Abstract
Patients with neurofibromatosis type 1 (NF1) develop benign plexiform neurofibromas that frequently progress to become malignant peripheral nerve sheath tumors (MPNSTs). A genetically engineered mouse model that accurately models plexiform neurofibroma–MPNST progression in humans would facilitate identification of somatic mutations driving this process. We previously reported that transgenic mice overexpressing the growth factor neuregulin-1 in Schwann cells (P0-GGFβ3 mice) develop MPNSTs. To determine whether P0-GGFβ3 mice accurately model human neurofibroma–MPNST progression, cohorts of these animals were monitored through death and were necropsied; 94% developed multiple neurofibromas, with 70% carrying smaller numbers of MPNSTs. Nascent MPNSTs were identified within neurofibromas, suggesting that these sarcomas arise from neurofibromas. Although neurofibromin expression was maintained, P0-GGFβ3 MPNSTs exhibited Ras hyperactivation, as in human NF1-associated MPNSTs. P0-GGFβ3 MPNSTs also exhibited abnormalities in the p16INK4A–cyclin D/CDK4–Rb and p19ARF–Mdm–p53 pathways, analogous to their human counterparts. Array comparative genomic hybridization (CGH) demonstrated reproducible chromosomal alterations in P0-GGFβ3 MPNST cells (including universal chromosome 11 gains) and focal gains and losses affecting 39 neoplasia-associated genes (including Pten, Tpd52, Myc, Gli1, Xiap, and Bbc3/PUMA). Array comparative genomic hybridization also identified recurrent focal copy number variations affecting genes not previously linked to neurofibroma or MPNST pathogenesis. We conclude that P0-GGFβ3 mice represent a robust model of neurofibroma-MPNST progression useful for identifying novel genes driving neurofibroma and MPNST pathogenesis.
Malignant peripheral nerve sheath tumors (MPNSTs) are highly aggressive Schwann cell–derived sarcomas. Although MPNSTs occur both sporadically and in individuals with neurofibromatosis type 1 (NF1),1–5 our current understanding of MPNST pathogenesis is derived largely from studies of NF1-associated MPNSTs. In NF1, MPNSTs arise from plexiform neurofibromas, which are benign tumors of peripheral nerve composed of NF1-null Schwann-like cells intermingled with NF1+/− mast cells, perineurial-like cells, and fibroblasts. The initiating event in plexiform neurofibroma pathogenesis is thought to be the loss of the remaining functional NF1 allele in an NF1+/− cell in the Schwann cell lineage. This loss sets into motion a complex series of changes, including enhanced production of growth factors that recruit the other NF1+/− cell types into the nascent neurofibroma. The subsequent mutation of additional tumor suppressor genes such as TP53, CDKN2A, and PTEN in the NF1−/− Schwann-like cells promotes malignant transformation and MPNST pathogenesis.6,7
The scenario outlined above is conceptually attractive and widely accepted. Nonetheless, there are several reasons to think that it is incomplete and understates the genomic diversity of plexiform neurofibromas and MPNSTs. For instance, specific genomic gains and losses affecting multiple chromosomes are common in neurofibromas,8,9 raising the question of whether additional somatic mutations act cooperatively with NF1 loss to promote neurofibroma pathogenesis. In addition, at least some sporadic MPNSTs lack NF1 mutations,10 indicating that these sarcomas can arise via genetic pathways that do not involve NF1 loss. There is also evidence suggesting that the pathways involved in neurofibroma–MPNST progression are heterogeneous. As an example, although it was initially reported that TP53 was mutated in a very high percentage of MPNSTs,11 more recent findings argue that only a minority of MPNSTs carry TP53 mutations.12,13 There is also reason to think that other genes, in addition to those noted above, promote neurofibroma–MPNST progression; for instance, an as yet unidentified tumor-suppressor gene on the short arm of chromosome 1 has been implicated in MPNST pathogenesis.14 Finally, it must be pointed out that MPNSTs have very complex karyotypes,15–26 in which specific gains and losses are repeatedly encountered, suggesting that important driver genes remain undiscovered in these tumors.
A more complete understanding of the various mutations contributing to plexiform neurofibroma and MPNST pathogenesis could identify novel molecular targets for therapeutic intervention. Ideally, this would be accomplished by sequencing the transcriptome and exome of a large cohort of human plexiform neurofibromas and MPNSTs, an approach whose effectiveness has been demonstrated by The Cancer Genome Atlas (TCGA) in glioblastomas27 and serous ovarian carcinomas.28 However, plexiform neurofibromas and MPNSTs are much less common than any of the tumor types thus far examined by TCGA, and it is difficult to obtain large numbers of these neoplasms for study. Consequently, it likely will be necessary to complement the sequencing of human tumors with other approaches, such as identifying the somatic mutations driving neurofibroma–MPNST progression in an appropriate mouse model. Indeed, analogous cross-species comparative oncogenomic studies using genetically engineered mouse cancer models29–32 or murine cancers produced by insertional mutagenesis with the Sleeping Beauty transposon system33–35 have proven quite useful for the identification of driver genes in other types of cancer. Unfortunately, it is not clear what mouse model can be used to study neurofibroma–MPNST progression. Krox20-Cre;Nf1flox/− mice develop numerous neurofibromas,36 but these lesions do not progress to become MPNSTs at an appreciable rate. On the other hand, mice with cis-linked Nf1 and p53 null alleles do develop MPNSTs,37,38 but these tumors arise de novo rather than from a pre-existing neurofibroma. Consequently, neither of these mouse models recapitulates the process of neurofibroma–MPNST progression seen in human NF1.
We have shown that the potent Schwann cell mitogen neuregulin-1 (NRG1) contributes to the pathogenesis of human neurofibromas and MPNSTs,39,40 and that transgenic mice overexpressing this growth factor in Schwann cells (P0-GGFβ3 mice) develop MPNSTs.41 These observations led us to ask whether P0-GGFβ3 mice accurately model human neurofibroma–MPNST progression and would thus be useful for identifying the driver gene mutations mediating this process. To address this question, we first determined whether P0-GGFβ3 mice develop neurofibromas and whether there is pathological evidence that these neurofibromas progress to become MPNSTs. We then asked whether the molecular abnormalities driving the pathogenesis of peripheral nerve sheath tumors in P0-GGFβ3 mice parallel those seen in their human counterparts. Finally, we used high-density array comparative genomic hybridization (aCGH) to identify additional previously unknown mutations potentially promoting the pathogenesis of P0-GGFβ3 MPNSTs.
Materials and Methods
Antibodies and Other Reagents
Rabbit polyclonal antibodies recognizing S100β (Z0311) and glial fibrillary acidic protein (GFAP; Z0334) and a mouse monoclonal anti-desmin (clone D33) antibody were purchased from Dako (Carpinteria, CA). A mouse monoclonal anti-neurofilament antibody (SMI34) was purchased from Covance–Sternberger Monoclonals (Lutherville, MD). Mouse monoclonal antibodies against collagen type IV (clone PHM-12), smooth muscle actin (SMA; clone 1A4), and GAPDH (clone 6C5) were purchased from Ventana Medical Systems (Tucson, AZ), NeoMarkers (Fremont, CA), and Advanced ImmunoChemical (Long Beach, CA), respectively. A rabbit polyclonal antibody recognizing neuron-specific enolase (NSE; AB591) and a mouse pan-Ras monoclonal antibody (clone RAS10) were purchased from Millipore (Billerica, MA). Antibodies against neurofibromin (sc-67), cyclin D2 (sc-593), p53 (sc-99), p16INK4A (sc-1207), Mdm2 (sc-965), Mdm4 (sc-28222), p21Cip1/Waf1 (sc-397), and p27Kip1 (sc-527) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal antibodies recognizing CDK2 (610145), CDK4 (610147), and cyclin D3 (610279) were purchased from BD Biosciences (San Jose, CA). Mouse monoclonal antibodies against cyclin D1 (2926) and Rb phosphorylated on Ser795 (9301) were purchased from Cell Signaling Technology (Danvers, MA). Horseradish peroxidase- and Cy3-conjugated secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA). The pan-ErbB inhibitor PD168393 was purchased from Millipore–EMD Chemicals (Billerica, MA).
Animal Care and Necropsies
Mice were cared for in accordance with the NIH Guide for the Care and Use of Laboratory Animals (8th ed., 2011). All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
P0-GGFβ3 mice were bred to C57BL/6J×SJL/J F1 hybrids or were backcrossed to purebred C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME). The resulting offspring were screened by polymerase chain reaction (PCR) as described previously.41 Mice were housed in standard cages with water and food available ad libitum. Moribund mice or mice suspected for other reasons of bearing tumors were anesthetized with isoflurane and sacrificed by decapitation. Tumor tissue was removed fresh, and a portion was used to establish early-passage cultures. The rest of the tumor was fixed in 4% paraformaldehyde, except for a small portion that was fixed with glutaraldehyde. Bodies were then immersion-fixed in 4% paraformaldehyde. After fixation, complete necropsies were performed, including a detailed examination of the central and peripheral nervous systems. To examine spinal cords and nerve roots in situ, vertebral columns together with adjacent soft tissue and ribs were decalcified by immersion in 0.3 mol/L EDTA/4% paraformaldehyde (pH 8.0) for 48 to 72 hours at 4°C. Tissues were dehydrated and paraffin-embedded. Sections (4 to 5 μm thick) were prepared, stained with H&E, mounted with Permount medium (Fisher Scientific, Pittsburgh, PA), and examined with light microscopy. All tumor diagnoses were established by a practicing neuropathologist (S.L.C.) certified by the American Board of Pathology in both anatomical pathology and neuropathology. Tumors were evaluated using World Health Organization (WHO) diagnostic criteria.42
Immunohistochemical and Histochemical Staining of Tumor Sections
Schwann cells and axons within peripheral nerve sheath tumors were identified by immunostaining paraffin sections (4 to 5 μm thick) according to our previously described protocol,41 with antibodies recognizing S100β or neurofilaments, respectively. Aberrant expression of p53 was examined by similarly incubating sections with an anti-p53 antibody. After incubation with horseradish peroxidase–conjugated secondary antibody, immunoreactive structures were visualized by diaminobenzidine deposition. To confirm specificity, control slides were treated identically, except that primary antibodies were replaced with nonimmune rabbit or mouse IgG; no staining was observed in these experiments. After immunostaining, slides were counterstained with hematoxylin, mounted with Permount, and coverslipped for light microscopic examination.
To examine expression of neuronal and muscle markers in MPNSTs, double-label immunohistochemistry was performed on tumor sections (4 to 5 μm thick) using a rabbit polyclonal anti-S100β antibody in combination with mouse monoclonal antibodies recognizing neurofilaments, desmin, or SMA. Immunoreactive cells were identified by then incubating the sections with Cy3-conjugated anti-rabbit and fluorescein isothiocyanate–conjugated anti-mouse secondary antibodies. Sections were counterstained with bisbenzimide before being mounted in 1:1 PBS:glycerol. To verify the specificity of the observed staining, primary antibodies were omitted from some slides, which were then otherwise identically processed.
Unna’s methylene blue staining for mast cells was performed as described previously.43
Establishment of Early-Passage Tumor Cultures and Schwann Cell Cultures
Tumor tissue was sterilely transferred to Hanks’ balanced salt solution, finely minced and then incubated in Dulbecco’s minimal essential medium supplemented with 10% fetal calf serum (DMEM10), 2 μmol/L forskolin, and 50 nmol/L NRG1β. (In preliminary experiments, we had found that the last two components facilitated the initial establishment of tumor cultures.) Cells were allowed to grow out from explants until confluent. Cultures were then split and maintained for no more than five to eight passages in DMEM10 without forskolin or NRG1β. Cultures of Schwann cells from wild-type (WT) and transgenic nerves were established identically, but were maintained continuously in DMEM10 supplemented with forskolin and NRG1β.
Examination of Tumor Suppressors and Lineage Markers in MPNST Cultures and Tumor Sections
For S100β and p53 immunostaining, early-passage tumor cells were plated at low density in chamber slides and allowed to adhere overnight. Slides were then fixed for 30 minutes at room temperature with 4% paraformaldehyde, followed by three rinses with PBS (5 minutes per wash). Fixed cells were incubated for 30 minutes in blocking buffer (PBS containing 1% bovine serum albumin, 0.2% nonfat dry milk, and 0.3% Triton X-100) and then incubated overnight at 4°C with anti-S100β antibody diluted 1:200 in blocking buffer. The next morning, cells were rinsed three times with PBS and then incubated for 1 hour at room temperature with Cy3-conjugated donkey anti-rabbit secondary antibody diluted 1:250 in blocking buffer. After three more rinses in PBS, slides were mounted using 1:1 PBS:glycerol and were examined with fluorescence microscopy.
Arrays of tumor cells were constructed as described previously.40 Sections (4 to 5 μm thick) of the arrays were prepared, deparaffinized, and rehydrated through graded ethanols to PBS. After 30 minutes in blocking buffer, rehydrated sections were incubated with primary antibody or nonimmune IgG overnight at 4°C. Dilutions of primary antibodies were as follows: collagen type IV, 1:50; GFAP, 1:500; NSE, 1:1000; SMA, 1:1000; and desmin, 1:50. Sections were rinsed three times with PBS and then incubated with horseradish peroxidase–conjugated secondary antibodies for 1 hour at room temperature. After three more PBS washes, immunoreactivity was detected by diaminobenzidine deposition. Sections were lightly counterstained with hematoxylin and mounted with Permount for light microscopic examination.
Primers for amplification of tumor cell markers were designed using Lasergene PrimerSelect software version 3.10 (DNASTAR, Madison, WI). GenBank accession numbers of the target sequences and the position of each primer are listed in Table 1. cDNA (2 μL) synthesized from total RNA was used in PCR reactions (35 cycles of 94°C for 1 minute, 55°C for 1 minute, and 72°C for 2 minutes).
Table 1.
PCR Primers Used for Amplification of Transcripts Encoding Differentiation Markers in MPNST Cultures
| Tumor marker | GenBank accession no. | Primer position |
|
|---|---|---|---|
| Forward | Reverse | ||
| S100α | BC005590 | 19–39 | 423–400 |
| S100β | NM_009115 | 28–51 | 197–175 |
| P0 | NM_008623 | 39–61 | 490–469 |
| MBP | M15060 | 339–359 | 767–744 |
| PMP22 | NM_008885 | 1049–1069 | 1457–1434 |
| p75LNTR | BC038365 | 521–542 | 935–912 |
| Sox10 | BC018551 | 1845–1864 | 2257–2237 |
| Pax3 | X59358 | 398–417 | 811–788 |
| Krox20 | X06746 | 376–399 | 817–794 |
| GFAP | NM_010277 | 777–798 | 1210–1187 |
| GAP43 | BC028288 | 362–383 | 822–799 |
| Neurofilament | BC029203 | 71–92 | 636–613 |
| Peripherin 1 | BC046291 | 131–151 | 607–585 |
| Calponin-1 | Z19542 | 546–569 | 976–953 |
| SM22α | U36588 | 79–97 | 480–458 |
| αSMA | BC064800 | 655–688 | 1162–1141 |
| Desmin | NM_010043 | 156–179 | 556–533 |
| MyoD1 | NM_010866 | 1313–1336 | 1799–1776 |
MBP, myelin basic protein; P0, myelin protein zero; p75LNTR, the low affinity neurotrophin receptor; PMP22, peripheral myelin protein 22; SM22α, transgelin; αSMA, α-smooth muscle actin.
Immunoblotting
Cells were homogenized in HES buffer (40 mmol/L HEPES, 2 mmol/L EDTA, 500 mmol/L sucrose) supplemented with protease (P8340; Sigma-Aldrich, St. Louis, MO) and phosphatase (P5726; Sigma-Aldrich) inhibitor cocktails at a 1:100 ratio. Protein concentrations were determined using a modified Lowry method (DC protein assay; Bio-Rad Laboratories, Hercules, CA). Equivalent quantities of protein lysates were resolved on 8% SDS-PAGE gels, immunoblotted according to our previously described protocol,40 and probed. Immunoreactive species were detected by enhanced chemiluminescence (Pierce; Thermo Fisher Scientific, Rockford, IL). As a loading control, membranes were reprobed with an anti-GAPDH antibody (1:20,000 dilution).
qPCR
Real-time quantitative PCR (qPCR) was performed using our previously described protocol44 with an ABI 7500 real-time PCR system (Life Technologies–Applied Biosystems, Foster City, CA). Assays of Cdkn2a and Nf1 transcripts were performed using FAM-labeled TaqMan MGB probes (ABI assays Mm00494449_m1 and Mm00812424_m1, respectively). The levels of cDNA encoding 18S ribosomal RNA were assayed in the same cDNAs using VIC-labeled TaqMan MGB probes (ABI 4319413E). Each specimen was assayed in triplicate and the levels of test cDNAs normalized to that of 18S rRNA cDNA in the same specimen. Assays were analyzed using Applied Biosystems Sequence Detection Software version 1.4.
Ras Activation Assays
Wild-type Schwann cells and early-passage P0-GGFβ3 MPNST cultures were lysed in magnesium-containing buffer [25 mmol/L HEPES (pH 7.5), 150 mmol/L NaCl, 10 mmol/L MgCl2, 1 mmol/L EDTA, 1% Igepal CA-630, 10% glycerol] supplemented with phosphatase (78420; 1:100 dilution; Thermo Scientific) and protease (P8340; 1:100 dilution; Sigma-Aldrich) inhibitor cocktails. Lysates were clarified by centrifugation at 20,000 × g for 10 minutes. After determination of their protein concentrations, lysates were diluted to 0.5 mg/mL protein; 250 μg (500 μL) of protein was then spiked with 20 μL Ras assay reagent beads (Raf-1 Ras-binding domain agarose beads; Millipore) and incubated for 30 minutes at 4°C with continuous rotation. Beads were washed three times in magnesium-containing lysis buffer supplemented with protease and phosphatase inhibitors and then were collected by centrifugation at 20,000 × g for 10 seconds. Beads were boiled for 15 minutes in 40 μL 2× stop buffer [250 mmol/L Tris-HCl (pH 6.8), 5 mmol/L EDTA, 5 mmol/L EGTA, 2% SDS, 10% glycerol, 25 mmol/L dithiothreitol, and 300 μmol/L bromophenol blue]. Material released from the beads and samples of the clarified lysates were resolved on 12% SDS-PAGE gels, immunoblotted as described above, and probed with a pan-Ras antibody.
Identification of Trp53 Mutations
Nested PCR was used to amplify Trp53 sequences for sequencing. The cDNA template was synthesized from total cellular RNA, as described above. The first round of PCR was performed with primers corresponding to nucleotides 268–287 and 1925–1947 of GenBank accession NM_011640.1. A portion of the initial PCR product was then further amplified with primers corresponding to nucleotides 357–375 and 1780–1802 of GenBank accession NM_011640.1. Both rounds of PCR amplification were performed with an initial 1-minute melt at 94°C, followed by 35 cycles of 94°C for 30 seconds, 50°C for 1 minute, and 72°C for 4 minutes, and then by a final 7-minute extension at 72°C. Final PCR products were gel-purified and sequenced with primers corresponding to nucleotides 485–503, 899–922, and 1275–1298 of GenBank accession NM_011640.1.
DNA Synthesis Assays
40,000 MPNST cells were plated per well in DMEM10 and allowed to attach overnight. The next morning, PD168393 or vehicle (dimethyl sulfoxide) was added to each well; 12 hours later, 1 μL of 1 μCi/mL [3H]thymidine was added. After 12 hours of radiolabeling, [3H]thymidine incorporation by MPNST cells was assayed using our previously described methodology.40 All experiments were performed in triplicate, with six replicates per condition performed in each experiment. A one-way analysis of variance, followed by a Tukey’s post hoc test, was used to analyze the resulting data. A P value of <0.05 was considered statistically significant.
Array Comparative Genomic Hybridization
Genomic DNA was isolated from early-passage P0-GGFβ3 MPNST cells and cultured WT (nontransgenic) mouse Schwann cells using a QIAamp DNA mini kit (Qiagen, Valencia, CA) according to the manufacturer’s recommendations. Test (MPNST) and reference (Schwann cell) genomic DNA was labeled with Cy5-dUTP and Cy3-dUTP, respectively, using a genomic DNA enzymatic labeling kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer’s recommendations. Whole-genome aCGH was performed using mouse 4x44k oligonucleotide arrays (Agilent Technologies), which have an overall median probe spacing of approximately 22 kb, and median probe spacing in RefSeq genes of approximately 13 kb. Slide hybridization, washing, and scanning were performed according to the manufacturer’s protocol. The arrays were scanned using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). The resulting data were analyzed using Feature Extraction version 9.5 and DNA Analytics version 4.0 software (Agilent Technologies). The analysis parameters used for the detection of genomic copy number variations (CNVs) included linear dye normalization, ADM-2 aberration detection algorithm, and aberration filter of four sequential probes with a log2 ratio cutoff of 0.25. All genomic breakpoints were mapped using mouse genome build NCBI37/mm9. The data sets resulting from these aCGH experiments have been deposited in the NCBI Gene Expression Omnibus under accession number GSE40212. The called CNVs identified in these experiments have also been deposited in the NCBI Database of Genomic Structural Variation (dbVar) under study ID number nstd74.
Results
P0-GGFβ3 Mice Develop Numerous Neurofibromas Throughout Their Peripheral Nervous System That Progress to Become MPNSTs at a High Frequency
If MPNSTs arising in P0-GGFβ3 mice develop via progression from pre-existing neurofibromas, neurofibromas should be evident in these animals. Because only a subset of these neurofibromas would be expected to accumulate the additional mutations required for progression, neurofibromas should also be more common than MPNSTs in P0-GGFβ3 mice. To test these postulates, we established a cohort of 44 mice (21 female and 23 male) carrying the P0-GGFβ3 transgene on an outbred C57BL/6J×SJL/J background and followed them until their death (mean age at death, 261.5 days; median, 220.5 days; range, 74 to 533 days). Complete necropsies were performed, including histological examination of longitudinal sections of the decalcified vertebral column and adjacent soft tissue. The peripheral nerve sheath tumors that were identified were evaluated using World Health Organization diagnostic criteria for neurofibromas and MPNSTs.42 In the vast majority of these animals (41/44 mice; 91%), virtually every dorsal spinal nerve root was markedly enlarged by intraneural tumor growth (Figure 1A). Histological examination of these neoplasms showed them to be moderately cellular lesions that did not demonstrate the hypercellularity, increased nuclear size, hyperchromasia, or increased mitotic activity seen in low-grade MPNSTs. The tumors were predominantly composed of elongated cells with spindled nuclei (Figure 1B) that diffusely infiltrated nerve roots and dorsal root ganglia, as demonstrated by the presence of entrapped neurons and axons (Figure 1, B and E). Prominent immunoreactivity for the Schwann cell marker S100β was evident in a major population of intratumoral cells that were intimately intermingled with S100β-negative cells (Figure 1C). These intraneural tumors additionally contained large numbers of mast cells, which were demonstrable by prominent metachromasia under methylene blue staining (Figure 1D). Similar neoplasms were present in the trigeminal ganglia and sympathetic nervous system (superior cervical, celiac, and superior mesenteric ganglia and associated nerves) of these P0-GGFβ3 mice. Considered in aggregate, the histopathological features of the low-grade neoplasms occurring in the dorsal spinal nerve roots, trigeminal ganglia, and sympathetic nervous system of P0-GGFβ3 mice are identical to those seen in human neurofibromas.
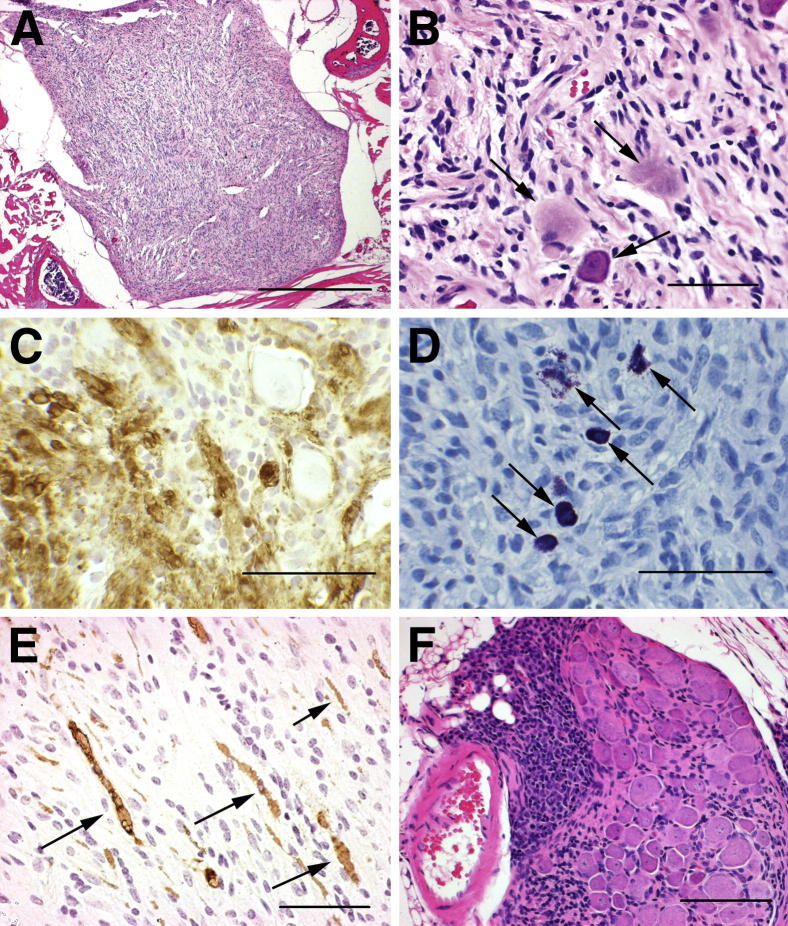
Figure 1.
P0-GGFβ3 mice develop multiple neurofibromas that may undergo malignant transformation. A: Low-power image of a representative neurofibroma developing on a dorsal spinal nerve root in a P0-GGFβ3 mouse. B: Higher-power view of the same tumor section, showing entrapped ganglionic neurons (arrows). C: In a section of the same tumor, immunostaining for S100β reveals coexistence of S100β-positive and -negative elements within this neoplasm. D: In a section of the same tumor, Unna’s methylene blue stain reveals numerous mast cells, evident by their prominent metachromasia (cells with dark purple granules) (arrows). E: In another tumor, immunostaining with the anti-neurofilament antibody SMI34 reveals axons (arrows) entrapped by infiltrating tumor cells. F: Microscopic focus of apparent malignant transformation, a markedly hypercellular region (left half of the field), arising in a spinal nerve root neurofibroma. Scale bars: 500 μm (A); 50 μm (B–F).
While evaluating the tumors described above, we found that 5 of the 44 mice had neurofibromas that contained small foci with a higher-grade appearance (Figure 1F). These foci were composed of closely packed, markedly atypical cells with angulated, hyperchromatic nuclei and multiple mitotic figures. A comparison of these foci to the MPNSTs that developed in P0-GGFβ3 mice (see below) demonstrated a striking similarity between the atypical foci and many of the higher-grade lesions found in these animals (eg, compare the focus illustrated in Figure 1F to the high-grade tumors in Figure 2, B and D). This observation suggests that the neurofibromas developing in P0-GGFβ3 mice are the precursors that ultimately give rise to MPNSTs in these animals.
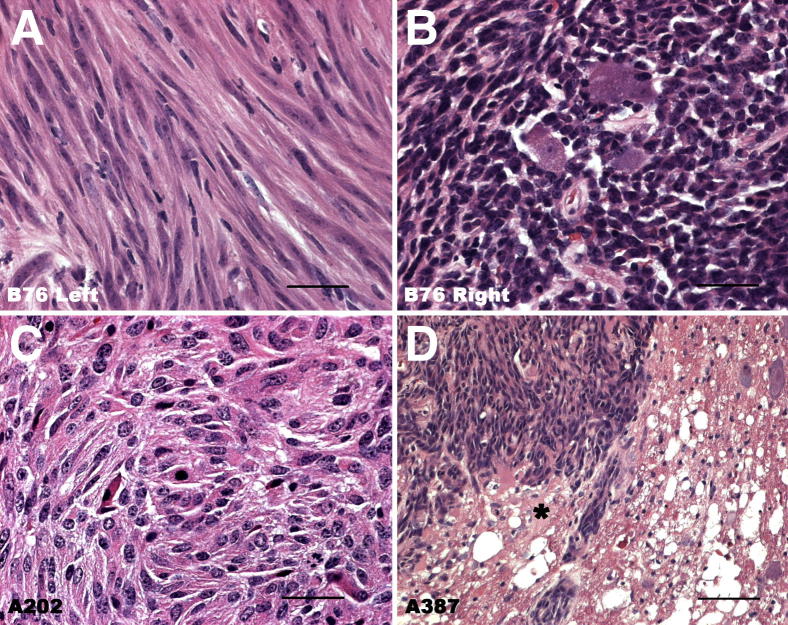
Figure 2.
Representative images of MPNSTs arising in P0-GGFβ3 mice, demonstrating the histological variability encountered in these neoplasms. A and B: Histology of tumors that arose independently in the left (A) and right (B) trigeminal nerves of mouse B76. Note the extreme spindled morphology of the tumor cells in A, which contrasts markedly with the densely packed, poorly differentiated cells composing the tumor seen in B. C: MPNST with an epithelioid morphology that arose in the sciatic nerve of mouse A202. D: A trigeminal MPNST from mouse A387 invading the undersurface of the brain. The overlying brain parenchyma (asterisk) is being invaded by this tumor. Scale bars: 50 μm (A–C); 200 μm (D).
MPNSTs were identified in 31 (71%) of the necropsied P0-GGFβ3 mice. Grossly, these large nerve-associated tumors had a fleshy appearance, with areas of hemorrhage and necrosis often evident. Microscopic examination showed them to be markedly hypercellular lesions with brisk mitotic activity that were histologically similar to human MPNSTs (Figure 2). Most of these MPNSTs were composed of closely packed, hyperchromatic cells with eosinophilic cytoplasm (Figure 2, B and D) and resembled the high-grade tumors seen in our initial studies of these animals.41 With this larger sampling of tumors, however, it became apparent that the MPNSTs developing in P0-GGFβ3 mice, like human MPNSTs,42 were morphologically diverse, with neoplasms that arose independently in the same animal sometimes differing markedly in appearance [eg, compare the extreme spindled morphology of the tumor cells in the neoplasm which arose in the left trigeminal nerve of mouse B76 (Figure 2A) to that of the densely packed, poorly differentiated cells evident in a tumor which arose in the right trigeminal nerve of this same mouse (Figure 2B)]. Occasional MPNSTs even had an epithelioid appearance (Figure 2C). Nonetheless, all of these tumors expressed S100β, as expected for MPNSTs. In contrast to the neurofibromas, which were confined to their nerve of origin, the MPNSTs were locally aggressive, commonly invading adjacent organs (Figure 2D), soft tissue, bone, and skeletal muscle.
In contrast to neurofibromas, which typically occurred in large numbers in tumor-bearing P0-GGFβ3 mice, MPNSTs were much less common. Of the 31 mice, 23 (74%) with these higher grade neoplasms had only a single MPNST, with 6 animals (19%) having two MPNSTs and 2 animals (6%) having three or five MPNSTs. In 24 (77%) of the 31 P0-GGFβ3 mice with MPNSTs, the neoplasms arose in trigeminal nerves. Spinal nerve roots and sciatic nerves were also common sites of MPNST occurrence [10/31 animals (32%)], with the remainder developing in the region of the superior cervical ganglion [2/31 mice (6.5%)]; the summed percentages exceed 100% because some animals developed multiple neoplasms. MPNSTs occurred with similar frequencies in male [16/23 (70%)] and female [15/21 (71%)] mice.
To further confirm these findings, we performed complete necropsies on a second cohort of 18 P0-GGFβ3 mice that had been backcrossed onto a C57BL/6J background for five to eight generations and followed until their death. The incidence and distribution of neurofibromas and MPNSTs in these animals was highly similar to that seen in mice carrying the P0-GGFβ3 transgene on a C57BL/6J×SJL/J background, except that the frequency with which they developed MPNSTs was higher (15/18 mice; 83%).
P0-GGFβ3 MPNSTs, Like Human MPNSTs, Demonstrate Ras Hyperactivation, Abnormalities in Cell-Cycle Regulatory Pathways, and a Continuing Dependence on Aberrant Growth-Factor Signaling
In human NF1, loss of the NF1 gene promotes the initial pathogenesis of neurofibromas, with the subsequent development of MPNSTs being driven, in part, by the accumulation of additional abnormalities in pathways regulating cell-cycle progression. The stepwise progression of tumorigenesis in P0-GGFβ3 mice suggested that this process similarly involves the sequential mutation of multiple driver genes. We therefore examined P0-GGFβ3 MPNSTs to determine whether they exhibit Nf1 and cell-cycle regulatory abnormalities analogous to those commonly found in human MPNSTs. We focused on MPNSTs in these experiments for two reasons. First, MPNSTs represent the culmination of the neoplastic process in P0-GGFβ3 mice and thus are the lesions most likely to demonstrate the full spectrum of driver mutations. Second, beyond a loss of NF1, mutations of tumor suppressors or other driver gene abnormalities have not yet been identified in human neurofibromas.
Because only a limited amount of MPNST tissue can be obtained from an individual P0-GGFβ3 mouse, we developed a methodology for establishing early-passage (no more than five to eight passages) cultures from P0-GGFβ3 MPNSTs for use in these experiments. In light of the histological diversity we had observed in P0-GGFβ3 MPNSTs, we first validated the identity of these cultures using an extensive panel of markers previously examined in human MPNSTs and/or in MPNSTs arising in other genetically engineered mouse models. Initial immunostaining verified that early-passage cultures established from 18 independently arising P0-GGFβ3 MPNSTs retained their immunoreactivity for S100β (Figure 3, A and B). Encouraged by this finding, we constructed arrays of these cells and probed sections of the arrays for additional Schwann cell markers (collagen type IV and GFAP) and markers of other lineages (NSE, SMA, and desmin). All of the cultures were collagen type IV immunoreactive (data not shown), and 12 of the 18 cultures expressed GFAP (Figure 3C). In contrast, NSE immunoreactivity was not found in any of these cells. Notably, however, 17 of the 18 cultures were SMA immunoreactive (Figure 3D), with 5 cultures (eg, A202 cells) labeling also for desmin. We therefore expanded our analyses to include an extensive panel of markers (Figure 3E) that has been previously examined in another genetically engineered mouse MPNST model (cis-Nf1+/−;p53 mice).37
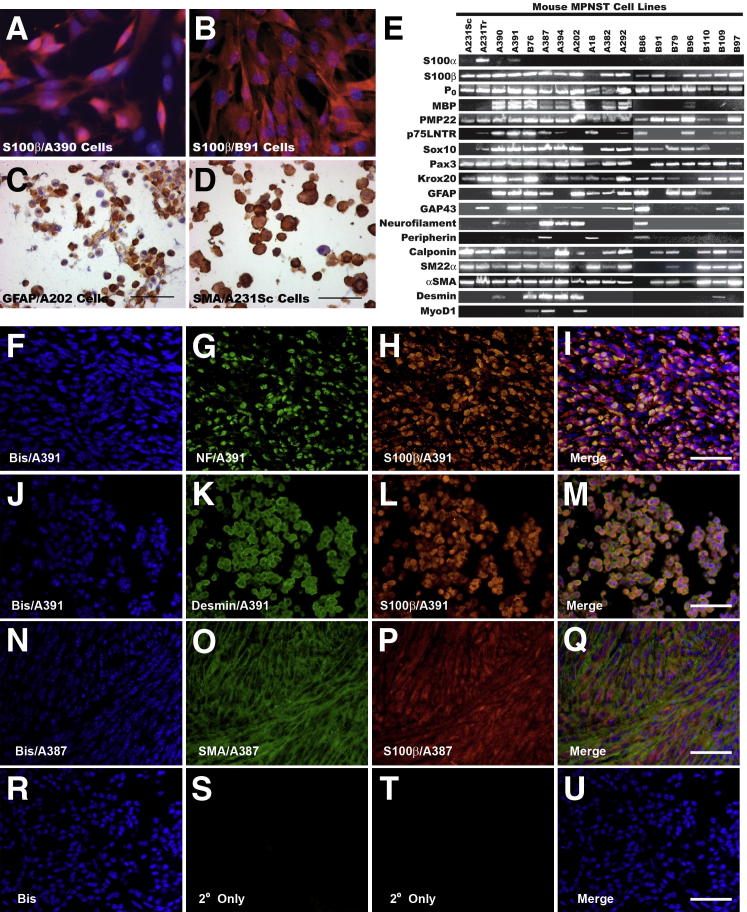
Figure 3.
Early-passage P0-GGFβ3 MPNST cells consistently express multiple Schwann cell markers and variably express neural and muscle markers. A and B: Early-passage cultures of tumors arising on mixed C57BL/6×SJL (A390 cells) (A) or inbred C57BL/6 (B91 cells) (B) backgrounds uniformly express the Schwann cell marker S100β (Cy3 immunoreactivity; red-orange staining). These cells have been counterstained with the nuclear stain bisbenzimide (blue). C and D: Immunostaining of array sections reveals the expression of other markers, such as GFAP (C) and SMA (D), that was observed in some lines. Sections have been lightly counterstained with hematoxylin. E: RT-PCR for Schwann cell (S100β, P0, MBP, PMP22, p75LNTR, Sox10, Pax3, Krox20, GFAP, GAP43), neural (neurofilament, peripherin), and muscle (calponin, SM22α, α-SMA, desmin, MyoD1) markers in early-passage cultures established from 18 independently arising P0-GGFβ3 MPNSTs. F-U: Double-label immunohistochemistry for S100β and neurofilaments (F–I), desmin (J–M), or SMA (N–Q) performed on the parent tumors (A391 and A387) used as the source of MPNST cultures that aberrantly expressed neuronal and/or muscle markers. For controls (R–U), immunostains were performed with secondary antibodies alone, in the absence of primary antibodies. Sections were counterstained with bisbenzimide (Bis).
Using RT-PCR, we found that the overwhelming majority of our early-passage cultures expressed transcripts encoding a series of classic Schwann cell markers [S100β, myelin protein zero (P0), peripheral myelin protein-22 (PMP22), the low-affinity neurotrophin receptor (p75LNTR), and the transcription factor genes Sox10, Pax3, and Egr2 (also known as Krox20)], with some other Schwann cell markers [myelin basic protein (MBP), GFAP, and GAP-43] evident in a subset of these cultures. Transcripts for several muscle markers (calponin, SM22α/transgelin, and α-SMA) were almost uniformly present in the P0-GGFβ3 MPNST cells, whereas the expression of neuronal markers was more restricted (light neurofilaments were present in 5/18 cultures, and peripherin was present in 3/18 cultures). PCR products of an identical size were detected for all these markers in control tissues and were absent when PCR reactions were performed using cDNA synthesis reactions in the absence of reverse transcriptase or with water blanks (Supplemental Figure S1A).
Thus, early-passage cultures derived from P0-GGFβ3 MPNSTs show clear evidence of Schwannian differentiation. However, these cells can also express muscle markers and, less frequently, neuronal markers, suggesting that they, like MPNSTs arising in humans and in cis-Nf1;p53 mice, are capable of expressing markers associated with other lineages. To verify that the expression of neuronal and muscle markers in our cultures was not a culture artifact, we performed double-label immunohistochemistry for S100β and neurofilaments, desmin, or SMA on sections of the MPNSTs that were used to derive cultures positive for these markers. S100β immunoreactive cells within these neoplasms were also labeled by antibodies recognizing neurofilaments, desmin, and SMA (Figure 3, F–U), indicating that our in vitro observations accurately reflected the phenotype of the parent tumor.
Having thus validated the phenotype of our early-passage cultures, we next examined neurofibromin expression in these cells. In these and subsequent analyses, we compared expression in MPNST cells to that in non-neoplastic transgenic Schwann cells, because this allowed us to identify changes in tumor suppressor and oncogene expression that are associated with tumor pathogenesis rather than with transgene expression. Lysates of MPNST cultures and non-neoplastic Schwann cells derived from the nerve type (trigeminal or sciatic) in which the tumors arose were immunoblotted and probed with an antibody that recognizes both of the major neurofibromin splice variants (220 and 250 kDa). Neurofibromin expression in P0-GGFβ3 MPNST cells was typically equal to or higher than that seen in non-neoplastic Schwann cells (Figure 4A). qPCR assays similarly revealed little evidence of a reduction in Nf1 mRNA levels (Supplemental Figure S1B).
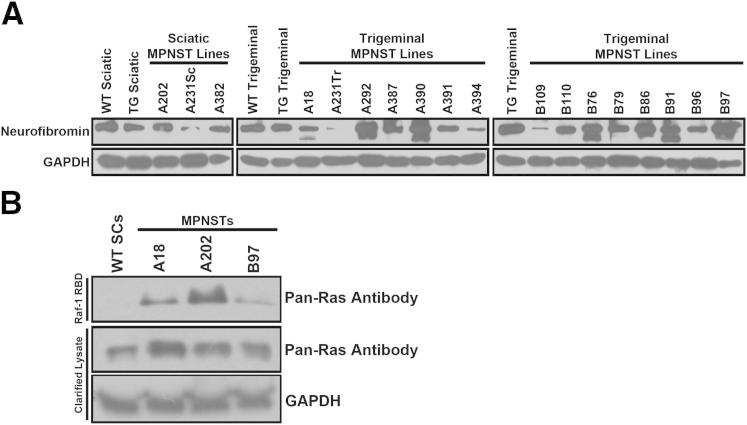
Figure 4.
Although neurofibromin expression is typically maintained in P0-GGFβ3 MPNSTs, Ras is hyperactivated in these neoplasms, compared with WT Schwann cells. A: Immunoblotted lysates of WT and non-neoplastic transgenic Schwann cells and 18 early-passage P0-GGFβ3 MPNST cultures probed for neurofibromin (1:500 antibody dilution). The 250-kDa form of neurofibromin is present in Schwann cells and the MPNST cells, with some MPNST cultures also exhibiting the 220-kDa splice variant of this protein. B: The Raf-1 Ras-binding domain (RBD) was used to pull down activated Ras protein in WT Schwann cells and three early-passage P0-GGFβ3 MPNST cultures. An immunoblot of the captured activated Ras proteins was then probed with a pan-Ras antibody that detects H-Ras, K-Ras, and N-Ras (upper panel). The middle and lower panels show immunoblots of the clarified lysate before Ras pull-down probed with the pan-Ras antibody and an anti-GAPDH antibody, respectively.
We next asked whether P0-GGFβ3 MPNST cells show evidence of the Ras hyperactivation characteristic of human NF1−/− MPNST cells.45,46 To compare the relative levels of activated Ras in non-neoplastic Schwann cells and in three representative P0-GGFβ3 MPNST cultures, a GST fusion protein containing the Raf1 Ras binding domain was used to pull down activated Ras proteins from lysates of these cells. Proteins captured with the Raf1 Ras binding domain were immunoblotted and probed with a pan-Ras antibody that recognizes all three members (H-Ras, N-Ras, and K-Ras) of the classic Ras subfamily. Although activated Ras was undetectable in non-neoplastic Schwann cells, it was easily identified in P0-GGFβ3 MPNST cells (Figure 4B). Therefore, although neurofibromin expression is maintained in P0-GGFβ3 MPNSTs, these tumors, like neurofibromin-null human MPNSTs,46,47 exhibit Ras hyperactivation.
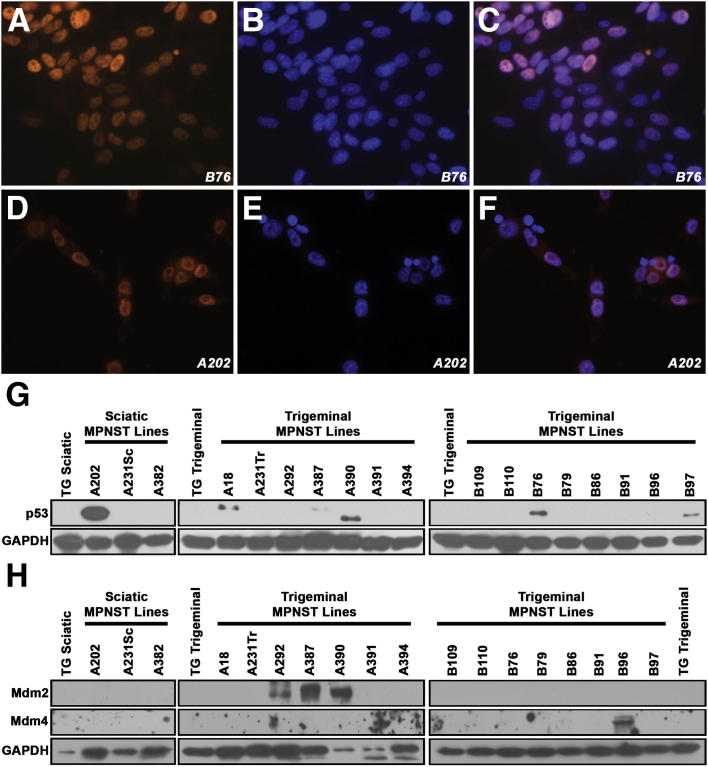
A number of laboratories have reported mutation or dysregulation of p53 in human MPNSTs.11–13,48,49 To determine whether p53 abnormalities are similarly present in P0-GGFβ3 MPNSTs, we first immunostained tumors and early-passage cultures derived from these tumors for p53. Although p53 immunoreactivity (indicative of p53 overexpression or increased p53 half-life) was not uniformly present, it was evident in six of the eighteen neoplasms and early-passage cultures derived from these neoplasms (Figure 5, A–F). This immunoreactivity was present predominantly in the nuclei of the tumor cells, with light staining at times evident also in the cytoplasm. To additionally determine whether the presence of p53 immunoreactivity in these MPNSTs reflects tumor progression, we also immunostained a series of 13 P0-GGFβ3 neurofibromas with an anti-p53 antibody. In contrast to the MPNSTs, none of the neurofibromas exhibited p53 immunoreactivity.
Figure 5.
Abnormalities in the p19ARF–Mdm–p53 signaling pathway occur in early-passage P0-GGFβ3 MPNST cultures. A–F: Immunocytochemistry reveals intranuclear accumulation of p53 in two representative p53-overexpressing P0-GGFβ3 MPNST early-passage cultures. p53 immunoreactivity is indicated by red-orange staining (A and D); bisbenzimide was used as a nuclear counterstain (blue) (B and E). Merged images of p53 immunoreactivity and bisbenzimide staining (C and F) demonstrate that p53 in these tumor cells accumulates within the nuclei. G: Immunoblotted lysates of non-neoplastic transgenic Schwann cells and early-passage cultures derived from 18 independently arising P0-GGFβ3 MPNSTs probed for p53 (1:1000 dilution). Blots were reprobed with GAPDH to verify equivalent loading. H: Immunoblotted lysates of non-neoplastic transgenic Schwann cells and early-passage cultures derived from 18 independently arising P0-GGFβ3 MPNSTs probed for Mdm2 or Mdm4 (each at 1:500 dilution). Blots were reprobed with GAPDH to verify equivalent loading. Original magnification, ×40 (A–F).
To compare the expression of p53 in P0-GGFβ3 MPNSTs to that in non-neoplastic Schwann cells, lysates of Schwann cells derived from the sciatic and trigeminal nerves of P0-GGFβ3 mice and early-passage MPNST cultures were immunoblotted and probed for p53. In both non-neoplastic transgenic Schwann cells (Figure 5G) and WT Schwann cells (data not shown), p53 was undetectable. In contrast, the MPNSTs with prominent nuclear p53 immunoreactivity (A202, A18, A387, A390, B76, and B97) all exhibited increased p53 expression in immunoblot analyses. The p53 immunoreactive species detected in these tumors had a mass of 53 kDa, with the exception of tumor A390; in this neoplasm, the p53 antibody recognized a species with a lower molecular weight, suggesting the occurrence of a truncating mutation or a deletion.
To identify potential p53 mutations in these tumors, nested PCR was used to amplify Trp53 sequences from the 18 early-passage P0-GGFβ3 MPNST cultures, and the resulting products were sequenced in their entirety. In keeping with the size shift seen in immunoblots, the Trp53 mRNA expressed in tumor A390 had a frameshift mutation in codon 314, which is within the p53 tetramerization domain. We also found that tumor A202 expressed a transcript with a small in-frame deletion (codons 164 to 172) within the region encoding the p53 DNA binding domain. In tumor B76, a point mutation (c.R207C) was identified. The online tool Mutation Assessor, which assesses the likely effect of a mutation based on the nature of the change and whether it occurs in a conserved region, predicts that the c.R207C mutation has a high probability of affecting p53 function (http://mutationassessor.org, release 1, last accessed July 2012). Although tumor B97 showed no evidence of mutations within Trp53 coding sequences, a 95-bp duplication was evident within the 3′ untranslated region of this mRNA. No Trp53 mutations were identified in the p53-overexpressing A18 and A387 tumors, nor in any of the tumors lacking p53 overexpression. Considered collectively, these findings indicate that 33% (6/18) of the P0-GGFβ3 MPNSTs we examined have abnormal expression or mutation of p53.
Dysregulation of the p19ARF–Mdm–p53 pathway can also result from overexpression of Mdm2 or Mdm4, two proteins that promote the ubiquitination of p53 and its degradation by proteasomes. To determine whether Mdm2 or Mdm4 overexpression occurs in P0-GGFβ3 MPNSTs, lysates of non-neoplastic Schwann cells from the sciatic and trigeminal nerves of P0-GGFβ3 mice and early-passage tumor cultures were lysed, immunoblotted, and probed for these two molecules. Mdm2 and Mdm4 were undetectable in non-neoplastic transgenic Schwann cells (Figure 5H). In contrast, three of the MPNST early-passage cultures (A292, A387, and A390) exhibited prominent Mdm2 overexpression; two of the positive specimens (A387 and A390) also had p53 overexpression. Mdm4 overexpression was evident in one tumor (B96). These results suggest that the p19ARF–Mdm–p53 pathway is dysregulated by Mdm2 or Mdm4 overexpression in some P0-GGFβ3 MPNSTs.
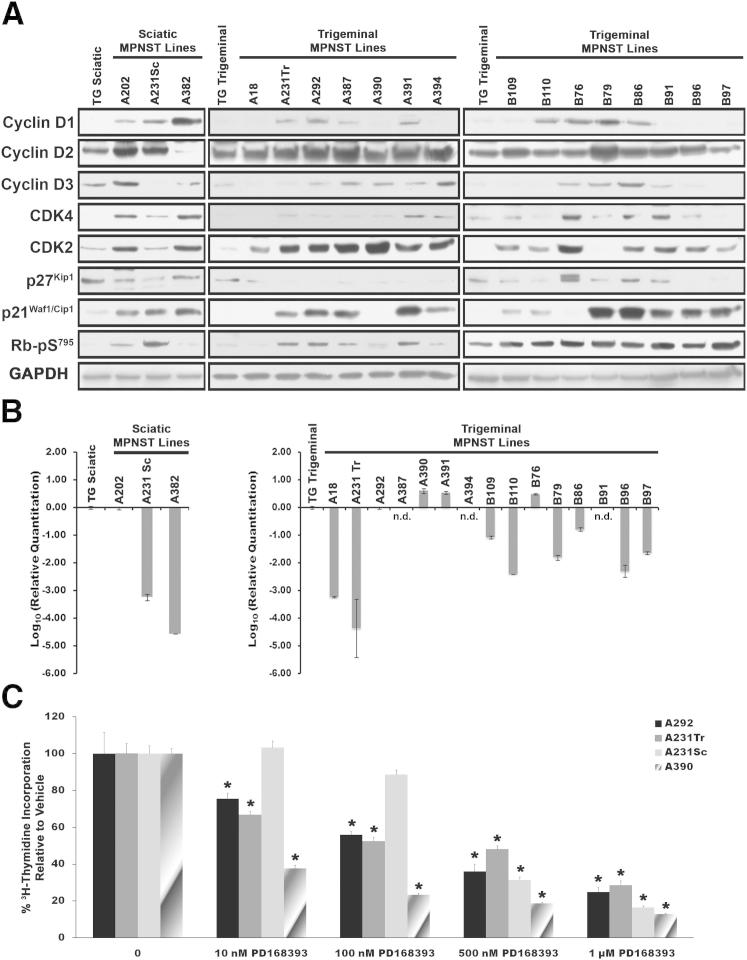
In human MPNSTs, the p16INK4A–cyclin D/CDK4–Rb signaling pathway, which controls S-phase entry, is often dysregulated.48,50–57 This disruption can occur via multiple mechanisms, including CDK4 overexpression, a loss of Rb expression or inappropriate Rb phosphorylation occurring as a result of p16INK4A mutation, CDK4 overexpression, or overexpression of D-cyclins. To determine whether abnormalities of this cell-cycle regulatory pathway occur in P0-GGFβ3 MPNSTs, lysates of non-neoplastic P0-GGFβ3 Schwann cell and early-passage P0-GGFβ3 MPNST cultures were immunoblotted and probed for proteins within this pathway. Cyclins D1, D2, and D3 were all detectable in at least some MPNSTs (Figure 6A), and often were expressed at levels higher than those evident in non-neoplastic Schwann cells. Overexpression of CDK4 was also seen in some tumors (eg, A202, A382, B76, and B91). Because our preliminary experiments indicated that detection of p16INK4A protein was problematic even in non-neoplastic Schwann cells, we instead used qPCR to quantify levels of the Cdkn2a mRNA encoding this protein in our cultures (Figure 6B). Compared with non-neoplastic transgenic Schwann cells, Cdkn2a mRNA expression was greatly decreased (10-fold to more than 10,000-fold) in 10 tumors, and was completely undetectable in three other tumors (A387, A394, and B91). Rb protein and mRNA levels were unchanged in these P0-GGFβ3 MPNSTs (data not shown). However, in keeping with the fact that D-cyclin overexpression, CDK4 overexpression and/or p16INK4A loss was present in most of the tumors, levels of Rb phosphorylated on Ser795 (a modification that promotes cell-cycle progression) were increased in the majority of these MPNSTs (Figure 6A). We conclude that dysregulation of the p16INK4A-cyclin D/CDK4–Rb pathway is quite common in P0-GGFβ3 MPNSTs, much as in their human counterparts.
Figure 6.
Abnormalities in cell-cycle regulatory proteins are common in early-passage cultures of P0-GGFβ3 MPNST cells, but these cells are persistently dependent on aberrant growth factor signaling. A: Immunoblotted lysates of non-neoplastic transgenic Schwann cells and early-passage cultures of P0-GGFβ3 MPNST cells probed for cyclin D1 (1:300 dilution), cyclin D2 (1:500 dilution), cyclin D3 (1:700 dilution), CDK4 (1:500 dilution), CDK2 (1:400 dilution), p27Kip1 (1:600 dilution), p21Cip1 (1:500 dilution), or Rb phosphorylated on Ser795 (1:700 dilution). Blots were reprobed with GAPDH to verify equivalent loading. B: qPCR analyses of Cdkn2a expression in non-neoplastic transgenic Schwann cells and early-passage cultures of P0-GGFβ3 MPNST cells. Transcript levels were normalized to levels detected in non-neoplastic transgenic Schwann cells. C: The ErbB inhibitor PD168393 inhibits DNA synthesis in early-passage cultures of P0-GGFβ3 MPNST cells in a concentration-dependent fashion. [3H]thymidine incorporation relative to vehicle is shown for four early-passage cultures (A292, A231Tr, A231Sc, and A390) derived from MPNSTs arising in P0-GGFβ3 mice. Data are expressed as means ± SEM. *P < 0.05 versus control. n.d., not detectable.
Expression of p21Cip1/Waf1 and p27Kip1, a pair of proteins that impede G1–S progression via their effects on CDK2, cyclin E, and Rb, is also lost in some human MPNSTs.55,58,59 We therefore examined our early-passage P0-GGFβ3 MPNST cultures to determine whether they had similarly lost expression of these cell-cycle inhibitors. Expression of p27Kip1, but not p21Cip1/Waf1, was commonly decreased, compared with non-neoplastic Schwann cells (Figure 6A). Interestingly, CDK2, a key target of p27Kip1 and p21Cip1/Waf1, was also overexpressed in 16/18 tumors (Figure 6A). These findings thus provide further evidence that cell-cycle progression is dysregulated in P0-GGFβ3 MPNSTs.
Abnormalities affecting cell-cycle regulatory proteins can render a tumor growth-factor independent. Consequently, we next asked whether P0-GGFβ3 MPNSTs are persistently dependent on NRG1/ErbB signaling. To address this question, early-passage cultures derived from four independently arising P0-GGFβ3 MPNSTs were challenged with 10 nmol/L to 1 μmol/L concentrations of the pan-ErbB inhibitor PD168393; [3H]thymidine incorporation was then assessed in these cultures. Although some variability in their sensitivity to PD168393 was noted (eg, A231Sc cells were less sensitive), all four of the early-passage cultures exhibited a decrease in DNA synthesis, which occurred in a concentration-dependent manner (Figure 6C). We conclude that, despite the occurrence of cell-cycle protein abnormalities in P0-GGFβ3 MPNSTs, these tumors remain persistently dependent on growth factor signaling.
Whole-Genome aCGH of P0-GGFβ3 MPNSTs Identifies Multiple Areas of Chromosomal CNVs Affecting Genes Implicated in Neoplasia
These findings indicated that MPNSTs arising in P0-GGFβ3 mice exhibit abnormalities in cell-cycle regulatory signaling cascades that parallel those in human MPNSTs. This in turn suggested that an examination of other genomic abnormalities evident in P0-GGFβ3 MPNSTs might identify additional, previously unknown driver genes involved in the pathogenesis of these sarcomas. To identify somatic CNVs potentially affecting key driver genes, genomic DNAs isolated from 11 P0-GGFβ3 MPNST early-passage cultures and cultured non-neoplastic nontransgenic Schwann cells were labeled with Cy5-dUTP and Cy3-dUTP, respectively. Equal amounts of MPNST and Schwann cell DNA were then hybridized to high-density aCGH microarrays, and the relative ratios of the signals from each genomic DNA were compared, to identify regions of unbalanced chromosomal gain or loss.
CNVs affecting whole chromosomes or entire chromosome arms were common in P0-GGFβ3 MPNSTs, occurring an average of 5.3 times (median, 4; range, 3 to 10) in each tumor genome (Table 2). Unbalanced gains predominated, with an average of four per genome (median, 3; range, 2 to 7), with whole chromosome gains evident in every MPNST examined. However, the distribution of these unbalanced gains was nonrandom. Strikingly, all 11 of the early-passage MPNST cultures exhibited gains of chromosome 11. Gains of chromosome 17 and the X chromosome were also common, occurring in 5 of the 11 cultures. In addition, gain of chromosomes 1, 3, 6, 7, 12, 15, 16, and 19 was observed in multiple tumors, albeit at a lower rate. In contrast, unbalanced losses of whole chromosomes or chromosome arms were less common, with an average of 2.8 per genome in affected tumors (median, 1; range, 1 to 7) and occurring in only five P0-GGFβ3 MPNSTs. These losses were also nonrandom, with unbalanced losses of chromosomes 9, 14, 16, and 19 identified in multiple tumors.
Table 2.
Whole Chromosome and Large Region Gains and Losses in P0-GGFβ3 MPNSTs
| Chromosome and gain or loss | Tumor cultures |
|---|---|
| Chromosome 1 | |
| Whole chromosome gain | B86, B91, B96 |
| Chromosome 3 | |
| Whole chromosome gain | A18, B76 |
| Chromosome 4 | |
| Whole chromosome loss | A202 |
| Chromosome 6 | |
| Whole chromosome gain | A18, B76, B86, B91 |
| Chromosome 7 | |
| Whole chromosome gain | A18, A292 |
| Chromosome 8 | |
| Whole chromosome loss | A202 |
| Whole chromosome gain | B76 |
| Chromosome 9 | |
| Whole chromosome loss | A202, A390 |
| Chromosome 10 | |
| Whole chromosome gain | A382 |
| Loss (3051,721–50,790,081) | B76 |
| Chromosome 11 | |
| Whole chromosome gain | A18, A202, A231Tr, A292, A382, A390, A394, B76, B86, B91, B96 |
| Chromosome 12 | |
| Whole chromosome gain | A231Tr, A202 |
| Loss (86,353,623–120,684,848) | A382 |
| Chromosome 13 | |
| Whole chromosome loss | A202 |
| Chromosome 14 | |
| Loss (12,738,652–125,076,290) | A18 |
| Loss (8760,452–125,076,290) | A202 |
| Gain (8794,768–125,076,290) | B76 |
| Chromosome 15 | |
| Whole chromosome gain | A18, A231Tr, B76, B91 |
| Chromosome 16 | |
| Whole chromosome loss | A202, A390 |
| Chromosome 17 | |
| Whole chromosome gain | A18, A292, A394, B76, B91 |
| Chromosome 18 | |
| Whole chromosome loss | A390 |
| Chromosome 19 | |
| Whole chromosome loss | A202, A390 |
| Whole chromosome gain | A382, B86, B96 |
| Gain (10,320,214–61,216,212) | B91 |
| Chromosome X | |
| Whole chromosome gain | A18, A202, A390, A394, B91 |
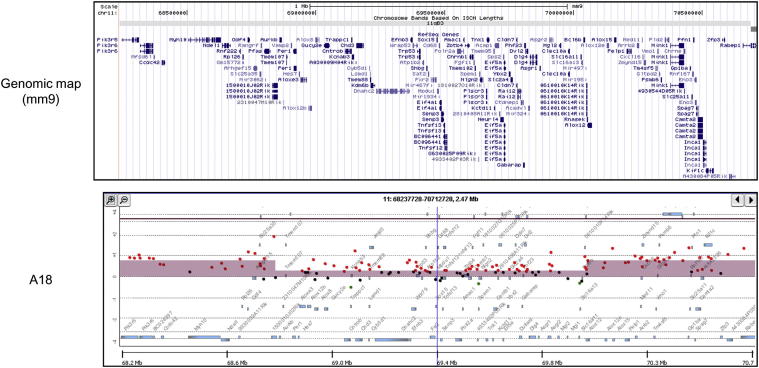
The observation that chromosome 11 was uniformly amplified in the P0-GGFβ3 MPNSTs we examined was curious, because this mouse chromosome carries the Nf1 and Trp53 genes, whose equivalents are commonly lost in human MPNSTs. This led us to examine chromosome 11 in more detail, to determine whether smaller areas of unbalanced loss are present, affecting either the Nf1 or the Trp53 gene. Consistent with our observation that neurofibromin expression is maintained in these tumors, we found no evidence of small losses affecting the Nf1 locus in any of the 11 tumors studied with aCGH. However, one tumor (A18) exhibited a relative loss of a 1.21-Mb segment (chr11:68,846,711–70,058,031) containing the Trp53 gene (Figure 7). This finding, combined with our mutational analyses of Trp53, suggests that the p19ARF–Mdm–p53 cascade in tumor A18 was impaired by both mutation of a Trp53 allele and an overall reduction in Trp53 copy number. Unbalanced losses affecting the Trp53 gene were not evident in any of the other 10 MPNSTs we studied.
Figure 7.
aCGH analysis of the 2.47-Mb region in chromosome 11 encoding the Trp53 tumor suppressor gene in P0-GGFβ3 MPNST A18. The genomic map indicates the location of the genes present within the region illustrated in the accompanying panel, which indicates the relative signals in tumor (red) and non-neoplastic Schwann cell (green) DNAs in the MPNST. The height of the colored bar indicates the relative difference in these two signals. Note the central 1.21-Mb region in which there is a relative copy number loss in the tumor.
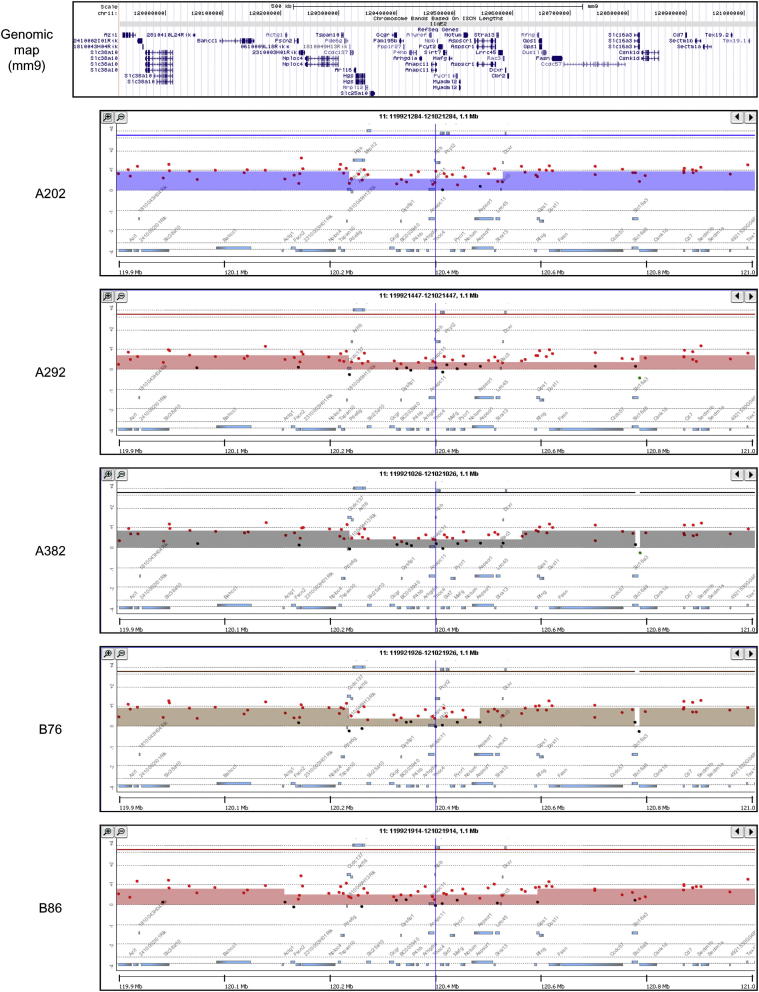
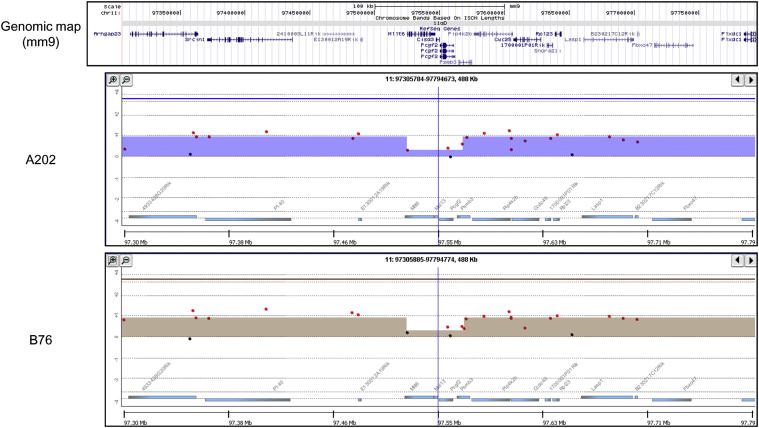
During the course of our examination of chromosome 11, we also noted two other small areas of relative loss that were present in multiple P0-GGFβ3 MPNSTs. Five tumors (A202, A292, A382, B76, and B86) exhibited a relative copy number loss that overlapped in a 225-kb region on chromosome 11 (chr11: 120,322,621–120,548,454; Figure 8); this region includes 24 known genes. To identify candidate driver genes within this interval, we compared these genes to the online comprehensive lists of driver genes available from the Atlas of Genetics and Cytogenetics in Oncology and Hematology,60 CANgenes,61 CIS,62 and the Sanger Cancer Gene Census.63 We found that two genes within this interval [Mafg (v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G [avian] and Aspscr1 (alveolar soft part sarcoma chromosome region, candidate 1 [human])] have been previously identified as genes that are affected in one or more cancer types. In addition to the region noted above, a 44-kb region (chr11: 97,526,341–97,570,620) containing four genes (Mllt6, Cisd3, Pcgf2, and Psmb3) also exhibited a relative reduction in copy number in tumors A202 and B76 (Figure 9). Examination of the databases noted above showed that one of these genes within this interval, Mllt6, has been previously implicated in the pathogenesis of acute myelocytic leukemias.64 Considered together, these observations suggest that multiple regions of relative copy number loss on chromosome 11 affect genes potentially relevant to MPNST pathogenesis.
Figure 8.
aCGH analyses of a 1.1-Mb region in chromosome 11 that surrounds a region of relative copy number loss present in five P0-GGFβ3 MPNSTs (A202, A292, A382, B76, and B86). The genomic map indicates the location of the genes present within the region illustrated in the accompanying panels, which indicate the relative signals in tumor (red) and non-neoplastic Schwann cell (green) DNAs in the MPNSTs. The height of the colored bar indicates the relative difference in these two signals. Note the variably sized central regions in which there is a relative copy number loss in these five MPNSTs.
Figure 9.
aCGH analysis of a 488-kb region in chromosome 11 that surrounds a region of relative copy number loss present in two P0-GGFβ3 MPNSTs (A202 and B76). The genomic map indicates the location of the genes present within the region illustrated in the accompanying panels, which indicate the relative signals in tumor (red) and non-neoplastic Schwann cell (green) DNAs in the MPNSTs. The height of the colored bar indicates the relative difference in these two signals. Note the central 44-kb region in which there is a relative copy number loss in these two MPNSTs.
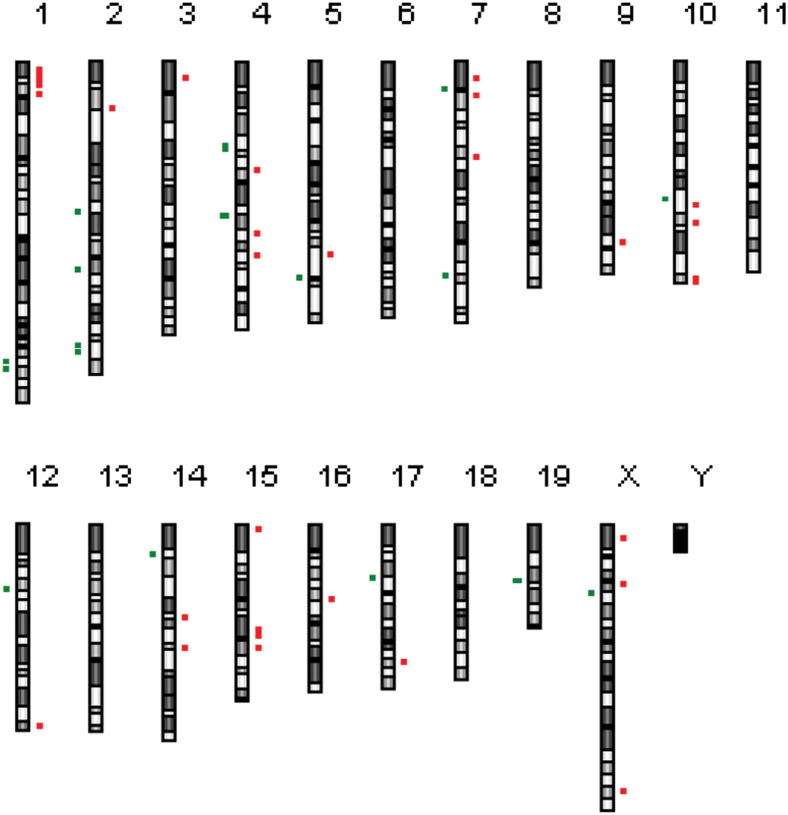
Unlike human cancers, in mouse cancer the genomes tend to have whole chromosome gains or losses with only a limited number of small CNVs. However, those small regions of unbalanced gain or loss that do occur in murine cancers develop under strong selective pressure, which implies that they contain important driver genes.65 Consequently, we next focused our attention on the small CNVs occurring in P0-GGFβ3 MPNSTs. In our 11 early-passage MPNST cultures, 44 focal regions of unbalanced chromosomal gain and loss were evident. These small CNVs, which included 26 unbalanced gains and 18 unbalanced losses, were spread across 15 chromosomes (Figure 10). Several of the small CNVs were evident in multiple P0-GGFβ3 MPNSTs (Table 3). For instance, a deletion on chromosome 4 was present in 6 of the 11 tumors; this region of chromosomal loss (chr4: 88,934,158–89,039,587) contains the Cdkn2a and Cdkn2b genes. A global examination of the focal CNVs present in these P0-GGFβ3 MPNSTs showed that genes represented in the Atlas of Genetics and Cytogenetics in Oncology and Hematology, CANgenes, CIS, or Sanger Cancer Gene Census databases were evident in 22 of the 44 regions; 13 were amplified and 9 were deleted (Table 3). Overall, 39 genes previously implicated in the pathogenesis of other human cancer types were identified within these regions (one to eight genes per focal CNV).
Figure 10.
Ideogram indicating the locations of focal CNVs detected by aCGH data in early-passage cultures established from P0-GGFβ3 MPNSTs. Regions of copy number gain are indicated in red, and regions of unbalanced loss are indicated in green. The lengths of the red and green bars are proportional to the size of the CNV.
Table 3.
Genes within Small and Minimal Critical Regions of Gain or Loss in P0-GGFβ3 MPNSTs
| Minimal critical region | Gain or loss | Cell lines | Genes |
|---|---|---|---|
| Chromosome 1 | |||
| 18,123,973–20,989,757 | Gain | A18 | Crisp4, Defb18, Defb49, Defb41, Gm15386, Tcfap2 days, Tcfap2b,Pkhd1, Mir206, Il17a, Il17f, AK205147, Mcm3, Paqr8, Efhc1 |
| 172,530,936–172,637,996 | Loss | A390 | Olfm12b, Atf6 |
| 173,444,742–173,494,144 | Loss | A292, B96, B91, B86, B76 | BC145376, Itln1, Cd244 |
| Chromosome 2 | |||
| 27,740,854–27,893,558 | Gain | A202 | Col15a1,BC152372 |
| 85,976,236–86,076,234 | Gain | A231Tr | Olfr1040, Olfr1042, Olfr1043, Olfr1044, Olfr52, Olfr1045, Olfr1046, Olfr1047, Olfr1048 |
| 86,061,726–86,087,429 | Loss | A202, A18 | Olfr1047, Olfr1048 |
| 118,994,902–119,055,137 | Loss | A18 | Gchfr, Dnajc17, Gm14137, Zfyve19, Ppp1r14 days |
| 163,824,466–163,964,059 | Loss | A18 | Ywhab, Pabpc1l, 1810053B01Rik, Tomm34,Stk4 |
| 165,860,752–165,971,779 | Loss | A18 | Ncoa3, Sulf2 |
| Chromosome 3 | |||
| 8,919,417–9,605,520 | Gain | A202 | Mrps28,Tpd52, AK016002, Zbtb10, C030034L19Rik, Zfp704 |
| Chromosome 4 | |||
| 47,961,806–52,868,964 | Loss | A390 | AK133250, Nr4a3, Stx17, Erp44, Invs, Tex10, 5730528L13Rik, Tmeff1, Murc, E130309F12Rik, Acnat2, Acnat1, AK018513, Baat, Mrp150, Aldob, Zfp189, Rnf20, Ppp3r2, 2810432L12Rik, Grin3a, Cylc2, S60135, S60130, AK012547, Smc2, 4930547C10Rik, Olfr275 |
| 62,162,092–62,182,694 | Gain | A231Tr, A202, A18, A390 | Hdhd3,Alad |
| 88,168,201–88,829,428 | Loss | A382 | Ifnb1, Gm12597, Ifna14, Ifna9, Ifna12,C87499, Ifna13, Gm13280, Ifna2, Ifnab, Klh19, Gm13271, Gm13285, Gm13276, Ifnz, Gm13275, Gm13287, Ifna7, Ifna11, Ifna6, Ifna5, Ifna4, Ifna1, Ifn3, Mir31, AK144962, Gm12603, Mtap |
| 88,934,158–89,039,587 | Loss | A231Tr, B96, B91, B86, A382, A292 | Cdkn2a, AK148321, Cdkn2b |
| 98,831,785–99,056,334 | Gain | A202 | Atg4c |
| 111,745,189–112,130,291 | Gain | A231Tr, B96, B91, B86, B76, A394, A382, A292, A202, A390 | Skint4, Skint3, Skint9 |
| Chromosome 5 | |||
| 110,540,802–111,792,099 | Gain | A202 | Zfp605,Chfr, AK053084, Golga3, Ankle2, Pgam5, Pxmp2, Pole, P2rx2, Gm1679, Fbrsl1, 2410025L10Rik, AK088579, AK041600, Galnt9, Ddx51, Ep400, Noc4l, AK044789, Pus1, Ulk1, Hscb, Chek2, Ttc28, Mir701, Pitpnb, AK039279 |
| 124,704,754–124,838,557 | Loss | A18 | Mphosph9, 281006K23Rik, Cdk2ap1, Sbno1 |
| Chromosome 7 | |||
| 12,807,582–14,904,447 | Gain | A18 | Vmn1r81, Vmn1r82, Vmn1r83, Vmn1r84, Zfp551, Zfp606, 290092C05Rik, Vmn2r53, Vmn2r54, Vmn2r55, Vmn2r56, Zscan18, Zfp329, Zfp110, Zfp128,Zscan22, Rp25, Zfp324, 2310014L17Rik, Zfp446, Zbtb45, Slc27a5, Trim28, Chmp2a, Ube2m, Mzf1, Vmn1r86, Vmn1r85, Vmn1r87, Vmn1r88, Vmn1r89, 6330408A02Rik, AK132923, Lig1, Pla2g4c, Cabp5, Bsph1, 9230107M04Rik, Bsph2, Sult2a5, Sult2a2, Sult2a1, Sult2a4, Sult2a3, Sult2a6, EG629219 |
| 16,685,001–17,087,560 | Loss | A18 | Napa, Kptn, Slc8a2, Meis3, mKIAA0134, Dhx34, Gpr77, C5ar1, Prr24, Ccdc9,Bbc3, Sae1, Zc3h4, Tmem160, Npas1, Grlf1 |
| 19,998,996–20,191,762 | Gain | B76 | Ckm,AK020867, mKIAA1860, Mark4, Exoc312, Bloc1s3, Trappc6a, Nkpd1, Lrrc68, AK195979, Gemin7, Zfp296, Sfrs16, Clasrp, Rel |
| 54,644,122–54,885,354 | Gain | B86 | Mrgpra2a, Mrgpra2b, Mrgpra3 |
| 123,174,938–123,176,223 | Loss | A292, A382 | Sox6 |
| Chromosome 9 | |||
| 103,906,930–103,968,229 | Gain | A18 | Nphp3, Uba5, Acad11 |
| Chromosome 10 | |||
| 80,128,600–80,381,401 | Loss | A394, B91 | Mknk2, Mnk2, Mobkl2a, Izumo4, Ap3d1, Dot1l, Plekhj1, Sf3a2, Amh, Jsrp1, Oaz1, Mir1982,AK087942, Lingo3, Lsm7, 3110056O03Rik, Spp12b, Tmprss9, Timm13, Lmnb2 |
| 80,993,669–83,063,521 | Gain | B76 | Gna11, Aes, Tle2, Tle6, Sirt6, BC025920, Ankrd24, Gm10778, BC064812, Zfp781, AK136722, BC062115, Zfp873, AU041133, B230315N10Rik, Mif1, Gm1553, 1190007I07Rik, Tdg, Glt8d2, Hcfc2, Nfyb, AK006461, Txnrd1, Eid3, Chst11, Slc41a2, D10Wsu102e, Aldh1l2, Appl |
| 93,173,564–93,303,322 | Gain | A202 | Ntn4,AK145310, DQ556351, Usp44 |
| 125,404,179–127,428,204 | Gain | A18 | Lrig3, AK005576, Xrcc6bp1, AK087024, Ctdsp2, Mir26a-2, Mir546, Avil, Tsfm, Mettl1, Cyp27b1, Cdk4, Tspan31, March9, Agap2, Os9, B4galnt1, Slc26a10, Arhgef25, Dtx3, Deltex3, AK077682, Pip4k2c, Kif5a, Dctn2, Mbd6, Ddit3, Mars, Arhgap9, Gli1, Inhbe, Inhbc, R3hdm2, Stac3, Ndufa4l2, Shmt2, Nxph4, Lrp1, Stat6, Nab2, BC029853, AK005897, Tmem194, Myo1a, Tac2, Zbtb39, Gpr182, Rdh1, Rdh9, Rdh16, Rdh18-ps, BC089597, Rdh7, Sdr9c7, Hsd17 |
| Chromosome 12 | |||
| 37,823,207–37,970,476 | Loss | A292 | Meox2, Tmem195 |
| 115,837,534–117,190,767 | Gain | A202, A18 | Antibody parts,X73024, AB345949 |
| Chromosome 14 | |||
| 21,275,133–22,200,898 | Loss | A18 | Anxa7, Zmynd17, Ppp3cb, 1810062O18Rik, Usp54, Myoz1, Synpo2l, Sec24c,AK018074, AK204442, 6230400D17Rik, 2310021P13Rik, Kiaa0913, Ndst2, Camk2g, AK039280, Plau, Vcl, Ap3m1, Adk |
| 53,008,490–54,874,949 | Gain | B86 | Olfr1511, Olfr1510,Tcra, Olfr1509, Olfr1508, Olfr1507, Gm10886, AJ311366, X98059, Z22845, Gm13948, Gm10905, X56719, D16605, Gm13960, Gm10895, Gm13980, ENSMUSG000000836, Vdelta6.4, GM16979, GM13907, AK138453, X02930, Gm10902, AK038197, EG547424, M21205, X56722, X02969, Gm16460, Gm16979, Gm13907, U07879, Gm16980, Gm13980, AK038197, EG547424, Gm17002, Gm13953, Gm13948, Gm16452, M34214and multiple TCR-alpha genes |
| 69,877,095–69,987,156 | Gain | A390, B76, B86, B91, B96 | Slc25a37, D930020E02Rik,AK046510, Entpd4, AK086749, BC086315 |
| Chromosome 15 | |||
| 3,228,882–4,824,497 | Gain | A390 | Sepp1, Ccdc152, Ghr, Fbxo4, AW549877, AK041537, BC037032, Oxct1, AK028088, Plcxd3, C6 |
| 60,562,187–62,065,471 | Gain | A202, A390 | Fam84b, 9930014A18Rik, DQ715236, AK015045, A1bg, EU234017, AK163289, AK132958, Myc, Pvt1, H2afy2 |
| 71,284,106–72,565,945 | Gain | A390 | Fam135b, Col22a1, Kcnk9, Trappc9 |
| Chromosome 16 | |||
| 43,620,829–43,773,291 | Gain | B76 | Zbtb20,AK038731, Mir568, Tigit, Drd3 |
| Chromosome 17 | |||
| 30,714,112–31,047,626 | Loss | A231Tr, A202, B86, A390, A382 | Glo1, Dnahc8, AK018977, Glp1r |
| 79,124,627–79,148,417 | Gain | B91 | Strn |
| Chromosome 19 | |||
| 32,866,825–32,944,421 | Loss | A202 | Pten, B430203M17Rik |
| Chromosome X | |||
| 7,276,714–7,415,340 | Gain | A394 | Gpkow, Wdr45, Praf2, Ccdc120, Mir684-1, Tcfe3, Gripap1, Kcnd1 |
| 34,183,857–34,755,807 | Gain | B76 | Akap17b, Slc25a43, Slc25a5, C330007P06Rik, Ube2a, Nkrf,Sept6, Ankrd58, Rp139, Snora69, Upf3b, Nkap, Akap14, Ndufa1, Rnf113a1, Gm9, Rhox1 |
| 39,447,121–40,770,156 | Loss | A292 | Xiap, Stag2, Xlp, Sh2d1a, Odz1, Ten-m1 |
| 153,911,519–153,991,073 | Gain | A202 | Sms, Mbtps2 |
Cancer driver genes represented in the Atlas of Genetics and Cytogenetics in Oncology and Haematology, CANgenes, CIS, and/or Sanger Cancer Gene Census databases are highlighted with boldface and underlining.
The functions of the candidate cancer driver genes present within focal CNVs in P0-GGFβ3 MPNSTs were varied, and affected multiple processes relevant to tumorigenesis (Table 4). A number of the affected loci encode proteins that control proliferation (Myc, Tpd52, Strn; all amplified), the cell cycle (Cdkn2a, Cdkn2b, Chfr, Chek2, Cdk2ap1, Cdk4), and chromatin remodeling (Ep400). Several genes regulating apoptosis (Bbc3/PUMA, Ddit3/GADD153, XIAP) were also present within focal areas of unbalanced gain or loss. Genes were identified that encode molecules in key cytoplasmic signaling pathways, including the Hippo (Stk4), Notch (Dtx3), Hedgehog (Gli1), Arf (Agap2), Rho (Arhgap9), PI3 kinase (Pten), and Myc (Myc) pathways; also affected were genes for growth factors, cytokines, and growth factor receptors potentially upstream of some of these signaling cascades (Ghr, Il17a, Inhbe). Other candidate cancer driver genes within these small CNVs function in key metabolic pathways (Aldob, Mars, Alad), transcriptional control (Ncoa3, Nr4a3, Zscan22), vesicular trafficking (Napa), and cell motility (Sept6, Vcl).
Table 4.
Functions of Candidate Driver Mouse Genes in Small Regions of Unbalanced Gain and Loss
| Gene symbol | Gene name | Function |
|---|---|---|
| Pkhd1 | polycystic kidney and hepatic disease 1 | Cilium assembly, centrosome duplication |
| Il17a | interleukin 17A | Inflammatory response |
| Stk4 | serine/threonine kinase 4 | Hippo signaling cascade |
| Ncoa3 | nuclear receptor coactivator 3 | Ligand-dependent transcription factor |
| Sulf2 | sulfatase 2 | Extracellular sulfatase |
| Tpd52 | tumor protein D52 | Promotes proliferation, survival and metastasis; regulates lysosomal trafficking |
| Nr4a3 | nuclear receptor subfamily 4, group A, member 3 | Ligand-dependent transcription factor |
| Aldob | aldolase B, fructose-biphosphate | Glycolysis |
| Alad | aminolevulinate, delta-, dehydratase | Heme biosynthesis |
| Cdkn2a | cyclin-dependent kinase inhibitor 2A | Cell cycle arrest |
| Cdkn2b | cyclin-dependent kinase inhibitor 2B | Cell cycle arrest |
| Chfr | checkpoint with forkhead and ring finger domains | Mitotic checkpoint protein, maintains chromosome integrity |
| Ep400 | E1A binding protein p400 (syn. mDomino) | ATP-dependent chromatin-remodeling protein |
| Chek2 | checkpoint kinase 2 (syn. Rad53) | DNA damage checkpoint control |
| Cdk2ap1 | CDK2 (cyclin-dependent kinase 2)-associated protein 1 | Negative regulator of Cdk2 function |
| Sbno1 | strawberry notch homolog 1 (Drosophila) | Unknown |
| Zscan22 | zinc finger and SCAN domain containing 22 | Gli–Kruppel family transcription factor |
| Napa | N-ethylmaleimide sensitive fusion protein attachment protein alpha | Trans-Golgi vesicle trafficking |
| Bbc3 | Bcl2 binding component 3 (syn. PUMA) | p53-regulated inducer of apoptosis |
| Mark4 | MAP/microtubule affinity-regulating kinase 4 | Serine-threonine kinase |
| Gna11 | guanine nucleotide binding protein, alpha 11 | G-protein coupled receptor signaling mediator |
| Zfp873 | zinc finger protein 873 | Unknown |
| Cdk4 | cyclin-dependent kinase 4 | Cell cycle progression |
| Agap2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | Regulator of ARF activity |
| Dtx3 | deltex 3 homolog (Drosophila) | Notch signaling pathway |
| Ddit3 | DNA damage-inducible transcript 3 (syn. GADD153, CHOP10) | Promotes cell cycle arrest, macroautophagy |
| Mars | methionine-tRNA synthetase | Charges Met-tRNA with methionine |
| Arhgap9 | Rho GTPase activating protein 9 | Inactivates Rho signaling |
| Gli1 | GLI-Kruppel family member GLI1 | Transcription factor that promotes proliferation, Hedgehog signaling pathway |
| Inhbe | inhibin beta E (syn. activin, activin betaE) | Growth factor |
| Vcl | vinculin | Adherens junction component, modulates focal adhesion |
| Tcra | T cell receptor alpha chain (gene cluster) | Antigen recognition |
| Ghr | growth hormone receptor | Growth hormone receptor |
| Myc | myelocytomatosis oncogene | Nuclear phosphoprotein regulating cell cycle progression, apoptosis and transformation |
| Pvt1 | plasmacytoma variant translocation 1 | Encodes multiple miRNAs |
| Strn | striatin, calmodulin binding protein | Cytoplasmic signaling molecule, inhibits proliferation |
| Pten | phosphatase and tensin homolog | Inhibits PI3 kinase activity |
| Sept6 | septin 6 | Cytoskeleton, regulates motility |
| Xiap | X-linked inhibitor of apoptosis | Inhibits apoptosis |
syn., synonym.
It was also notable, however, that some small CNVs that did not contain genes previously implicated in the pathogenesis of any tumor type were repeatedly identified in P0-GGFβ3 MPNSTs (Table 3). The most common of these CNVs was a region of copy number gain on chromosome 4 (chr4: 111,745,189–112,130,291) that was present in 10/11 tumors examined. This region contains three genes (Skint4, Skint3, and Skint9) with poorly understood function; Skint1, the prototypic member of this group of molecules, has been implicated in the maturation of epidermal γδ T cells.66 Other recurrent CNVs lacking genes previously implicated in the pathogenesis of human cancers included a region of deletion on chromosome 17 that occurred in 5/11 tumors (chr17: 30,714,112–31,047,626, encompassing Glo1, Dnahc8, AK018977, and Glp1R), a focal gain on chromosome 14 that was present in 5/11 tumors (chr14: 69,877,095–69,987,156, encompassing Slc25a37 and Entpd4), and a small deletion on chromosome 1 that was evident in five tumors (chr1: 173,444,742–173,494,144, encompassing Itln1 and Cd244). Considered together, these observations suggest that several of the relatively small regions of chromosomal copy number gain or loss evident in P0-GGFβ3 MPNSTs contain important driver genes that have not been previously implicated in carcinogenesis.
Discussion
We have previously shown that the NRG1/ErbB signaling pathway is constitutively activated in human neurofibromas and MPNSTs,40 that this pathway promotes the proliferation and migration of neoplastic Schwann cells isolated from these human tumors,39,40 and that transgenic mice expressing the NRG1 isoform GGFβ3 in Schwann cells develop MPNSTs.41 Building on these earlier findings, in the present study we have demonstrated that P0-GGFβ3 mice develop large numbers of neurofibromas and that these lesions potentially progress to become MPNSTs at a high frequency. This suggests that neurofibroma–MPNST progression in P0-GGFβ3 mice, in contrast to that in Nf1 knockout models, occurs commonly and recapitulates the pathological process seen in human NF1. Furthermore, the molecular abnormalities evident in P0-GGFβ3 MPNSTs, which include Ras hyperactivation and defects in the p19ARF–Mdm–p53, p16INK4A–cyclin D/CDK4–Rb, and p27Kip1/CDK2 pathways, parallel the alterations often encountered in human MPNSTs. These findings led us to hypothesize that P0-GGFβ3 mice could be used to identify previously unknown driver genes mediating neurofibroma pathogenesis and the subsequent progression of these tumors to become MPNSTs. Consistent with this postulate, our aCGH studies identified multiple chromosomal gains and losses that affected a number of driver genes that had been previously implicated in the pathogenesis of other types of human cancers, but not in MPNSTs. We also identified several recurrent CNVs that affected genes that have never been implicated in the pathogenesis of any type of human cancer, suggesting that P0-GGFβ3 mice can be useful for the identification of previously unknown driver genes with novel mechanisms of action.
More than 90% of the P0-GGFβ3 mice that we studied developed multiple low-grade nerve sheath tumors with histological and immunohistochemical features identical to those of human neurofibromas. Our previous necropsy examinations of younger (0 to 4 months old) P0-GGFβ3 mice showed no evidence of such lesions,41 which indicates that these neurofibromas are not congenital, but instead develop later in life. This delay in the development of neurofibromas, along with the fact that a small percentage of P0-GGFβ3 mice do not develop neurofibromas, implies that GGFβ3 overexpression alone is not sufficient for the pathogenesis of these benign neoplasms and that additional abnormalities, such as the mutation of as yet unknown driver genes, are required for their development. Consequently, P0-GGFβ3 mice can be a useful model system for identifying novel driver genes mediating neurofibroma pathogenesis.
In addition, the distribution of neurofibromas in P0-GGFβ3 mice raises interesting questions about the origin of these tumors. We observed that neurofibromas arising in P0-GGFβ3 mice typically develop within ganglia associated with sensory (spinal dorsal nerve root), sympathetic (celiac, superior mesenteric, and superior cervical ganglia), and motor (trigeminal) nerves. This suggests that neurofibroma development within ganglia in P0-GGFβ3 mice is a result of these neoplasms being derived from a progenitor intrinsic to these structures, or that it occurs because the intraganglionic microenvironment is particularly conducive to tumorigenesis. Of note, embryonic dorsal root ganglia have been shown to contain a progenitor pool that expands and becomes hypersensitive to growth factors after Nf1 loss, and it has been proposed that neurofibromas may be derived from these progenitors.67 Further studies would be warranted to determine whether NRG1 overexpression similarly expands this intraganglionic progenitor pool, whether these cells give rise to neurofibromas in P0-GGFβ3 mice, and whether the expansion of this progenitor pool and subsequent neurofibroma pathogenesis in Nf1 knockout models is also dependent on NRG1 signaling.
MPNSTs were also present in P0-GGFβ3 mice carrying multiple neurofibromas but the MPNSTs most often occurred as single lesions. Furthermore, we noted that some neurofibromas contained microscopic foci morphologically identical to the MPNSTs that we found in these animals. Considered together, these findings suggest that MPNSTs arise via progression from pre-existing neurofibromas in P0-GGFβ3 mice, much as is seen in human NF1. The molecular abnormalities present in P0-GGFβ3 MPNSTs also largely parallel those typically seen in their human counterparts. We did not find evidence of a loss of neurofibromin protein or Nf1 mRNA in all of our MPNSTs. Although these initial studies did not exclude the possibility that point mutations might be present in the Nf1 transcripts expressed in P0-GGFβ3 MPNSTs, we have since performed RNA-Seq high-throughput sequencing on two of these sarcomas and found no evidence of Nf1 mutations (data not shown). The absence of Nf1 mutations but continued neurofibromin expression in P0-GGFβ3 MPNSTs raises the question of whether the sarcomas seen in these mice are analogous to human sporadic MPNSTs, at least some of which lack NF1 mutations.10 However, we also found that Ras proteins, which are negatively regulated by neurofibromin, are hyperactivated in P0-GGFβ3 MPNSTs, indicating that these tumors demonstrate the biochemical abnormality classically associated with neurofibromin loss. Consequently, these findings argue that class II NRG1 overexpression results in neurofibroma and MPNST pathogenesis, because this growth factor acts largely within the same biochemical pathways that are affected by NF1 loss. Furthermore, disruption of the p16INK4A–cyclin D/CDK4–Rb pathway was evident in every P0-GGFβ3 MPNST we examined. Lost or diminished Cdkn2a expression, which often correlated with copy number loss affecting this locus, was particularly common, occurring in 72% (13/18) of the MPNSTs. This parallels observations in human MPNSTs, where 50% to 86% of these tumors have been shown to have CDKN2A loss.51,52,55
Our finding that 33% (6/18) of P0-GGFβ3 MPNSTs demonstrate abnormal p53 expression or mutation is also consistent with multiple earlier studies demonstrating that p53 abnormalities are present in a major subset of human MPNSTs.11,12,48,49,58 Mdm2 overexpression, which we found in 17% of the P0-GGFβ3 MPNSTs we examined, is likewise present in 20% to 25% of human MPNSTs.48,68 To our knowledge, the possibility that Mdm4 is overexpressed in human MPNSTs has not yet been examined. Finally, p27Kip1 abnormalities, which we identified in P0-GGFβ MPNSTs, have also been identified in human MPNSTs.58 Considered together, this pathological and molecular evidence indicates that tumorigenesis in the peripheral nervous system of P0-GGFβ3 mice occurs via a process that is highly similar to that operant in human NF1.
Mouse cancer models can greatly facilitate the rapid identification of driver mutations relevant to the pathogenesis of their human counterpart. In part, this advantage is due to intrinsic differences in the stability of human and mouse cancer genomes.69 Human cancer genomes are usually structurally complex, with numerous regions of focal chromosomal gain and loss; however, only a small subset of these focal chromosomal gains and losses contain cancer driver genes (genes that promote cancer pathogenesis), with the remainder resulting in passenger mutations (mutations that do not confer an advantage on the tumor cells). In contrast, mouse cancer genomes have relatively few structural abnormalities; they tend rather to have whole chromosome gains or losses, with a much more limited number of focal gains and losses. These patterns are borne out in both human MPNSTs and the MPNSTs that arise in P0-GGFβ3 mice. Human MPNSTs have complex hypodiploid or near-triploid karyotypes, with numerous and highly diverse regions of chromosomal gain or loss.15–22,70,71 However, our aCGH studies revealed a more restricted set of chromosomal abnormalities in the MPNSTs that arose in P0-GGFβ3 mice, with whole chromosome or whole chromosome arm CNVs being particularly common. The whole chromosome gains and losses evident in these mouse cancer genomes were not random, because some events occurred repeatedly. Of particular note, copy number gains affecting chromosome 11 were uniformly present in all 18 MPNSTs. Mouse chromosome 11 contains regions that are syntenic with segments of human chromosomes 1, 2, 5, 7, 16, 17, and 22. Although comparative genomic hybridization analyses of human MPNSTs have not identified CNVs affecting chromosomes 1, 2, 5, 16, or 22, gains affecting the short arm of chromosome 7 (7p) and the long arm of chromosome 17 (17q) are reproducibly present in human MPNSTs.20,21,72,73 Although the region on chromosome 7p that is affected in human MPNSTs is not syntenic with mouse chromosome 11, the 17q amplicon occurring in human MPNSTs is syntenic with mouse chromosome 11. Genes within this region of copy number gain that have been suggested to potentially promote neurofibroma–MPNST progression in humans include BIRC5/survivin, ERBB2, TOP2A, PTDSR/Jmjd6, SEPT9, and SOSC3.73 Going forward, it should be of great interest to establish the functional significance of these genes in human and mouse MPNST pathogenesis and to determine whether there are additional genes on mouse chromosome 11 that promote MPNST pathogenesis.
The intrinsic stability of mouse cancer genomes also ensures that those focal gains or losses that do occur in these tumors arise as a result of strong selective pressure; this selective pressure often reflects the presence of a driver gene in the affected interval.18 Moreover, because key driver genes (and their associated pathways) are usually evolutionarily conserved, driver genes within focal CNVs occurring in mouse cancers are potentially relevant to the pathogenesis of their human counterparts. We identified a total of 44 focal gains and losses in the 18 P0-GGFβ3 MPNSTs; this limited number of focal CNVs affected 39 genes that had previously been implicated in the pathogenesis of human cancer types other than MPNSTs. Several of these CNVs occurred in multiple tumors, emphasizing their importance to the pathogenesis of P0-GGFβ3 MPNSTs. Other CNVs, however, were less common, with some identified on only a single MPNST. This does not mean that these rarer CNVs are irrelevant to MPNST pathogenesis, because at least some of these gains and losses affected genes that have already been linked to the development of MPNSTs. For instance, we found a focal loss on chromosome 19 (chr19; 32,866,825–32,944,421) in only one of the tumors we examined with aCGH; however, this loss affected the Pten tumor suppressor gene, which has been implicated in the pathogenesis of human MPNSTs74,75 and which, when deleted in the setting of K-Ras activation, leads to MPNST pathogenesis in genetically engineered mice.75 Consequently, further studies are warranted, performing aCGH on a larger series of P0-GGFβ3 MPNSTs to identify additional focal CNVs relevant to MPNST pathogenesis, determine how frequently each of these focal gains and losses occur, and investigate whether specific gains and losses are also associated with distinct molecular subtypes of MPNSTs.
To date, only NF1 loss has been implicated in the development of neurofibromas, whereas studies of the genes responsible for neurofibroma–MPNST progression have focused almost exclusively on proteins within cell-cycle regulatory pathways. Although we identified abnormalities in these same molecules in P0-GGFβ3 MPNSTs, our aCGH findings suggest that abnormalities affecting a much more diverse variety of cellular functions also occur in these sarcomas. Potentially affected functions include proliferative signaling (eg, Stk4, Tpd52, Agap2, Gna11, Dtx3), transcription (eg, Ncoa3, Gli1), tumor cell survival (eg, Xiap, Bbc3, Ddit3), cytoskeletal structure and motility (eg, Vcl, Sept6), immune function (eg, Il17a), and metabolism (eg, Aldob, Alad, Mars). The occurrence of abnormalities affecting a variety of cellular functions in P0-GGFβ3 MPNSTs is consistent with observations in numerous other human cancer types.70 However, the observation that some recurrent focal gains and losses did not contain any genes previously implicated in the pathogenesis of other human cancer types is even more intriguing, because this suggests that these intervals may contain novel driver genes. In future studies, it would be of great interest to identify these novel driver genes, establish their mechanism of action, and determine whether they are similarly affected in human neurofibromas and/or MPNSTs. Having said that, we also emphasize that the abnormalities that we have detected with aCGH likely represent only a subset of the abnormalities driving the pathogenesis of peripheral nerve sheath tumors in P0-GGFβ3 mice. This is because aCGH analyses identify relatively large CNVs and do not detect some quite common types of mutations that are highly relevant to tumorigenesis (eg, relatively small deletions, frameshift mutations, missense mutations, and nonsense mutations). Approaches such as sequencing the transcriptome and the exome would be required to identify these latter types of mutations.
In summary, we have shown that the vast majority of transgenic mice overexpressing the NRG1 isoform GGFβ3 in Schwann cells develop numerous neurofibromas and smaller numbers of MPNSTs which potentially arise from pre-existing neurofibromas. These observations, considered together with our finding that molecular abnormalities occurring in human neurofibromas and MPNSTs are present also in P0-GGFβ3 MPNSTs, argue that P0-GGFβ3 mice appropriately model the process of neurofibroma–MPNST progression seen in human NF1. A number of previous reports have commented on the fact that chromosomal gains and losses are highly variable in human MPNSTs and that tumor suppressors such as p53 are not uniformly mutated in these neoplasms.2 These observations suggest that there may be more than one pathway leading to MPNST pathogenesis. Because neurofibroma and MPNST formation in P0-GGFβ3 mice results from growth factor overexpression (rather than from ablation of specific tumor suppressor genes), tumorigenesis in these animals, unlike in previous knockout models, is not necessarily dependent on the mutation of a restricted collection of tumor suppressors; this suggestion is consistent with our observation that key tumor suppressor mutations (eg, p53) and focal CNVs were variably present in P0-GGFβ3 MPNSTs. Consequently, P0-GGFβ3 mice represent a novel model system that could be used to identify alternative pathways mediating neurofibroma–MPNST progression. We intend to conduct future studies directed toward expanding our aCGH experiments to include a larger cohort of P0-GGFβ3 neurofibromas and MPNSTs and examining these neoplasms with additional approaches, such as RNA-Seq and exome sequencing, so that more subtle mutations promoting peripheral nervous system tumorigenesis can be identified and considered as potential therapeutic targets.
Acknowledgment
We thank the Alabama Neuroscience Blueprint Initiative Cellular and Molecular Neuropathology Core and the UAB Neuroscience Core for technical assistance.
Footnotes
Supported by the NIH National Cancer Institute grants R01 CA122804 (S.L.C.) and R01 CA134773 (K.A.R. and S.L.C.), National Institute of Neurological Diseases and Stroke grant R01 NS048353 (S.L.C.) and F30 NS063626 (N.M.B.), and the Department of Defense grants X81XWH-09-1-0086 and W81XWH-12-1-0164 (S.L.C.). The University of Alabama at Birmingham Neuroscience Blueprint Core Centers, which provided technical assistance, were supported by NIH grants P30 NS057098 and P30 NS474466.
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Defense.
No person at The University of Alabama at Birmingham was involved in the peer review process or final disposition for this article.
Supplemental Data
A: Representative RT-PCR control experiments supporting the specificity of the results illustrated in Figure 3. PCR was performed on reverse transcription reactions performed in the presence (+) and absence (−) of reverse transcriptase for each of the markers indicated to the right of each panel. The positive control tissues and cultures [cultured WT mouse Schwann cells (cultured s.c.), WT mouse sciatic nerve (nerve), WT mouse brain (brain), WT aorta (aorta), and WT gastrocnemius muscle (muscle)] were selected because they are known to express the markers examined in these experiments. Also shown are PCR reactions performed in the presence (+) and absence (−) of reverse transcriptase using mRNA isolated from representative P0-GGFβ3 MPNST early-passage cultures that were positive for Schwann cell (S100β, P0, MBP, PMP22, p75LNTR, Sox10, Pax3, Krox20, GFAP, GAP43), neural (neurofilament, peripherin), and muscle (calponin, SM22α, α-SMA, desmin, MyoD1) markers. PCR reactions were also performed on water blanks (blk) to confirm an absence of contamination in these experiments. B: qPCR analyses of Nf1 expression in non-neoplastic transgenic Schwann cells and early-passage cultures of P0-GGFβ3 MPNST cells. Transcript levels were normalized to levels detected in non-neoplastic transgenic Schwann cells; Nf1 mRNA was detected in all of the tumors examined.
References
- 1.Gutmann D.H., Aylsworth A., Carey J.C., Korf B., Marks J., Pyeritz R.E., Rubenstein A., Viskochil D. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA. 1997;278:51–57. [PubMed] [Google Scholar]
- 2.Carroll S.L., Stonecypher M.S. Tumor suppressor mutations and growth factor signaling in the pathogenesis of NF1-associated peripheral nerve sheath tumors. I. The role of tumor suppressor mutations. J Neuropathol Exp Neurol. 2004;63:1115–1123. doi: 10.1093/jnen/63.11.1115. [DOI] [PubMed] [Google Scholar]
- 3.Carroll S.L., Ratner N. How does the Schwann cell lineage form tumors in NF1? Glia. 2008;56:1590–1605. doi: 10.1002/glia.20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans D.G., Baser M.E., McGaughran J., Sharif S., Howard E., Moran A. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet. 2002;39:311–314. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCaughan J.A., Holloway S.M., Davidson R., Lam W.W. Further evidence of the increased risk for malignant peripheral nerve sheath tumour from a Scottish cohort of patients with neurofibromatosis type 1. J Med Genet. 2007;44:463–466. doi: 10.1136/jmg.2006.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brossier N.M., Carroll S.L. Genetically engineered mouse models shed new light on the pathogenesis of neurofibromatosis type I-related neoplasms of the peripheral nervous system. Brain Res Bull. 2012;88:58–71. doi: 10.1016/j.brainresbull.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll S.L. Molecular mechanisms promoting the pathogenesis of Schwann cell neoplasms. Acta Neuropathol. 2012;123:321–348. doi: 10.1007/s00401-011-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koga T., Iwasaki H., Ishiguro M., Matsuzaki A., Kikuchi M. Frequent genomic imbalances in chromosomes 17, 19, and 22q in peripheral nerve sheath tumours detected by comparative genomic hybridization analysis. J Pathol. 2002;197:98–107. doi: 10.1002/path.1101. [DOI] [PubMed] [Google Scholar]
- 9.Koga T., Iwasaki H., Ishiguro M., Matsuzaki A., Kikuchi M. Losses in chromosomes 17, 19, and 22q in neurofibromatosis type 1 and sporadic neurofibromas: a comparative genomic hybridization analysis. Cancer Genet Cytogenet. 2002;136:113–120. doi: 10.1016/s0165-4608(02)00527-7. [DOI] [PubMed] [Google Scholar]
- 10.Miller S.J., Rangwala F., Williams J., Ackerman P., Kong S., Jegga A.G., Kaiser S., Aronow B.J., Frahm S., Kluwe L., Mautner V., Upadhyaya M., Muir D., Wallace M., Hagen J., Quelle D.E., Watson M.A., Perry A., Gutmann D.H., Ratner N. Large-scale molecular comparison of human Schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res. 2006;66:2584–2591. doi: 10.1158/0008-5472.CAN-05-3330. [DOI] [PubMed] [Google Scholar]
- 11.Legius E., Dierick H., Wu R., Hall B.K., Marynen P., Cassiman J.J., Glover T.W. TP53 mutations are frequent in malignant NF1 tumors. Genes Chromosomes Cancer. 1994;10:250–255. doi: 10.1002/gcc.2870100405. [DOI] [PubMed] [Google Scholar]
- 12.Holtkamp N., Atallah I., Okuducu A.F., Mucha J., Hartmann C., Mautner V.F., Friedrich R.E., Mawrin C., von Deimling A. MMP-13 and p53 in the progression of malignant peripheral nerve sheath tumors. Neoplasia. 2007;9:671–677. doi: 10.1593/neo.07304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verdijk R.M., den Bakker M.A., Dubbink H.J., Hop W.C., Dinjens W.N., Kros J.M. TP53 mutation analysis of malignant peripheral nerve sheath tumors. J Neuropathol Exp Neurol. 2010;69:16–26. doi: 10.1097/NEN.0b013e3181c55d55. [DOI] [PubMed] [Google Scholar]
- 14.Van Roy N., Van Gele M., Vandesompele J., Messiaen L., Van Belle S., Sciot R., Mortéle K., Gyselinck J., Michiels E., Forsyth R., Van Marck E., De Paepe A., Speleman F. Evidence for involvement of a tumor suppressor gene on 1p in malignant peripheral nerve sheath tumors. Cancer Genet Cytogenet. 2003;143:120–124. doi: 10.1016/s0165-4608(02)00853-1. [DOI] [PubMed] [Google Scholar]
- 15.Mertens F., Rydholm A., Bauer H.F., Limon J., Nedoszytko B., Szadowska A., Willén H., Heim S., Mitelman F., Mandahl N. Cytogenetic findings in malignant peripheral nerve sheath tumors. Int J Cancer. 1995;61:793–798. doi: 10.1002/ijc.2910610609. [DOI] [PubMed] [Google Scholar]
- 16.Forus A., Weghuis D.O., Smeets D., Fodstad O., Myklebost O., van Kessel A.G. Comparative genomic hybridization analysis of human sarcomas: I. Occurrence of genomic imbalances and identification of a novel major amplicon at 1q21-q22 in soft tissue sarcomas. Genes Chromosomes Cancer. 1995;14:8–14. doi: 10.1002/gcc.2870140103. [DOI] [PubMed] [Google Scholar]
- 17.Lothe R.A., Karhu R., Mandahl N., Mertens F., Saeter G., Heim S., Borresen-Dale A.L., Kallioniemi O.P. Gain of 17q24-qter detected by comparative genomic hybridization in malignant tumors from patients with von Recklinghausen’s neurofibromatosis. Cancer Res. 1996;56:4778–4781. [PubMed] [Google Scholar]
- 18.Plaat B.E., Molenaar W.M., Mastik M.F., Hoekstra H.J., te Meerman G.J., van den Berg E. Computer-assisted cytogenetic analysis of 51 malignant peripheral-nerve-sheath tumors: sporadic vs. neurofibromatosis-type-1-associated malignant schwannomas. Int J Cancer. 1999;83:171–178. doi: 10.1002/(sici)1097-0215(19991008)83:2<171::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 19.Mechtersheimer G., Otaño-Joos M., Ohl S., Benner A., Lehnert T., Willeke F., Möller P., Otto H.F., Lichter P., Joos S. Analysis of chromosomal imbalances in sporadic and NF1-associated peripheral nerve sheath tumors by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;25:362–369. [PubMed] [Google Scholar]
- 20.Schmidt H., Würl P., Taubert H., Meye A., Bache M., Holzhausen H.J., Hinze R. Genomic imbalances of 7p and 17q in malignant peripheral nerve sheath tumors are clinically relevant. Genes Chromosomes Cancer. 1999;25:205–211. [PubMed] [Google Scholar]
- 21.Schmidt H., Taubert H., Meye A., Wurl P., Bache M., Bartel F., Holzhausen H.J., Hinze R. Gains in chromosomes 7, 8q, 15q and 17q are characteristic changes in malignant but not in benign peripheral nerve sheath tumors from patients with Recklinghausen’s disease. Cancer Lett. 2000;155:181–190. doi: 10.1016/s0304-3835(00)00426-2. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt H., Taubert H., Würl P., Bache M., Bartel F., Holzhausen H.J., Hinze R. Cytogenetic characterization of six malignant peripheral nerve sheath tumors: comparison of karyotyping and comparative genomic hybridization. Cancer Genet Cytogenet. 2001;128:14–23. doi: 10.1016/s0165-4608(01)00393-4. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa Y., Yoshida A., Numoto K., Kunisada T., Wai D., Ohata N., Takeda K., Kawai A., Ozaki T. Chromosomal imbalances in malignant peripheral nerve sheath tumor detected by metaphase and microarray comparative genomic hybridization. Oncol Rep. 2006;15:297–303. [PubMed] [Google Scholar]
- 24.Kresse S.H., Skarn M., Ohnstad H.O., Namløs H.M., Bjerkehagen B., Myklebost O., Meza-Zepeda L.A. DNA copy number changes in high-grade malignant peripheral nerve sheath tumors by array CGH. Mol Cancer. 2008;7:48. doi: 10.1186/1476-4598-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantripragada K.K., Spurlock G., Kluwe L., Chuzhanova N., Ferner R.E., Frayling I.M., Dumanski J.P., Guha A., Mautner V., Upadhyaya M. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin Cancer Res. 2008;14:1015–1024. doi: 10.1158/1078-0432.CCR-07-1305. [DOI] [PubMed] [Google Scholar]
- 26.Mantripragada K.K., Díaz de Ståhl T., Patridge C., Menzel U., Andersson R., Chuzhanova N., Kluwe L., Guha A., Mautner V., Dumanski J.P., Upadhyaya M. Genome-wide high-resolution analysis of DNA copy number alterations in NF1-associated malignant peripheral nerve sheath tumors using 32K BAC array. Genes Chromosomes Cancer. 2009;48:897–907. doi: 10.1002/gcc.20695. [DOI] [PubMed] [Google Scholar]
- 27.McLendon R., Friedman A., Bigner D., Van Meir E.G., Brat D.J., Mastrogianakis G.M., Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell D., Berchuck A., Birrer M., Chien J., Cramer D., Dao F., Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma [Erratum appeared in Nature 2012, 490:298] Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Mu D., Lucito R., Powers S., Lowe S.W. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varela I., Klijn C., Stephens P.J., Mudie L.J., Stebbings L., Galappaththige D., van der Gulden H., Schut E., Klarenbeek S., Campbell P.J., Wessels L.F., Stratton M.R., Jonkers J., Futreal P.A., Adams D.J. Somatic structural rearrangements in genetically engineered mouse mammary tumors. Genome Biol. 2010;11:R100. doi: 10.1186/gb-2010-11-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson R.A., Wright K.D., Poppleton H., Mohankumar K.M., Finkelstein D., Pounds S.B., Rand V., Leary S.E., White E., Eden C., Hogg T., Northcott P., Mack S., Neale G., Wang Y.D., Coyle B., Atkinson J., DeWire M., Kranenburg T.A., Gillespie Y., Allen J.C., Merchant T., Boop F.A., Sanford R.A., Gajjar A., Ellison D.W., Taylor M.D., Grundy R.G., Gilbertson R.J. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim M., Gans J.D., Nogueira C., Wang A., Paik J.H., Feng B., Brennan C., Hahn W.C., Cordon-Cardo C., Wagner S.N., Flotte T.J., Duncan L.M., Granter S.R., Chin L. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Uren A.G., Kool J., Matentzoglu K., de Ridder J., Mattison J., van Uitert M., Lagcher W., Sie D., Tanger E., Cox T., Reinders M., Hubbard T.J., Rogers J., Jonkers J., Wessels L., Adams D.J., van Lohuizen M., Berns A. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starr T.K., Allaei R., Silverstein K.A., Staggs R.A., Sarver A.L., Bergemann T.L., Gupta M., O’Sullivan M.G., Matise I., Dupuy A.J., Collier L.S., Powers S., Oberg A.L., Asmann Y.W., Thibodeau S.N., Tessarollo L., Copeland N.G., Jenkins N.A., Cormier R.T., Largaespada D.A. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupuy A.J., Rogers L.M., Kim J., Nannapaneni K., Starr T.K., Liu P., Largaespada D.A., Scheetz T.E., Jenkins N.A., Copeland N.G. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y., Ghosh P., Charnay P., Burns D.K., Parada L.F. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel K.S., Klesse L.J., Velasco-Miguel S., Meyers K., Rushing E.J., Parada L.F. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cichowski K., Shih T.S., Schmitt E., Santiago S., Reilly K., McLaughlin M.E., Bronson R.T., Jacks T. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 39.Eckert J.M., Byer S.J., Clodfelder-Miller B.J., Carroll S.L. Neuregulin-1 beta and neuregulin-1 alpha differentially affect the migration and invasion of malignant peripheral nerve sheath tumor cells. Glia. 2009;57:1501–1520. doi: 10.1002/glia.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stonecypher M.S., Byer S.J., Grizzle W.E., Carroll S.L. Activation of the neuregulin-1/ErbB signaling pathway promotes the proliferation of neoplastic Schwann cells in human malignant peripheral nerve sheath tumors. Oncogene. 2005;24:5589–5605. doi: 10.1038/sj.onc.1208730. [DOI] [PubMed] [Google Scholar]
- 41.Huijbregts R.P., Roth K.A., Schmidt R.E., Carroll S.L. Hypertrophic neuropathies and malignant peripheral nerve sheath tumors in transgenic mice overexpressing glial growth factor beta3 in myelinating Schwann cells. J Neurosci. 2003;23:7269–7280. doi: 10.1523/JNEUROSCI.23-19-07269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheithauer B.W., Louis D.N., Hunter S., Woodruff J.M., Antonescu C.R. Malignant peripheral nerve sheath tumour (MPNST). WHO Classification of Tumours of the Central Nervous System, ed 4. In: Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K., editors. World Health Organization Classification of Tumours. WHO Press; Geneva: 2007. pp. 160–162. [Google Scholar]
- 43.Luna L. ed 3. McGraw-Hill; New York: 1968. Unna’s method for mast cells. Manual of Histologic Staining Methods of the AFIP. pp 115–116. [Google Scholar]
- 44.Stonecypher M.S., Chaudhury A.R., Byer S.J., Carroll S.L. Neuregulin growth factors and their ErbB receptors form a potential signaling network for schwannoma tumorigenesis. J Neuropathol Exp Neurol. 2006;65:162–175. doi: 10.1097/01.jnen.0000199575.93794.2f. [DOI] [PubMed] [Google Scholar]
- 45.Basu T.N., Gutmann D.H., Fletcher J.A., Glover T.W., Collins F.S., Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 46.DeClue J.E., Papageorge A.G., Fletcher J.A., Diehl S.R., Ratner N., Vass W.C., Lowy D.R. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell. 1992;69:265–273. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- 47.Guha A., Lau N., Huvar I., Gutmann D., Provias J., Pawson T., Boss G. Ras-GTP levels are elevated in human NF1 peripheral nerve tumors. Oncogene. 1996;12:507–513. [PubMed] [Google Scholar]
- 48.Birindelli S., Perrone F., Oggionni M., Lavarino C., Pasini B., Vergani B., Ranzani G.N., Pierotti M.A., Pilotti S. Rb and TP53 pathway alterations in sporadic and NF1-related malignant peripheral nerve sheath tumors. Lab Invest. 2001;81:833–844. doi: 10.1038/labinvest.3780293. [DOI] [PubMed] [Google Scholar]
- 49.Menon A.G., Anderson K.M., Riccardi V.M., Chung R.Y., Whaley J.M., Yandell D.W., Farmer G.E., Freiman R.N., Lee J.K., Li F.P., Barker D.F., Ledbetter D.H., Kleider A., Martuza R.L., Gusella J.F., Seizinger B.R. Chromosome 17p deletions and p53 gene mutations associated with the formation of malignant neurofibrosarcomas in von Recklinghausen neurofibromatosis. Proc Natl Acad Sci USA. 1990;87:5435–5439. doi: 10.1073/pnas.87.14.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berner J.M., Sørlie T., Mertens F., Henriksen J., Saeter G., Mandahl N., Brogger A., Myklebost O., Lothe R.A. Chromosome band 9p21 is frequently altered in malignant peripheral nerve sheath tumors: studies of CDKN2A and other genes of the pRB pathway. Genes Chromosomes Cancer. 1999;26:151–160. [PubMed] [Google Scholar]
- 51.Kourea H.P., Orlow I., Scheithauer B.W., Cordon-Cardo C., Woodruff J.M. Deletions of the INK4A gene occur in malignant peripheral nerve sheath tumors but not in neurofibromas. Am J Pathol. 1999;155:1855–1860. doi: 10.1016/S0002-9440(10)65504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen G.P., Stemmer-Rachamimov A.O., Ino Y., Moller M.B., Rosenberg A.E., Louis D.N. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. Am J Pathol. 1999;155:1879–1884. doi: 10.1016/S0002-9440(10)65507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perry A., Kunz S.N., Fuller C.E., Banerjee R., Marley E.F., Liapis H., Watson M.A., Gutmann D.H. Differential NF1, p16, and EGFR patterns by interphase cytogenetics (FISH) in malignant peripheral nerve sheath tumor (MPNST) and morphologically similar spindle cell neoplasms. J Neuropathol Exp Neurol. 2002;61:702–709. doi: 10.1093/jnen/61.8.702. [DOI] [PubMed] [Google Scholar]
- 54.Perrone F., Tabano S., Colombo F., Dagrada G., Birindelli S., Gronchi A., Colecchia M., Pierotti M.A., Pilotti S. p15INK4b, p14ARF, and p16INK4a inactivation in sporadic and neurofibromatosis type 1-related malignant peripheral nerve sheath tumors. Clin Cancer Res. 2003;9:4132–4138. [PubMed] [Google Scholar]
- 55.Agesen T.H., Flørenes V.A., Molenaar W.M., Lind G.E., Berner J.M., Plaat B.E., Komdeur R., Myklebost O., van den Berg E., Lothe R.A. Expression patterns of cell cycle components in sporadic and neurofibromatosis type 1-related malignant peripheral nerve sheath tumors. J Neuropathol Exp Neurol. 2005;64:74–81. doi: 10.1093/jnen/64.1.74. [DOI] [PubMed] [Google Scholar]
- 56.Sabah M., Cummins R., Leader M., Kay E. Loss of p16 (INK4A) expression is associated with allelic imbalance/loss of heterozygosity of chromosome 9p21 in microdissected malignant peripheral nerve sheath tumors. Appl Immunohistochem Mol Morphol. 2006;14:97–102. doi: 10.1097/01.pai.0000143787.80564.f5. [DOI] [PubMed] [Google Scholar]
- 57.Endo M., Kobayashi C., Setsu N., Takahashi Y., Kohashi K., Yamamoto H., Tamiya S., Matsuda S., Iwamoto Y., Tsuneyoshi M., Oda Y. Prognostic significance of p14ARF, p15INK4b and p16INK4a inactivation in malignant peripheral nerve sheath tumors. Clin Cancer Res. 2011;17:3771–3782. doi: 10.1158/1078-0432.CCR-10-2393. [DOI] [PubMed] [Google Scholar]
- 58.Kourea H.P., Cordon-Cardo C., Dudas M., Leung D., Woodruff J.M. Expression of p27(kip) and other cell cycle regulators in malignant peripheral nerve sheath tumors and neurofibromas: the emerging role of p27(kip) in malignant transformation of neurofibromas. Am J Pathol. 1999;155:1885–1891. doi: 10.1016/S0002-9440(10)65508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou H., Coffin C.M., Perkins S.L., Tripp S.R., Liew M., Viskochil D.H. Malignant peripheral nerve sheath tumor: a comparison of grade, immunophenotype, and cell cycle/growth activation marker expression in sporadic and neurofibromatosis 1-related lesions. Am J Surg Pathol. 2003;27:1337–1345. doi: 10.1097/00000478-200310000-00006. [DOI] [PubMed] [Google Scholar]
- 60.Huret J.L., Minor S.L., Dorkeld F., Dessen P., Bernheim A. Atlas of genetics and cytogenetics in oncology and haematology, an interactive database. Nucleic Acids Res. 2000;28:349–351. doi: 10.1093/nar/28.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sjöblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S.D., Willis J., Dawson D., Willson J.K., Gazdar A.F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B.H., Bachman K.E., Papadopoulos N., Vogelstein B., Kinzler K.W., Velculescu V.E. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 62.Akagi K., Suzuki T., Stephens R.M., Jenkins N.A., Copeland N.G. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32(Suppl 1):D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prasad R., Leshkowitz D., Gu Y., Alder H., Nakamura T., Saito H., Huebner K., Berger R., Croce C.M., Canaani E. Leucine-zipper dimerization motif encoded by the AF17 gene fused to ALL-1 (MLL) in acute leukemia. Proc Natl Acad Sci USA. 1994;91:8107–8111. doi: 10.1073/pnas.91.17.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chin L., Hahn W.C., Getz G., Meyerson M. Making sense of cancer genomic data [Erratum appeared in Genes Dev 2012;26:1003] Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boyden L.M., Lewis J.M., Barbee S.D., Bas A., Girardi M., Hayday A.C., Tigelaar R.E., Lifton R.P. Skint1, the prototype of a newly identified immunoglobulin superfamily gene cluster, positively selects epidermal gammadelta T cells. Nat Genet. 2008;40:656–662. doi: 10.1038/ng.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams J.P., Wu J., Johansson G., Rizvi T.A., Miller S.C., Geiger H., Malik P., Li W., Mukouyama Y.S., Cancelas J.A., Ratner N. Nf1 mutation expands an EGFR-dependent peripheral nerve progenitor that confers neurofibroma tumorigenic potential. Cell Stem Cell. 2008;3:658–669. doi: 10.1016/j.stem.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guillery R.W., Herrup K. Quantification without pontification: choosing a method for counting objects in sectioned tissues. J Comp Neurol. 1997;386:2–7. doi: 10.1002/(sici)1096-9861(19970915)386:1<2::aid-cne2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 69.Maser R.S., Choudhury B., Campbell P.J., Feng B., Wong K.K., Protopopov A., O’Neil J., Gutierrez A., Ivanova E., Perna I., Lin E., Mani V., Jiang S., McNamara K., Zaghlul S., Edkins S., Stevens C., Brennan C., Martin E.S., Wiedemeyer R., Kabbarah O., Nogueira C., Histen G., Aster J., Mansour M., Duke V., Foroni L., Fielding A.K., Goldstone A.H., Rowe J.M., Wang Y.A., Look A.T., Stratton M.R., Chin L., Futreal P.A., DePinho R.A. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abercrombie M. Estimation of nuclear population from microtome sections. Anat Rec. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- 71.Wallace M.R., Rasmussen S.A., Lim I.T., Gray B.A., Zori R.T., Muir D. Culture of cytogenetically abnormal Schwann cells from benign and malignant NF1 tumors. Genes Chromosomes Cancer. 2000;27:117–123. [PubMed] [Google Scholar]
- 72.Upadhyaya M., Spurlock G., Thomas L., Thomas N.S., Richards M., Mautner V.F., Cooper D.N., Guha A., Yan J. Microarray-based copy number analysis of neurofibromatosis type-1 (NF1)-associated malignant peripheral nerve sheath tumors reveals a role for Rho-GTPase pathway genes in NF1 tumorigenesis. Hum Mutat. 2012;33:763–776. doi: 10.1002/humu.22044. [DOI] [PubMed] [Google Scholar]
- 73.Storlazzi C.T., Brekke H.R., Mandahl N., Brosjö O., Smeland S., Lothe R.A., Mertens F. Identification of a novel amplicon at distal 17q containing the BIRC5/SURVIVIN gene in malignant peripheral nerve sheath tumours. J Pathol. 2006;209:492–500. doi: 10.1002/path.1998. [DOI] [PubMed] [Google Scholar]
- 74.Kawaguchi K., Oda Y., Saito T., Takahira T., Yamamoto H., Tamiya S., Iwamoto Y., Tsuneyoshi M. Genetic and epigenetic alterations of the PTEN gene in soft tissue sarcomas. Hum Pathol. 2005;36:357–363. doi: 10.1016/j.humpath.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 75.Gregorian C., Nakashima J., Dry S.M., Nghiemphu P.L., Smith K.B., Ao Y., Dang J., Lawson G., Mellinghoff I.K., Mischel P.S., Phelps M., Parada L.F., Liu X., Sofroniew M.V., Eilber F.C., Wu H. PTEN dosage is essential for neurofibroma development and malignant transformation. Proc Natl Acad Sci USA. 2009;106:19479–19484. doi: 10.1073/pnas.0910398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Representative RT-PCR control experiments supporting the specificity of the results illustrated in Figure 3. PCR was performed on reverse transcription reactions performed in the presence (+) and absence (−) of reverse transcriptase for each of the markers indicated to the right of each panel. The positive control tissues and cultures [cultured WT mouse Schwann cells (cultured s.c.), WT mouse sciatic nerve (nerve), WT mouse brain (brain), WT aorta (aorta), and WT gastrocnemius muscle (muscle)] were selected because they are known to express the markers examined in these experiments. Also shown are PCR reactions performed in the presence (+) and absence (−) of reverse transcriptase using mRNA isolated from representative P0-GGFβ3 MPNST early-passage cultures that were positive for Schwann cell (S100β, P0, MBP, PMP22, p75LNTR, Sox10, Pax3, Krox20, GFAP, GAP43), neural (neurofilament, peripherin), and muscle (calponin, SM22α, α-SMA, desmin, MyoD1) markers. PCR reactions were also performed on water blanks (blk) to confirm an absence of contamination in these experiments. B: qPCR analyses of Nf1 expression in non-neoplastic transgenic Schwann cells and early-passage cultures of P0-GGFβ3 MPNST cells. Transcript levels were normalized to levels detected in non-neoplastic transgenic Schwann cells; Nf1 mRNA was detected in all of the tumors examined.