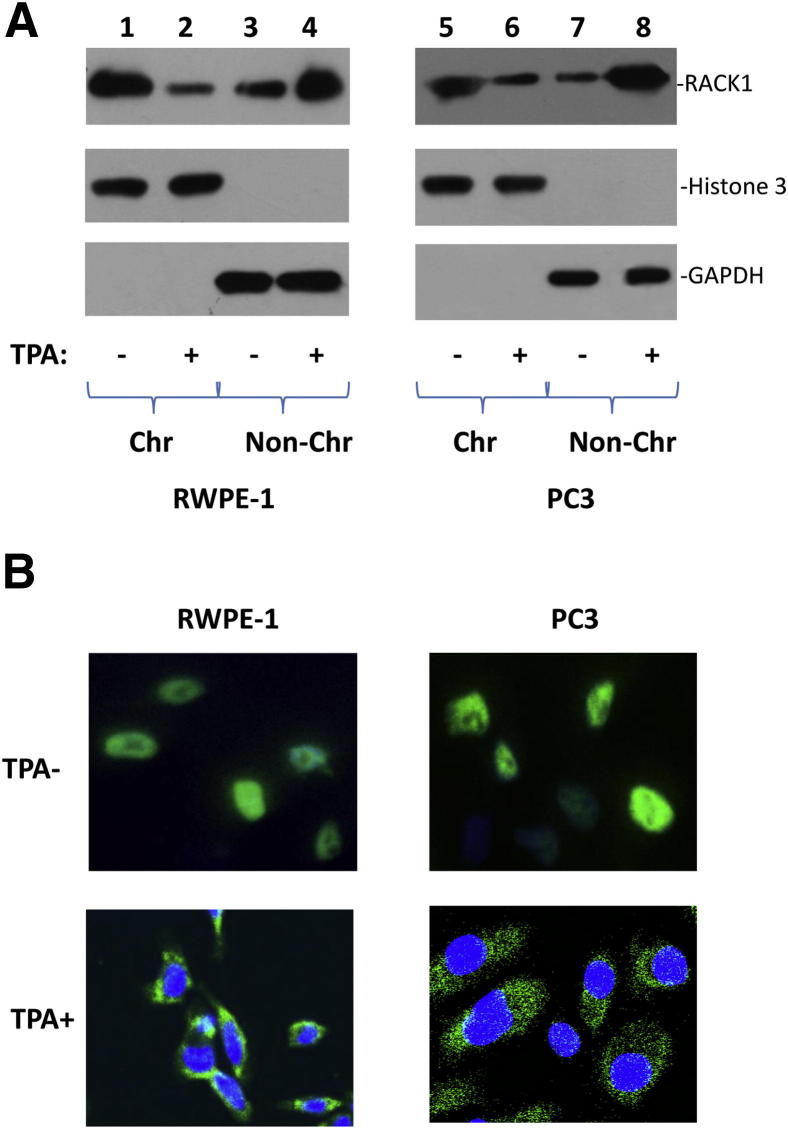
To investigate whether RACK1 binding of MCM7 leads to RACK1 chromatin association, chromatin association analysis of RACK1 was analyzed. As shown in Figure 6, a significant amount of RACK1 was found in chromatin fraction. On stimulation of TPA, however, dramatic dissociation of RACK1 from chromatin occurred in both RWPE1 and PC3 cells. These findings are accompanied with an increase of RACK1 in the cytoplasm after treatment of TPA (Figure 6B). These findings suggest that PKC activation prevents entry of RACK into the nucleus, and thus the binding with MCM7.
Figure 6.
Activation of PKC inhibits RACK1 association with chromatin. A: Phorbol ester treatment reduced RACK1 chromatin association. RWPE1 or PC3 cells were treated with or without 1 mg/mL TPA for 24 hours. These cells were harvested. The chromatin (Chr) and non-chromatin fractions (Non-Chr) were purified and immunoblotted with antibodies specific for RACK1. Antibodies against histone 3 and glyceraldehydes-3-phosphate dehydrogenase were used as purity controls. Lanes 1 to 4: RWPE1 cells treated with or without TPA. Lanes 5 to 8: PC3 cells treated with or without TPA. B: RACK1 redistribution from the nucleus to cytoplasm on treatment of phorbol ester. RWPE1 or PC3 cells were treated with or without 1 mg/mL TPA for 24 hours. Immunofluorescence staining of these cells using antibodies specific for RACK1 was performed. Representative images were shown.
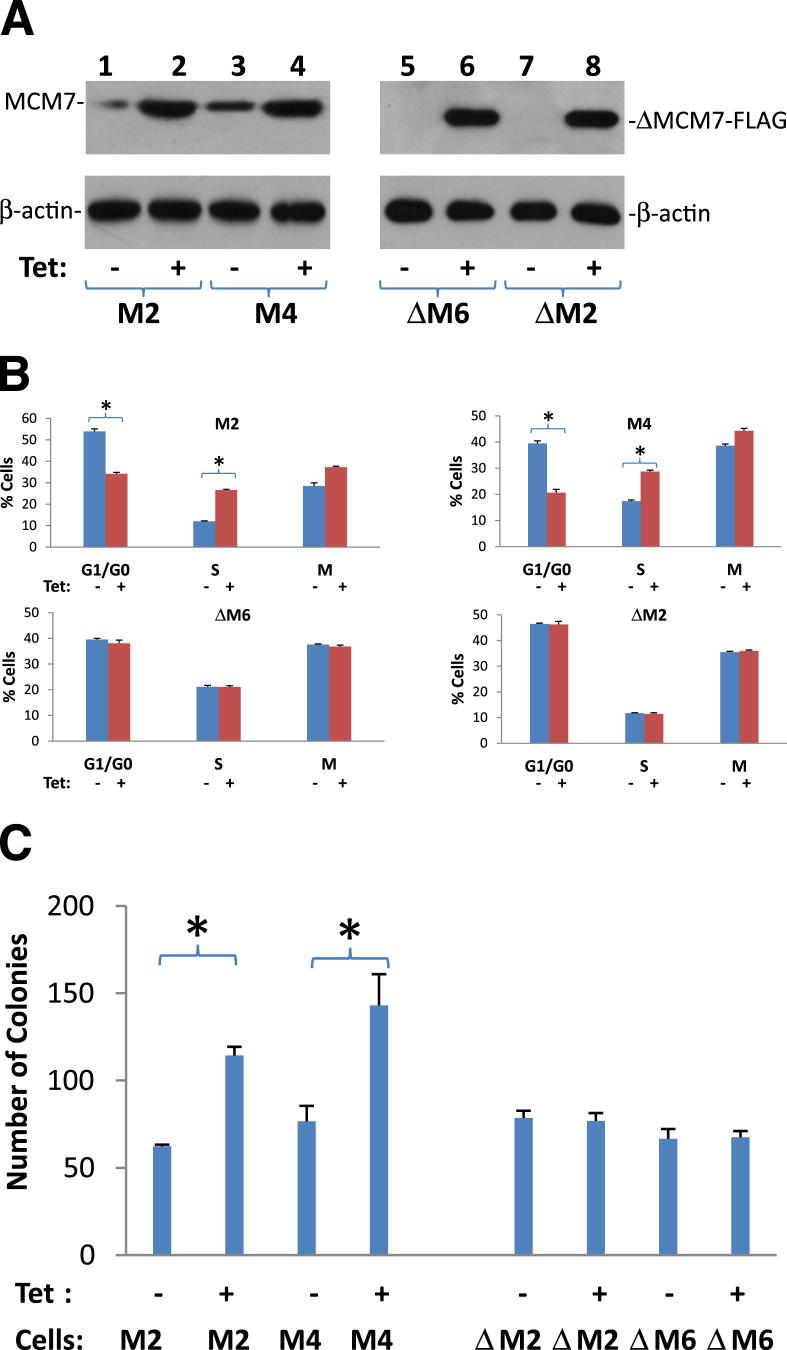
To investigate whether RACK1-MCM7 interaction is essential for chromatin association activity of MCM7, a MCM7 mutant with 28 amino acid RACK1 interaction motifs (amino acids 221-248) deleted, was inserted into pCDNA4-TO to create pCDNA4-TO-ΔMCM7. This vector was subsequently transfected into PC3 cells. Two clones (ΔM2 and ΔM6) with tetracycline inducible expression of mutant MCM7 were selected for further analyses. As shown in Figure 7, mutant MCM7 did not generate additional S phase entry in cell cycle analysis, even though its wild-type counterpart significantly increased the S phase and decreased G0/G1 phases, supporting that RACK1 interaction is necessary for MCM7 DNA replication licensing. Overall, these results support the fact that interaction of RACK1/MCM7 plays an important role in DNA replication licensing and activation of PKC leads to redistribution of RACK1 and inhibition of RACK1/MCM7 mediated replication licensing.
Figure 7.
Binding with RACK1 is essential for MCM7 DNA replication and transformation activity. A: Immunoblot analysis of mutant MCM7 that does not bind RACK1 in pCDNA4-TO-ΔMCM7Δ221-248/pCDNA6 transformed PC3 cells (lanes 7 and 8, ΔM2; lanes 5 and 6, ΔM6) and MCM7 wild-type controls (lanes 1 and 2, M2; lanes 3 and 4, M4). These cells were induced to express mutant or wild-type MCM7 with 5 μg/mL tetracycline, and immunoblotted with antibody specific for FLAG-tag. Immunoblots with antibodies specific for β-actin were used for normalization. B: Cell cycle analysis of duplicate experiments from A. *P < 0.05. C: Colony formation analysis of mutant MCM7 (ΔM2 and ΔM6) or its wild-type counterpart (M2 and M4) from A. *P < 0.05.
Discussion
MCM7 is one of the major components of DNA replication licensing complex. Numerous studies suggest that MCM7 is a pivotal point in regulating DNA replication licensing activity by interacting with several signaling molecules,28 including retinoblastoma gene, androgen receptor, and integrin linked kinase. The present study indicates that RACK1 interacts and activates MCM7 both in vivo and in vitro. Several lines of evidence support that MCM7 binds with RACK1. First, both full length and the N-terminus of MCM7 binds RACK1 in the yeast two-hybrid system and cell free in vitro binding assays. The cell-free recombinant MCM7 N-terminus and Histag-RACK1 binding suggests that no bridge protein is involved in MCM7/RACK1 interaction. Second, both MCM7 and RACK1 are readily co-immunoprecipitated from prostate epithelial cells and prostate cancer cells by antibodies either against RACK1 or MCM7, suggesting MCM7/RACK1 interaction occurring intracellularly. Third, MCM7 and RACK1 are co-localized in the nucleus of immortalized prostate epithelial and prostate cancer cells. Interestingly, RACK1/MCM7 interaction appears to occur only in nucleus because no colocalization MCM7/RACK1 was detected in the cytoplasm of these cells. By identifying MCM7/RACK1 interaction, this study reveals a critical role of MCM7 in relaying signaling from stimulation of PKC to DNA replication. PKC is a family of kinases regulated by calcium, diacylglycerol and phorbol ester.29 Depending on their response to calcium and phorbol ester, PKC is generally divided into three different classes: classical PKC that responds to both calcium and phorbol ester; novel PKC that responds to lipid ligands such as diacylglycerol and phorbol ester but not calcium; atypical PKC responds to none of these stimulants. Depending on the circumstances, PKC can be either an oncogene or a tumor suppressor gene.30 Indeed, several novel and classical PKC members have been shown to be anti-proliferative in prostate epithelial cells.31,32 The mechanism for PKC-mediated anti-proliferative activity in prostate cancer and prostate epithelial cells has been elusive. This study, to our best knowledge, provides the first direct link of PKC signaling in regulating cell proliferation by revealing a novel signaling pathway of PKC-RACK1-MCM7.
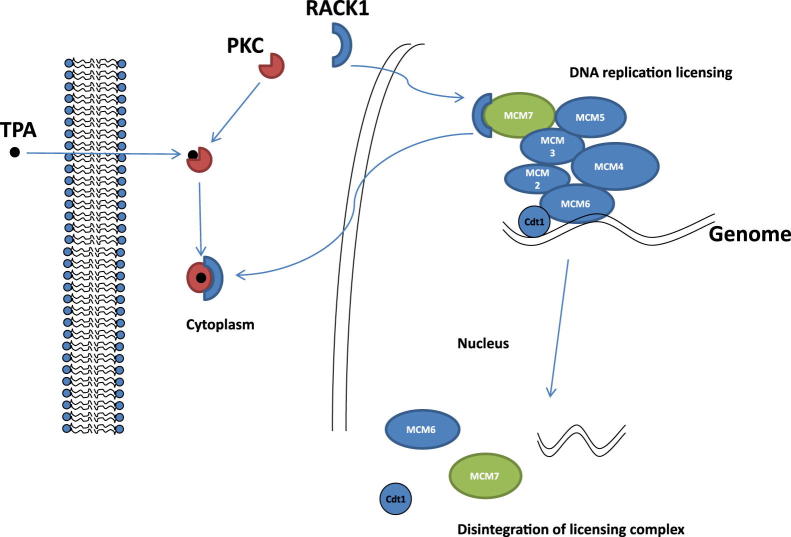
RACK1 has been shown to be an important adaptor for several signaling molecules, including PKC,33 integrin,34 and c-abl.35 RACK1 also plays an important role in several cellular physiological processes. In contrast to previous studies, our analysis reveals that RACK1 is a versatile protein, located in both cytoplasm and the nucleus. A significant amount of RACK1 is located in the nucleus and is associated with chromatin. Removal of RACK1 from the chromatin complex by knocking down RACK1 protein expression or removal of RACK1-binding motif from MCM7 severely compromised the DNA replication licensing activity, and results in stalling of entry into S phase. As a result, RACK1 appears to play a critical role in DNA replication licensing, far beyond an adaptor role for other signaling molecules. It is not entirely clear what drives RACK1 redistribution from the nucleus to cytoplasm on PKC activation. Because quiescent PKCs do not bind RACK1, it is possible that the binding of RACK1 by activated PKC alters the structural conformation and leads to shifting RACK1 from the nucleus to cytoplasm. Alternatively, competitive binding of activated PKC and MCM7 to RACK1 is a possible mechanism. Based on these findings, we argue that one of the natural physiological roles of RACK1 is a replication co-factor in DNA replication machinery (Figure 8). The DNA replication licensing factors may not be limited to factors of a core complex, such as Cdt1, geminin, MCM components, but also to factors dictating the function of this complex, such as AR, Rb, and RACK1. Interestingly, the AR and RACK1 binding motif in MCM7 overlap. While RACK1 binding with MCM7 appears to enhance the DNA replication licensing, the binding of AR with MCM7 appears inhibitory (with a high dosage of AR ligand).36 AR recruits MCM7 for its transcription activity.36 This analysis suggests that AR and RACK1 cross talk through MCM7 to balance cellular DNA synthesis and RNA transcription; an AR-bound MCM7 primarily functions as a co-transcription factor, whereas a RACK1-bound MCM7 functions as a DNA helicase.
Figure 8.
Schematic diagram of interplay of PKC, RACK1, and MCM7. PKC binding with TPA leads to activation of PKC and PKC/RACK1 binding. This event redistributes RACK1 from nucleus to cytoplasm, inactivation of MCM7, and disintegration of DNA replication licensing complex.
MCM7 overexpression and amplification were found in multiple malignancies. Amplification and a high level of expression of MCM7 were found to associate poor clinical outcomes, probably due to misfiring of MCM7 that results in hyperploidy and other genome abnormalities.9,14 Intron 13 of MCM7 genome contains a cluster of miRNA that was found to down-regulate several important tumor suppressors, including p21, BIM, and pTEN.9,37–40 This makes MCM7 gene cluster an intriguing oncogenic complex. There is a growing body of literature suggesting that MCM7 plays a gate-keeping role in communicating DNA replication licensing with both anti-proliferative and pro-proliferative signaling pathways. The unique pivotal role of MCM7 opens the door for future therapeutic intervention on human malignancies. Indeed, our previous study showed that treatment using shRNA specific for MCM7 dramatically reduced tumor size, metastasis, and mortality in xenografted PC3, DU145 tumors in severe combined immunodeficiency mice.41 This analysis implies that targeted molecule upstream of MCM7 is a possible approach to control the activity of DNA replication licensing, and thus proliferation, possibly migration, and metastasis of human malignancies.
Footnotes
Supported by National Cancer Institute grants RO1 CA098249 and R56 CA098249 (J.H.L.) and the American Cancer Society (RSG-08-137-01-CNE to Y.P.Y.).
References
- 1.Kearsey S.E., Maiorano D., Holmes E.C., Todorov I.T. The role of MCM proteins in the cell cycle control of genome duplication. Bioessays. 1996;18:183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- 2.Chong J.P., Thommes P., Blow J.J. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- 3.Coxon A., Maundrell K., Kearsey S.E. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992;20:5571–5577. doi: 10.1093/nar/20.21.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalton S., Whitbread L. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc Natl Acad Sci USA. 1995;92:2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 6.You Z., Komamura Y., Ishimi Y. Biochemical analysis of the intrinsic Mcm4-Mcm6-mcm7 DNA helicase activity. Mol Cell Biol. 1999;19:8003–8015. doi: 10.1128/mcb.19.12.8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S.S., Xue W.C., Khoo U.S., Ngan H.Y., Chan K.Y., Tam I.Y., Chiu P.M., Ip P.P., Tam K.F., Cheung A.N. Replicative MCM7 protein as a proliferation marker in endometrial carcinoma: a tissue microarray and clinicopathological analysis. Histopathology. 2005;46:307–313. doi: 10.1111/j.1365-2559.2005.02069.x. [DOI] [PubMed] [Google Scholar]
- 8.Gambichler T., Shtern M., Rotterdam S., Bechara F.G., Stucker M., Altmeyer P., Kreuter A. Minichromosome maintenance proteins are useful adjuncts to differentiate between benign and malignant melanocytic skin lesions. J Am Acad Dermatol. 2009;60:808–813. doi: 10.1016/j.jaad.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 9.Kan T., Sato F., Ito T., Matsumura N., David S., Cheng Y., Agarwal R., Paun B.C., Jin Z., Olaru A.V., Selaru F.M., Hamilton J.P., Yang J., Abraham J.M., Mori Y., Meltzer S.J. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishihara K., Shomori K., Fujioka S., Tokuyasu N., Inaba A., Osaki M., Ogawa T., Ito H. Minichromosome maintenance protein 7 in colorectal cancer: implication of prognostic significance. Int J Oncol. 2008;33:245–251. [PubMed] [Google Scholar]
- 11.Feng C.J., Li H.J., Li J.N., Lu Y.J., Liao G.Q. Expression of Mcm7 and Cdc6 in oral squamous cell carcinoma and precancerous lesions. Anticancer Res. 2008;28:3763–3769. [PubMed] [Google Scholar]
- 12.Facoetti A., Ranza E., Grecchi I., Benericetti E., Ceroni M., Morbini P., Nano R. Immunohistochemical evaluation of minichromosome maintenance protein 7 in astrocytoma grading. Anticancer Res. 2006;26:3513–3516. [PubMed] [Google Scholar]
- 13.Kebebew E., Peng M., Reiff E., Duh Q.Y., Clark O.H., McMillan A. Diagnostic and prognostic value of cell-cycle regulatory genes in malignant thyroid neoplasms. World J Surg. 2006;30:767–774. doi: 10.1007/s00268-005-0308-2. [DOI] [PubMed] [Google Scholar]
- 14.Ren B., Yu G., Tseng G.C., Cieply K., Gavel T., Nelson J., Michalopoulos G., Yu Y.P., Luo J.H. MCM7 amplification and overexpression are associated with prostate cancer progression. Oncogene. 2006;25:1090–1098. doi: 10.1038/sj.onc.1209134. [DOI] [PubMed] [Google Scholar]
- 15.Sterner J.M., Dew-Knight S., Musahl C., Kornbluth S., Horowitz J.M. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18:2748–2757. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schechtman D., Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene. 2001;20:6339–6347. doi: 10.1038/sj.onc.1204778. [DOI] [PubMed] [Google Scholar]
- 17.Koehler J.A., Moran M.F. RACK1, a protein kinase C scaffolding protein, interacts with the PH domain of p120GAP. Biochem Biophys Res Commun. 2001;283:888–895. doi: 10.1006/bbrc.2001.4889. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez M.M., Ron D., Touhara K., Chen C.H., Mochly-Rosen D. RACK1, a protein kinase C anchoring protein, coordinates the binding of activated protein kinase C and select pleckstrin homology domains in vitro. Biochemistry. 1999;38:13787–13794. doi: 10.1021/bi991055k. [DOI] [PubMed] [Google Scholar]
- 19.Chang B.Y., Conroy K.B., Machleder E.M., Cartwright C.A. RACK1, a receptor for activated C kinase and a homolog of the beta subunit of G proteins, inhibits activity of src tyrosine kinases and growth of NIH 3T3 cells. Mol Cell Biol. 1998;18:3245–3256. doi: 10.1128/mcb.18.6.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hibbs M.L., Jakes S., Stacker S.A., Wallace R.W., Springer T.A. The cytoplasmic domain of the integrin lymphocyte function-associated antigen 1 beta subunit: sites required for binding to intercellular adhesion molecule 1 and the phorbol ester-stimulated phosphorylation site. J Exp Med. 1991;174:1227–1238. doi: 10.1084/jem.174.5.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mourton T., Hellberg C.B., Burden-Gulley S.M., Hinman J., Rhee A., Brady-Kalnay S.M. The PTPmu protein-tyrosine phosphatase binds and recruits the scaffolding protein RACK1 to cell-cell contacts. J Biol Chem. 2001;276:14896–14901. doi: 10.1074/jbc.M010823200. [DOI] [PubMed] [Google Scholar]
- 22.Yarwood S.J., Steele M.R., Scotland G., Houslay M.D., Bolger G.B. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J Biol Chem. 1999;274:14909–14917. doi: 10.1074/jbc.274.21.14909. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y.V., Baek J.H., Zhang H., Diez R., Cole R.N., Semenza G.L. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol Cell. 2007;25:207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Y.P., Luo J.H. Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 2006;66:7414–7419. doi: 10.1158/0008-5472.CAN-06-0227. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Z.H., Yu Y.P., Shi Y.K., Nelson J.B., Luo J.H. CSR1 induces cell death through inactivation of CPSF3. Oncogene. 2009;28:41–51. doi: 10.1038/onc.2008.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y.C., Yu Y.P., Nelson J., Wu C., Wang H., Michalopoulos G.K., Luo J.H. Interaction of integrin-linked kinase and miniature chromosome maintenance 7-mediating integrin {alpha}7 induced cell growth suppression. Cancer Res. 2010;70:4375–4384. doi: 10.1158/0008-5472.CAN-09-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing L., Liu L., Yu Y.P., Dhir R., Acquafondada M., Landsittel D., Cieply K., Wells A., Luo J.H. Expression of myopodin induces suppression of tumor growth and metastasis. Am J Pathol. 2004;164:1799–1806. doi: 10.1016/S0002-9440(10)63738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo JH: Oncogenic activity of MCM7 transforming cluster. World J Clin Oncol 2:120-124 [DOI] [PMC free article] [PubMed]
- 29.Parker P.J., Coussens L., Totty N., Rhee L., Young S., Chen E., Stabel S., Waterfield M.D., Ullrich A. The complete primary structure of protein kinase C–the major phorbol ester receptor. Science. 1986;233:853–859. doi: 10.1126/science.3755547. [DOI] [PubMed] [Google Scholar]
- 30.Bosco R, Melloni E, Celeghini C, Rimondi E, Vaccarezza M, Zauli G: Fine tuning of protein kinase C (PKC) isoforms in cancer: shortening the distance from the laboratory to the bedside. Mini Rev Med Chem 11:185-199 [DOI] [PubMed]
- 31.Bae K.M., Wang H., Jiang G., Chen M.G., Lu L., Xiao L. Protein kinase C epsilon is overexpressed in primary human non-small cell lung cancers and functionally required for proliferation of non-small cell lung cancer cells in a p21/Cip1-dependent manner. Cancer Res. 2007;67:6053–6063. doi: 10.1158/0008-5472.CAN-06-4037. [DOI] [PubMed] [Google Scholar]
- 32.Black J.D. Protein kinase C-mediated regulation of the cell cycle. Front Biosci. 2000;5:D406–D423. doi: 10.2741/black. [DOI] [PubMed] [Google Scholar]
- 33.Ron D., Chen C.H., Caldwell J., Jamieson L., Orr E., Mochly-Rosen D. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc Natl Acad Sci USA. 1994;91:839–843. doi: 10.1073/pnas.91.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liliental J., Chang D.D. Rack1, a receptor for activated protein kinase C, interacts with integrin beta subunit. J Biol Chem. 1998;273:2379–2383. doi: 10.1074/jbc.273.4.2379. [DOI] [PubMed] [Google Scholar]
- 35.Huang C.C., Liu C.H., Chuang N.N. An enhanced association of RACK1 with Abl in cells transfected with oncogenic ras. Int J Biochem Cell Biol. 2008;40:423–431. doi: 10.1016/j.biocel.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Shi Y.K., Yu Y.P., Zhu Z.H., Han Y.C., Ren B., Nelson J.B., Luo J.H. MCM7 Interacts with Androgen Receptor. Am J Pathol. 2008;173:1758–1767. doi: 10.2353/ajpath.2008.080363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrocca F., Vecchione A., Croce C.M. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- 38.Ambs S., Prueitt R.L., Yi M., Hudson R.S., Howe T.M., Petrocca F., Wallace T.A., Liu C.G., Volinia S., Calin G.A., Yfantis H.G., Stephens R.M., Croce C.M. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrocca F., Visone R., Onelli M.R., Shah M.H., Nicoloso M.S., de Martino I., Iliopoulos D., Pilozzi E., Liu C.G., Negrini M., Cavazzini L., Volinia S., Alder H., Ruco L.P., Baldassarre G., Croce C.M., Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Poliseno L., Salmena L., Riccardi L., Fornari A., Song M.S., Hobbs R.M., Sportoletti P., Varmeh S., Egia A., Fedele G., Rameh L., Loda M., Pandolfi P.P. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y.K., Yu Y.P., Tseng G.C., Luo J.H. Inhibition of prostate cancer growth and metastasis using small interference RNA specific for minichromosome complex maintenance component 7. Cancer Gene Ther. 2010;17:694–699. doi: 10.1038/cgt.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]