Abstract
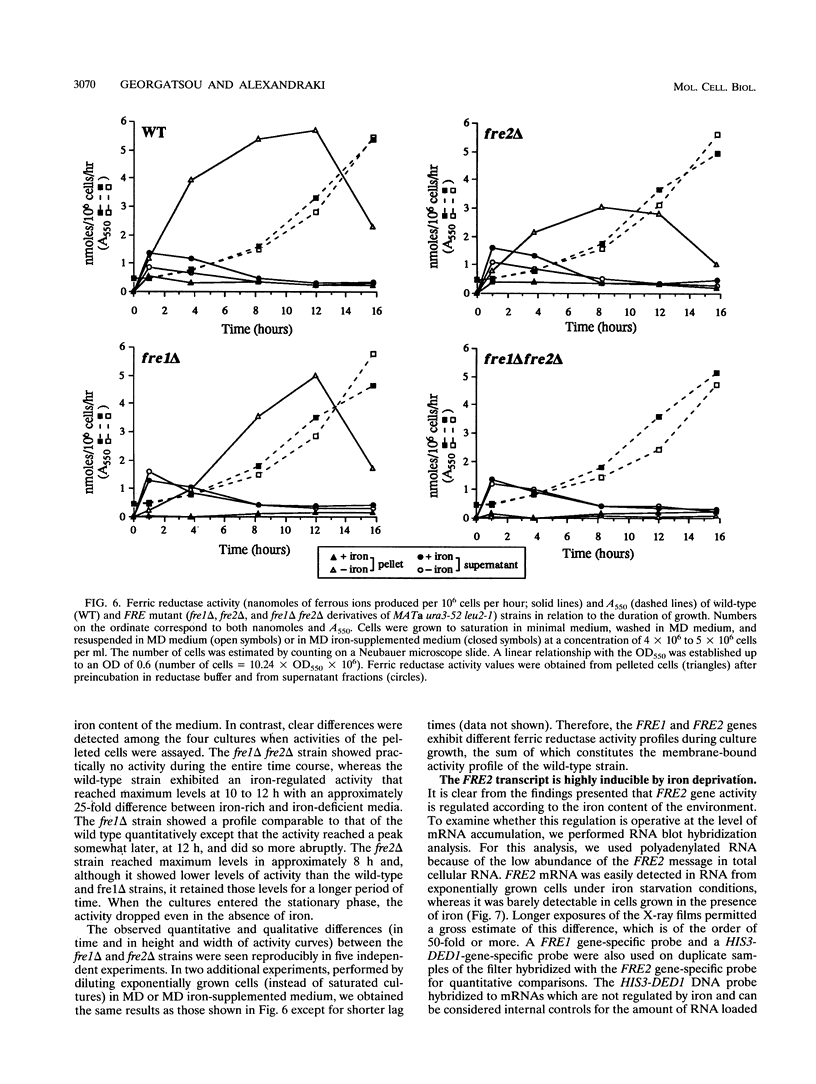
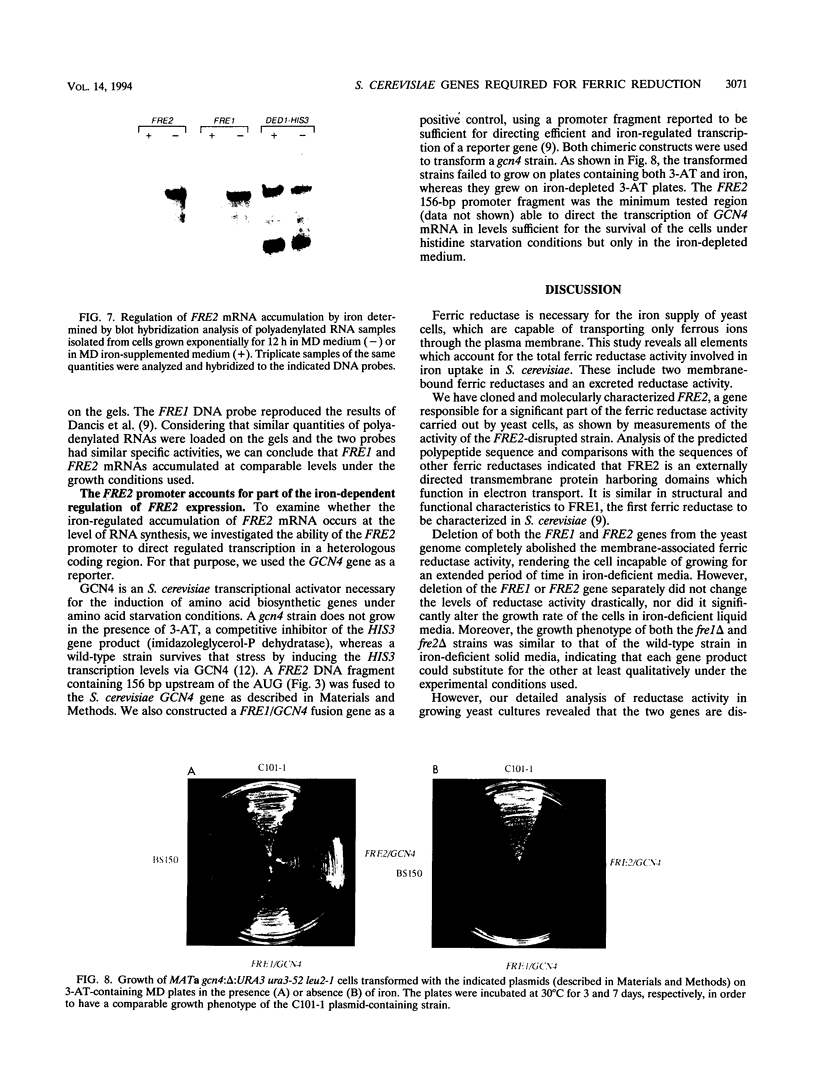
Iron uptake in Saccharomyces cerevisiae involves at least two steps: reduction of ferric to ferrous ions extracellularly and transport of the reduced ions through the plasma membrane. We have cloned and molecularly characterized FRE2, a gene which is shown to account, together with FRE1, for the total membrane-associated ferric reductase activity of the cell. Although not similar at the nucleotide level, the two genes encode proteins with significantly similar primary structures and very similar hydrophobicity profiles. The FRE1 and FRE2 proteins are functionally related, having comparable properties as ferric reductases. FRE2 expression, like FRE1 expression, is induced by iron deprivation, and at least part of this control takes place at the transcriptional level, since 156 nucleotides upstream of the initiator AUG conferred iron-dependent regulation when fused to a heterologous gene. However, the two gene products have distinct temporal regulation of their activities during cell growth.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boorstein W. R., Craig E. A. Transcriptional regulation of SSA3, an HSP70 gene from Saccharomyces cerevisiae. Mol Cell Biol. 1990 Jun;10(6):3262–3267. doi: 10.1128/mcb.10.6.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane F. L., Roberts H., Linnane A. W., Löw H. Transmembrane ferricyanide reduction by cells of the yeast Saccharomyces cerevisiae. J Bioenerg Biomembr. 1982 Jun;14(3):191–205. doi: 10.1007/BF00745020. [DOI] [PubMed] [Google Scholar]
- Crichton R. R., Charloteaux-Wauters M. Iron transport and storage. Eur J Biochem. 1987 May 4;164(3):485–506. doi: 10.1111/j.1432-1033.1987.tb11155.x. [DOI] [PubMed] [Google Scholar]
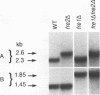
- Dancis A., Klausner R. D., Hinnebusch A. G., Barriocanal J. G. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1990 May;10(5):2294–2301. doi: 10.1128/mcb.10.5.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A., Roman D. G., Anderson G. J., Hinnebusch A. G., Klausner R. D. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc Natl Acad Sci U S A. 1992 May 1;89(9):3869–3873. doi: 10.1073/pnas.89.9.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D., Davis-Kaplan S., Jordan I., Sipe D., Kaplan J. Regulation of iron uptake in Saccharomyces cerevisiae. The ferrireductase and Fe(II) transporter are regulated independently. J Biol Chem. 1992 Oct 15;267(29):20774–20781. [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Kaplan J., Jordan I., Sturrock A. Regulation of the transferrin-independent iron transport system in cultured cells. J Biol Chem. 1991 Feb 15;266(5):2997–3004. [PubMed] [Google Scholar]
- Karplus P. A., Daniels M. J., Herriott J. R. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991 Jan 4;251(4989):60–66. [PubMed] [Google Scholar]
- Klausner R. D., Rouault T. A., Harford J. B. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993 Jan 15;72(1):19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Köhrer K., Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Lesuisse E., Labbe P. Reductive and non-reductive mechanisms of iron assimilation by the yeast Saccharomyces cerevisiae. J Gen Microbiol. 1989 Feb;135(2):257–263. doi: 10.1099/00221287-135-2-257. [DOI] [PubMed] [Google Scholar]
- Lesuisse E., Raguzzi F., Crichton R. R. Iron uptake by the yeast Saccharomyces cerevisiae: involvement of a reduction step. J Gen Microbiol. 1987 Nov;133(11):3229–3236. doi: 10.1099/00221287-133-11-3229. [DOI] [PubMed] [Google Scholar]
- Marck C. 'DNA Strider': a 'C' program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988 Mar 11;16(5):1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Toh-e A. Isolation and characterization of two novel ras superfamily genes in Saccharomyces cerevisiae. Gene. 1992 May 1;114(1):43–49. doi: 10.1016/0378-1119(92)90705-t. [DOI] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Orkin S. H. Molecular genetics of chronic granulomatous disease. Annu Rev Immunol. 1989;7:277–307. doi: 10.1146/annurev.iy.07.040189.001425. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Thireos G., Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984 Mar;4(3):520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Stotz A., Scherf C. DNA of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- Roman D. G., Dancis A., Anderson G. J., Klausner R. D. The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol Cell Biol. 1993 Jul;13(7):4342–4350. doi: 10.1128/mcb.13.7.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Schwob E., Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993 Jul;7(7A):1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Segal A. W., Abo A. The biochemical basis of the NADPH oxidase of phagocytes. Trends Biochem Sci. 1993 Feb;18(2):43–47. doi: 10.1016/0968-0004(93)90051-n. [DOI] [PubMed] [Google Scholar]
- Segal A. W., West I., Wientjes F., Nugent J. H., Chavan A. J., Haley B., Garcia R. C., Rosen H., Scrace G. Cytochrome b-245 is a flavocytochrome containing FAD and the NADPH-binding site of the microbicidal oxidase of phagocytes. Biochem J. 1992 Jun 15;284(Pt 3):781–788. doi: 10.1042/bj2840781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstensen K., Romslo I. Uptake of iron from transferrin by isolated rat hepatocytes. A redox-mediated plasma membrane process? J Biol Chem. 1988 Jun 25;263(18):8844–8850. [PubMed] [Google Scholar]
- Yamashoji S., Kajimoto G. Decrease of NADH in yeast cells by external ferricyanide reduction. Biochim Biophys Acta. 1986 Nov 5;852(1):25–29. doi: 10.1016/0005-2728(86)90052-6. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]