X-linked intellectual disability (XLID), defined as clinical ID combined with a pedigree consistent with X-linked inheritance, is a genetically heterogeneous condition that affects more than 10% of males with ID. Currently there are at least 92 genes known to cause XLID,1–3 yet a large proportion of XLID cases remain unexplained, as each of the XLID genes identified so far only accounts for a small fraction (< 1%) of affected individuals. Given that about one third of mutations affect gene expression levels,4 we reasoned that transcriptome profiling of lymphoblast cell lines from XLID patients may highlight genes harboring disease-causing mutations and may be an efficient follow-up method for rare sequence variants of unknown functional significance.
We analyzed expression profiles of lymphoblast cell lines from 64 XLID patients, including 13 cases that were part of a recent X-chromosome exon re-sequencing study5 (Supplementary Methods, Supplementary Table 1). We found polyglutamine-binding protein 1 (PQBP1), a gene previously implicated in XLID,6,7 to be significantly downregulated in two cases (Supplementary Table 2), and confirmed an exon 4 (AG)2 deletion as the cause of mRNA downregulation in both instances. PQBP1 mutations cause a sydromic form of XLID commonly referred to as Renpenning syndrome.8 The specific mutation we describe here has been previously shown to cause XLID,6 and has also been proven to decrease mRNA levels by nonsense-mediated mRNA decay,7 thus being likely to be detected by assessment of gene expression. The cases for which we identified PQBP1 mutations were not part of the cohort studied by Tarpey et al. (Supplementary Table 1).
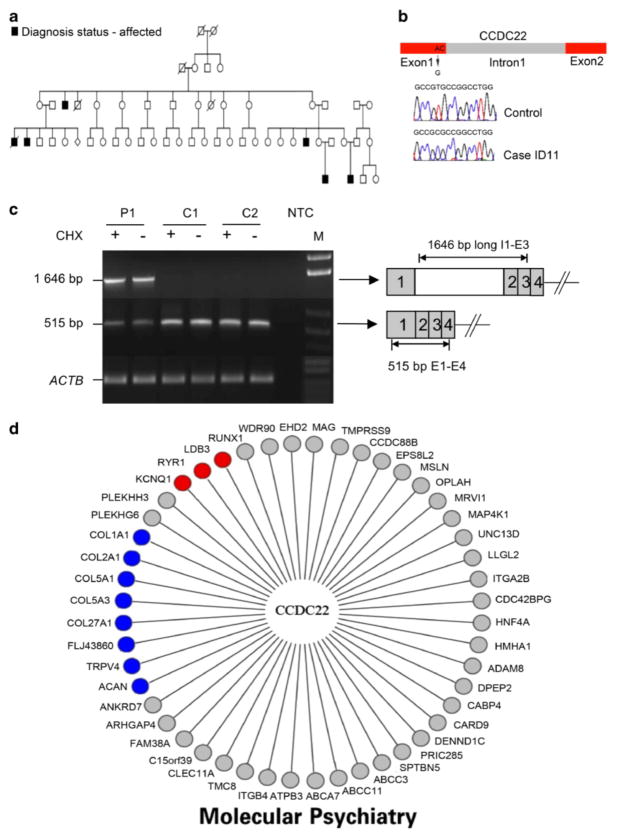
We further asked whether any of the non-recurrent sequence variants identified by the exon re-sequencing study in the 13 overlapping cases was associated with a significant alteration of mRNA levels. We found that CCDC22, which encodes a coiled-coil domain protein of unknown function, was significantly downregulated (Figure 1c, Supplementary Figure 1) in one of our XLID patients. A CCDC22 non-recurrent sequence variant c.49A > G/p.T17A had been identified in a proband from the same family by the Sanger re-sequencing study, but its functional significance had not been determined.5 The proband belongs to a large family (55 individuals) with six affected males over three generations (Figure 1a). Notably, female carriers in this family show highly skewed X-chromosome inactivation (data not shown). Linkage analysis delineated a 58.4-Mb linkage interval (logarithm in base 10 of odds (LOD) score = 2.7). Whole X-chromosome exon re-sequencing identified four non-recurrent mis-sense variants within the linkage interval: CCDC22 c.49A> G/p.T17A, FAM121A (APOOL) c.40G> T/p.A14S, LOC402414 c.453T> A/p.S151R, ABCB7 c.941G> A/p.R314Q.5 Although these sequence variants were predicted to be neutral for protein function (Polyphen, data not shown), we found that the CCDC22 change was associated with a fivefold decrease in mRNA level (Figure 1c, Supplementary Figure 1 and Supplementary Table 3). No other gene in the 58.4-Mb linkage interval showed significant downregulation of mRNA levels (Supplementary Figure 2a). We sequenced CCDC22 in all available family members and found that the c.49A>G change segregates with the disease consistent with its location within the linkage interval.
Figure 1.
CCDC22 mutation causes X-linked intellectual disability (XLID) in a large pedigree. (a) The pedigree of the IGOLD #586 family shows six affected males over three generations. The case analyzed by expression profiling in this study (ID11) is the case from the first affected generation. (b) CCDC22 mutation in ID11. Top: schematic representation of the location of the mutation. The c.49A > G/p.T17A mutation is located close to the first 5′ splice site of CCDC22. Bottom: chromatograms showing the CCDC22 mutation on the minus strand. (c) Expression of the normal CCDC22 mRNA (E1–E4, 515 bp product) was decreased in the patient (P1), whereas expression of the abnormal CCDC22 mRNA-retaining intron 1 (long I1-E3 1646 bp product) was only visible in P1. Actin B (ACTB) quantitative reverse transcriptase was used as reference control. Cells were treated with (+) or without (−) cyclohexamide (CHX). C, control; P, patient; NTC, no template control. The right panel shows the abnormal transcript with intron 1 retention and the normally spliced transcript of CCDC22. Gray box, exons; white box, intron 1. (d) Genes co-expressed with CCDC22 in the fetal brain, as identified by multi-node topological overlap measure. Two groups of functionally related genes are highlighted: in blue, genes involved in multiple skeletal abnormalities and in red, genes associated with cardiac abnormalities, the two main pathological changes characteristic of the syndromic XLID in the IGOLD #586 family.
The c.49A >G sequence variant is located within exon 1, two base pairs away from the 5′ splice site of intron 1 of CCDC22 (Figure 1b) and is predicted to significantly decrease the splicing efficiency at the corresponding 5′ splice site (splice site score decrease from 0.9 to 0.7, using NNsplice9; http://www.fruitfly.org/seq_tools/splice.html). We found abnormally spliced transcripts retaining intron 1 to be much more abundant in cases harboring the c.49A >G mutation than in controls (Figure 1c, Supplementary Methods, Supplementary Figure 1), demonstrating that the mutation impedes efficient alternative splicing at the 5′ splice site. The abnormally spliced transcripts with intron 1 retention contain several in-frame premature terminal codons, which would likely cause the transcript to be degraded by nonsense-mediated mRNA decay surveillance (NMD). Alternatively, the splicing defect could have a negative impact on the transcription efficiency of CCDC22.10–12 Inhibition of NMD by cyclohexamide did not significantly affect the total CCDC22 mRNA level or the abundance of the abnormally spliced isoform (Supplementary Figure 1), indicating that NMD did not have a major role in CCDC22 mRNA downregulation. Recent studies have suggested that abnormal transcripts with retained intron(s), having failed the nuclear quality control mechanism, are not exported into the cytoplasm and thus are not degraded by NMD.13–17 In addition, splicing events proximal to the transcription start sites have been shown to be important for efficient recruitment of basal transcription factors.10–12 We thus suspect that the c.49A >G change near the first splice site removes the positive feedback necessary for efficient transcription of CCDC22. To test whether CCDC22 downregulation occurred frequently in the general population we analyzed CCDC22 expression level in 52 age-matched control males using genome-wide expression data from lymphoblast cell lines from an AGRE cohort (Methods). None of the controls showed significant downregulation of CCDC22 mRNA level (Supplementary Figure 2b).
The phenotype of the IGOLD #586 family is consistent with syndromic XLID. In addition to intellectual disability, affected individuals have cardiac abnormalities (atrial septal defect, ventricular septal defect, dextrocardia), skeletal abnormalities (hypoplastic distal phalanges, syndactyly, hip subluxation, scoliosis) and specific facial features (Table 1).
Table 1.
Phenotype characterization of the IGOLD 586 family
| CNS | Intellectual disability (3/5) |
| Defects of posterior fossa (1/5): Dandy–Walker malformation, cerebellar hypoplasia, arachnoid cyst. | |
| Cardiac | ASD (1/5), VSD (2/5), PDA (1/5), (2/5), hypoplastic right pulmonary artery (1/5). |
| Skeletal | Hypoplastic distal phalanges (3/5), syndactyly (1/5), hip subluxation (1/5), scoliosis (1/5), absent distal creases of fingers with overriding fingers and toes (1/5). |
| Facial features | Hypertelorism (4/5), beaked nose and broad nasal tip (3/5), ear abnormalities (3/5), high arched palate (1/5). |
| Others | Undescended testis (3/5), hypoplastic lung (2/5), intestinal malrotation (1/5), absent kidney (1/5). |
Abbreviations: ASD, atrial septal defect; CNS, central nervous system; PDA, patent ductus arteriosus; VSD, ventricular septal defect.
The pedigree of the IGOLD#586 family is shown in Figure 1 and comprises six affected males, one of which was an unviable male fetus and is not included in the phenotype description.
CCDC22 is a ubiquitously expressed coiled-coil domain protein (Supplementary Figure 3). Although the function of CCDC22 is currently poorly defined, CCDC22 has been shown to interact in vitro with copines, a family of calcium-dependent membrane-binding proteins, via its coiled-coil domain18 as well as the Nance–Horan syndrome protein.19 In the rat brain, CCDC22 is expressed in multiple regions including the prefrontal and somatosensory cortex, dentate gyrus and thalamus,20 and CCDC22-specific antibodies stain primarily axons.20,21 In the rat spinal cord, CCDC22 is primarily expressed in the dorsal columns, as well as in ipsilateral motor neurons after sciatic nerve trans-section,21 suggesting a role for this gene in neuronal injury response. To gain further insight into the function of CCDC22, we took a bioinformatics approach. To identify genes that are functionally related to, or potentially interact with CCDC22 in the developing human brain, we used a recently published human fetal brain transcriptome dataset22 and queried which are the nearest neighbors of CCDC22 by co-expression topological overlap.23 Figure 1d shows the genes co-expressed with CCDC22 in the human fetal brain. Remarkably, this module contains genes that have been implicated in hereditary cardiac and skeletal disorders, the main classes of extra-central nervous system pathological changes observed in family IGOLD #586.
The IGOLD #586 case was the only one in the X-chromosome re-sequencing cohort of 208 with a CCDC22 mutation, suggesting that mutations of CCDC22 are a rare cause of XLID. Here we highlight CCDC22 as a novel XLID candidate gene for future targeted re-sequencing studies and propose that the mRNA downregulation associated with the described mutation likely results from reduced transcriptional efficiency rather than nonsense-mediated mRNA decay.
Supplementary Material
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Gecz J, Shoubridge C, Corbett M. Trends Genet. 2009;25:308–316. doi: 10.1016/j.tig.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Giannandrea M, Bianchi V, Mignogna ML, Sirri A, Carrabino S, D’Elia E, et al. Am J Hum Genet. 2010;86:185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoubridge C, Tarpey PS, Abidi F, Ramsden SL, Rujirabanjerd S, Murphy JA, et al. Nat Genet. 2010;42:486–488. doi: 10.1038/ng.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell JT, Dietz HC. Cell. 2001;107:411–414. doi: 10.1016/s0092-8674(01)00583-9. [DOI] [PubMed] [Google Scholar]
- 5.Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, et al. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalscheuer VM, Freude K, Musante L, Jensen LR, Yntema HG, Gecz J, et al. Nat Genet. 2003;35:313–315. doi: 10.1038/ng1264. [DOI] [PubMed] [Google Scholar]
- 7.Musante L, Kunde SA, Sulistio TO, Fischer U, Grimme A, Frints SG, et al. Hum Mutat. 2010;31:90–98. doi: 10.1002/humu.21146. [DOI] [PubMed] [Google Scholar]
- 8.Germanaud D, Rossi M, Bussy G, Gerard D, Hertz-Pannier L, Blanchet P, et al. Clin Genet. 2010;79:225–235. doi: 10.1111/j.1399-0004.2010.01551.x. [DOI] [PubMed] [Google Scholar]
- 9.Reese MG, Eeckman FH, Kulp D, Haussler D. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 10.Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. Mol Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 11.Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore MJ, Proudfoot NJ. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Fasken MB, Corbett AH. Nat Struct Mol Biol. 2005;12:482–488. doi: 10.1038/nsmb945. [DOI] [PubMed] [Google Scholar]
- 14.Fasken MB, Corbett AH. RNA Biol. 2009;6:237–241. doi: 10.4161/rna.6.3.8330. [DOI] [PubMed] [Google Scholar]
- 15.Isken O, Maquat LE. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 16.Nakai K, Sakamoto H. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 17.Schmid M, Jensen TH. Chromosoma. 2008;117:419–429. doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- 18.Tomsig JL, Snyder SL, Creutz CE. J Biol Chem. 2003;278:10048–10054. doi: 10.1074/jbc.M212632200. [DOI] [PubMed] [Google Scholar]
- 19.Coccia M. Doctoral Thesis. University College; London: 2010. [Google Scholar]
- 20.Mulder J, Bjorling E, Jonasson K, Wernerus H, Hober S, Hokfelt T, et al. Mol Cell Proteomics. 2009;8:1612–1622. doi: 10.1074/mcp.M800539-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder J, Wernerus H, Shi TJ, Ponten F, Hober S, Uhlen M, et al. Neuroscience. 2007;146:1689–1703. doi: 10.1016/j.neuroscience.2007.02.054. [DOI] [PubMed] [Google Scholar]
- 22.Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, et al. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li A, Horvath S. Bioinformatics. 2007;23:222–231. doi: 10.1093/bioinformatics/btl581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.