Abstract
Autism Spectrum Disorder (ASD) has a heterogeneous etiology that is genetically complex. It is defined by deficits in communication and social skills and the presence of restricted and repetitive behaviors. Genetic analyses of heritable quantitative traits that correlate with ASD may reduce heterogeneity. With this in mind, deficits in nonverbal communication (NVC) were quantified based on items from the Autism Diagnostic Interview Revised. Our previous analysis of 228 families from the Autism Genetics Research Exchange (AGRE) repository reported 5 potential quantitative trait loci (QTL). Here we report an NVC QTL replication study in an independent sample of 213 AGRE families. One QTL was replicated (P < 0.0004). It was investigated using a targeted-association analysis of 476 haplotype blocks with 708 AGRE families using the Family Based Association Test (FBAT). Blocks in two QTL genes were associated with NVC with a P-value of 0.001. Three associated haplotype blocks were intronic to the Nerve Growth Factor (NGF) gene (P= 0.001, 0.001, 0.002), and one was intronic to KCND3 (P= 0.001). Individual haplotypes within the associated blocks drove the associations (0.003, 0.0004 and 0.0002) for NGF and 0.0001 for KCND3. Using the same methods, these genes were tested for association with NVC in an independent sample of 1517 families from an Autism Genome Project (AGP). NVC was associated with a haplotype in an adjacent NGF block (P= 0.0005) and one 46 kb away from the associated block in KCND3 (0.008). These analyses illustrate the value of QTL and targeted association studies for genetically complex disorders such as ASD. NGF is a promising risk gene for NVC deficits.
Keywords: autism spectrum disorders, GWAS, nerve growth factor, nonverbal communication, QTL
Introduction
Autism spectrum disorders (ASDs) are a heterogeneous group of neurodevelopemental disorders defined by impairments in language and nonverbal communication, deficits in reciprocal social interactions and an excess of restricted and repetitive behaviors. The onset of developmental disabilities occurs before the age of 3 years and persists throughout life.1,2 ASD prevalence estimates have been increasing, and it has recently been reported that approximately 9 children per 1000 are affected, as reported on the website of the Centers for Disease Control. ASD symptoms vary among individuals, and it is likely that this wide range in phenotypes along with inconsistent genetic results derive, at least partially, from a substantial amount of genetic heterogeneity.
Prior studies established that ASDs are heritable genetically complex disorders with associated quantitative traits exhibiting familiality. A recent twin study estimates the probandwise concordance rate for strict autism in male monozygotic twin pairs as 0.58, with a 95% confidence interval estimate of 0.42–0.74 and in male dizygotic twin pairs as 0.21, with a 95% confidence interval estimate of 0.09–0.43,3 implicating a substantial heritability and an important role for environmental factors in Autism. Although, a monozygotic prenatal environment can make those pairs more concordant than dizygotic pairs, thereby inflating the heritability estimate, such an effect is unlikely to be large enough to preclude an important role for genes.
Research indicates that siblings and parents of affected children are more likely to show deficits in quantitative measures of social skills and their abilities to communicate,4,5 providing evidence that some quantitative ASD traits are familial and likely to be heritable. Analyzing a single quantitative heritable ASD phenotype can provide a powerful and complementary avenue to gene identification when combined with the analysis of the binary ASD diagnosis.6–8 Quantitative traits measured in affected and unaffected individuals in families can lead to genes that contribute to the ASD directly. Alternatively, quantitative traits that only exhibit substantial variation among those with ASD can also provide important genetic information. Genes influencing such traits will contribute to particular deficits in those with ASD, and will not cause the disorder directly. However, they may have a marked impact on to its severity and that severity may allow the patient to satisfy ASD diagnostic criteria. Appropriate traits are heritable, exhibit variation in those with ASD, and correlate with the presence or absence of ASD, although variation in the unaffected can be minimal or unmeasured.9 In our previous study,10 a trait quantifying deficits in nonverbal communication (NVC) was shown to satisfy these three essential criteria, making it a good candidate for quantitative trait locus (QTL) studies in those with ASD.
NVC was designed to capture the degree of deficits in nonverbal communication in those with ASD. It was quantified by summing the ordinal values between 0 and 3 that reflect severity on seven ADI-R items in the Autism Diagnostic Interview Revised (ADIR).11 Our previous study shows that NVC exhibits familiality and is correlated with deficits in verbal communication, allowing assessment in those with ASD, regardless of their verbal status, and providing additional power to detect genes with smaller effect sizes because everyone in the sample can be included in the analysis. Nonparametric quantitative linkage analyses of NVC in 228 Autism Genetics Resource Exchange (AGRE) families ascertained for at least two children with ASD identified five chromosomal loci or QTL that might harbor genes contributing to variation in NVC.10 Among the five QTL, one genome wide significant locus on chromosome 1 with Z score of 3.6 (P < 0.0001) was replicated in the current analyses. Here, we report the genome-wide QTL replication study, along with the follow-up QTL targeted-association analysis and follow-up study in the associated genes.
Four approaches were taken to improve the odds of identifying a true NVC-risk gene. The first was to identify a quantitative trait that was familial, the second was to conduct replication analyses for the initial QTL, the third was to conduct targeted-association studies (QTL-targeted association) in the replicated chromosome region, mitigating the reduction in power necessitated by a very stringent correction for multiple testing when performing genome wide association and the fourth was to conduct replication studies for the associated genes in the targeted QTL.
Patients and methods
Overview of the study design
The first aim of the present study was to conduct a replication analysis of the original NVC QTL study10 in an independent AGRE sample. The replication sample consisted of 213 families with 274 sibpairs having at least two ASD-affected children, and using the same ADI-R NVC score composed of the sum of reported values on the seven items given in Table 1, and the same QTL analysis method using the Nonparametric option of the Genehunter software.12 In addition, the 400 multiallelic markers used for QTL analysis that were genotyped at the Center for Inherited Disease Research have a significant overlap with those used in the original study. Following QTL replication, the two AGRE samples were combined to conduct a QTL analysis of the replicated region using the same analytic methods.
Table 1.
Nonverbal communication (NVC) score quantified by seven ADI-R items
| Lack of, or delay in, spoken language and failure to compensate through gesture |
| 1. Pointing to express interest |
| 2. Conventional instrumental gestures |
| 3. Nodding |
| 4. Headshaking |
| Lack of varied spontaneous make-believe or social imitative play |
| 5. Spontaneous imitation of actions |
| 6. Imaginative play |
| 7. Imitative social play |
For the replicated Chromosome-1 QTL, a sample of 708 AGRE families from the original and replication QTL samples, along with additional AGRE families,13 was available for a targeted-association analysis of that QTL locus, as they had been genotyped for a genome-wide association study (GWAS) with 550K Illumina SNP Chips (San Diego, CA, USA). The association analysis consisted of three parts. First, the QTL was divided into haplotype blocks for analysis. Then, a global association analysis of each haplotype block was conducted, followed by association testing of the individual haplotypes within the significantly associated blocks. This analytic approach provides an efficient method for association testing that capitalizes on the likely structure of the genetic contributions to the QTL. That is, individual haplotypes within some of the blocks within the QTL are expected to tag one or more predisposing variants in the genes that contribute to variation in NVC. This approach allows us to assess more than one variant in a single overall analysis of a block. The Haploview software was used to define haplotype blocks in the replicated QTL, and haplotype option (HBAT) of the Family-Based Association Test (FBAT) software was used to test the blocks for the distorted transmission of haplotypes to those with high or low NVC values more than one would expect by chance alone.14 Significant blocks, defined as those with a global P-value < 0.001, were used to identify the associated genes by their proximity. After results were obtained in the AGRE sample, the 29 haplotype blocks of two associated genes were each tested for association in an independent sample of 1517 families from the Autism Genome Project (AGP)15 using the same ADI-R score and the same HBAT option of the FBAT software.
Study samples
The AGRE repository
AGRE is a publicly accessible resource for genetic studies of autism.13 The families are ascertained for at least two affected siblings with an autism, Asperger or pervasive developmental disorder. AGRE has IRB (institutional review board) approval from the Western Institutional Review Board and this study also obtained the approval of the UCLA IRB. Diagnoses of ASD are performed using the ADI-R, a standardized and semi-structured clinical protocol for caregivers of children and adults.16 The ADI-R algorithm assesses deficits in the three ASD behavioral domains of verbal, social interaction and restricted, repetitive and stereotyped interests and behaviors. The 93-item ADI-R is based on ICD-1017 and DSM-IV2 criteria for diagnoses of autistic disorder. To meet the strict diagnostic criteria of autism, children meet, or exceed the specified cutoffs in scores in the three domains, and their onset of the disorder is before 36 months of age. Here we allow other diagnoses that fall within the category of ASD (see AGRE website). For the analyses, monozygotic twins, those with known ASD genetic etiologies, those with clear cytogenetic abnormalities and those with dysmorphic features were removed from the sample. 93% of the families had complete genotype information on both parents in the trios.
The AGP sample
The AGP is a large-scale collaborative pooled resource for genetic research comprised of families collected at more than 50 Autism centers in North America and Europe.15 Diagnoses of ASD and measures of the three ASD behavioral domains are based on the ADI-R and the Autism Diagnostic Observation Schedule.18 Nuclear families with two affected individuals and parent/child trios with one affected individual have been ascertained through these Centers. Similar to AGRE, subjects with known karyotypic abnormalities, Fragile-X mutations or other genetic abnormalities have been screened and excluded. The analyses reported here were conducted on 1517 AGP families from the Phase-2, Stage-1 sample that were independent of the AGRE families and were composed of 1500 complete trios from simplex and multiplex families and 17 complete multiplex families. Both parents and children were genotyped using the Illumina Human 1M-single Infinium BeadChip SNP array. Greater than 99% of the trios had both parents genotyped.
NVC trait definition
The NVC score is derived from the sum of scores from 7 ADI-R items listed in Table 1. Each item is scored in the range of 0 to 3, where the higher score represents greater deficits. The age of the child is considered in the score in the following way. The response labeled ‘CURRENT’ is used for a child under the age of 4, which gives his/her manifestation of symptom at the time of interview. The response labeled ‘MOST ABNORMAL AT 4 to 5’ is used for a child beyond the specific age period. In those with ASD, NVC has a unimodal distribution that is skewed and not normal (P < 0.0001). Thus, nonparametric statistics or those robust to non-normality are used throughout. NVC ranges from 1–18, with a median of 13. There is no significant difference in NVC median when the sample is stratified by sex, however, there is a positive correlation with verbal ability as assessed by an ordinal variable that addresses severity of verbal deficits in the ADIR (P < 0.0001).
QTL replication: study sample, genotyping, data cleaning and analysis
The AGRE QTL replication sample includes 213 families with 274 affected sibling pairs who were genotyped for 400 multiallelic markers. Mendelian inconsistencies were identified and those individuals were zeroed out for that genotype. The sex-averaged genetic map distances were estimated from the Rutgers map database in the original and replication QTL studies. Since the time of the first publication, a map from Rutgers has become available for marker localization,19 and we capitalized on this more recent map. QTL analyses were performed using the Non-parametric option of the Genehunter 2.1 software.20 This model-free QTL linkage-mapping method is based on the Wilcoxon rank sum statistic21 and tests for evidence of a QTL using ranked trait differences among all sibling pairs and their allele sharing estimates at every centiMorgan. A Z-score exceeding 2.33 (P < 0.01) at the same QTL region was considered a QTL replication.22 Since our publication of the original results,10 a manuscript reporting the power and Type-1 errors of 12 different algorithms for QTL analysis has been published.23 For their analyses conducted on independent sibships, ascertained for two affected individuals and with a non-normal trait distribution, the Type-1 statistical error rate was as expected and the power was comparable or superior to all other methods examined.
Targeted association: study sample, genotyping, data cleaning and analysis
The AGRE association sample is composed of 708 families with 1371 ASD children that have both NVC and GWAS data. Most of the families from the original and replication AGRE QTL studies are included. Single-nucleotide polymorphism (SNP) genotyping was done using the Illumina 550 chip. SNP quality was addressed using the exclusion criteria of more than a 10% missingness per SNP, a minor-allele frequency < 1%, and Hardy–Weinberg Equilibrium, as assessed in parents, with P < 0.0001 using the PLINK software. Individuals with greater than 10% missing genotypes were excluded.
A Z-score drop of one unit restricted the targeted association region to one between 100 000 000 and 120 000 000 base pairs containing 2815 SNPs in the replicated linked region on 1p21.2–12. To identify the haplotype blocks in this region, the SNPs were partitioned into segments of strong linkage disequilibrium based on the confidence interval criteria derived by Gabriel.24 The default setting of the Haploview 4.2 software25,26 was used to identify the blocks for analysis, with an upper bound of D′ >0.98, a lower bound >0.7, an upper confidence interval maximum for strong recombination of 0.9, and the fraction of strong linkage disequilibrium in information comparison ≥0.95. 476 blocks of varying sizes were defined. These blocks encompass 1564 tagging SNPs with the number of tagging SNPs in each block ranging between 2 to 13.
Association of haplotype blocks and NVC was tested using the haplotype option (HBAT) of the FBAT software.14 This software provides statistically conservative haplotype tests for family-based studies, which are not based directly on the EM algorithm, like a number of other methods. Instead, they rely on conditioning on the observed genotypes in the parents and child and have been found to provide analyses that are robust to the Type-1 statistical errors caused by population admixture, the phenotype distribution, and ascertainment based on phenotypes. The software can handle missing parental genotypes and/or missing phase in both offspring and parents. Briefly, parental genotype data is used to phase compatible haplotypes in the offspring. A conditional statistic is derived from these observed haplotypes. Weights, based on estimated haplotype frequencies, are applied to the HBAT test statistic in the fraction of families with phase ambiguities. The explicit steps used to calculate the HBAT statistic are clearly delineated and illustrated in a paper by Horvath et al.27 The paper also reports the results of a simulation study that supports the use of HBAT in the current AGRE sample that is 85% Caucasian, and potentially vulnerable to the effects of population stratification using most non-family based approaches to association. The study simulates an admixed sample, similar to the one reported here, and the Type-1 statistical error is not inflated for any of the simulation models used.
Global HBAT P-values from the distribution were used to assess association between NVC and a block with H-observed haplotypes. Once a block was associated using the criterion of P < 0.001, individual haplotypes within the blocks were tested separately for association with NVC, also using the HBAT option of the FBAT software. The test statistic is computed from the distribution of offspring genotypes conditioned on the parental genotypes under the null hypothesis of ‘no association in the presence of linkage.’ The genetic effect was assumed to be additive and the test statistic was calculated using the software option −e, which uses the empirical variance–covariance estimator (the Huber–White sandwich estimator) that allows siblings to be included in the same analysis and accounts for their correlation.28 The estimator is also robust against heteroscedasticity in the distribution of the trait, precluding the need for a transformation to normality in this analysis.
Replication of associated genes: study sample, genotyping, data cleaning and analysis
An independent sample of 1517 families from the AGP15 was studied using the same ADI-R score as that used in the AGRE samples. Both parents and one child from each nuclear family were genotyped for the Illumina Human 1M-single Infinium BeadChip SNP array. Quality control for SNPs from this existing GWAS was addressed using the same exclusion criteria as that used in the AGRE association sample. The AGRE and AGP samples were genotyped on two different microarray SNP platforms, and whereas most SNPs genotyped in the AGRE sample were also genotyped on the AGP platform, in the 29 haplotype blocks tested, 4 or 2% of the 189 tagging SNPs in the 29 hpalotype blocks tested were not genotyped in the AGP sample. They were located in 4 of the 22 blocks of the associated potassium voltage-gated channel subfamily D member 3 (KCND3) that were not associated with NVC in the AGRE sample. Two were imputed successfully using the IMPUTE229 software using a reference panel from HapMap phase-3 and the MaCH software30 was used for cross-validation, where both methods resulted in the same minor allele frequency and a Mendelian error rate of < 0.005 for those SNPs. The other two were replaced by SNPs that tagged them in the AGRE sample. The AGP blocks were screened using the same criteria as that used for the AGRE association sample. This was followed by tests of individual haplotypes in the blocks that met a 0.05 level of significance.
Results
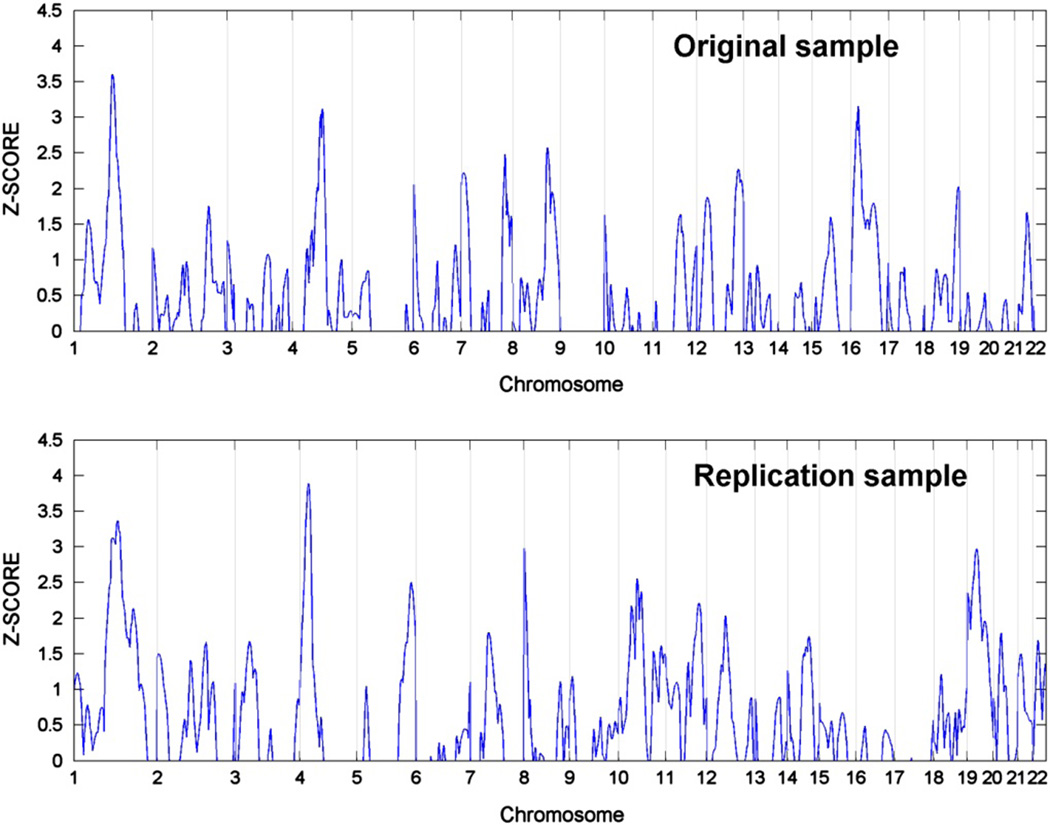
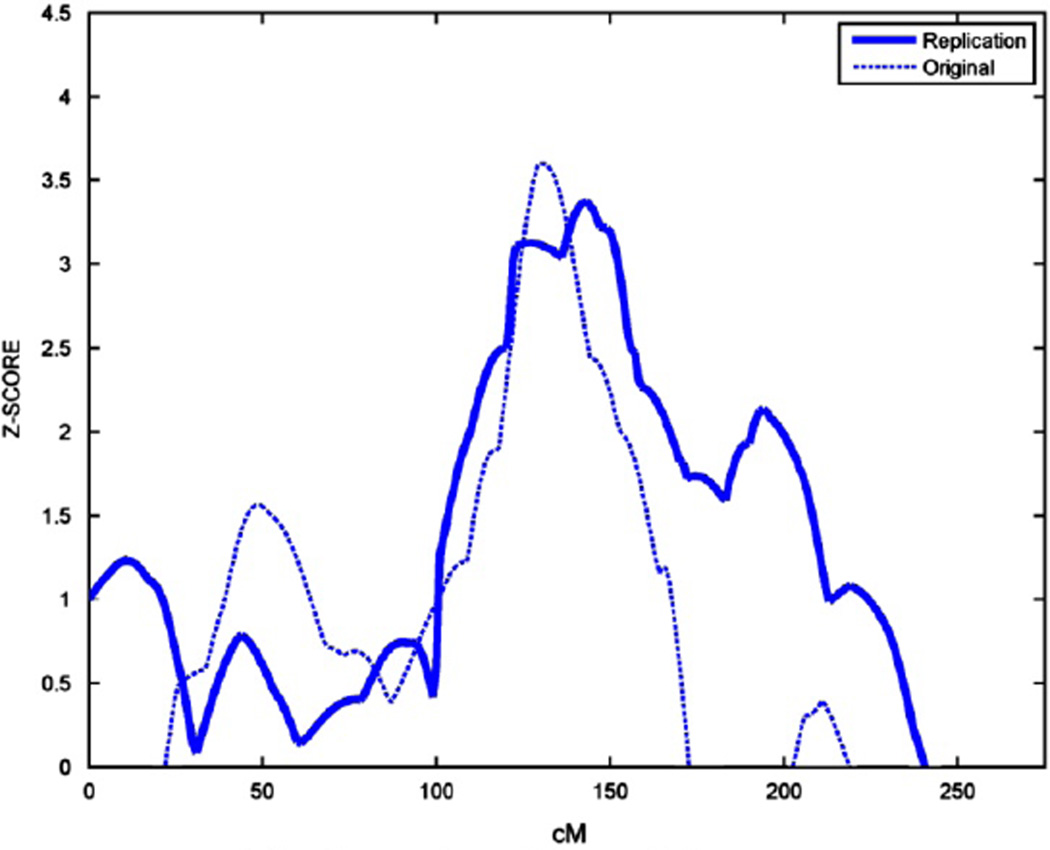
Figure 1 presents the results from the original QTL study in the top panel and the replication QTL study in the bottom panel. In the replication cohort, QTL signals are seen on chromosomes 1 (Z = 3.36, P < 0.0004) and 4 (Z = 3.9, P < 0.00005). However, formal replication is only observed on chromosome 1p21.2–1p12, where the QTL from the prior and current study are coincident (Figure 2). The linkage peaks on chromosome 4 are separated by 70 centi-Morgans. The combined sample increased the Z-score to 4.1 on chromosome 1 (P < 0.00002), but did not narrow the range of the QTL.
Figure 1.
Nonverbal communication (NVC) quantitative trait locus (QTL) analysis in original and replication samples.
Figure 2.
Replicated nonverbal communcation (NVC) quantitative trait locus (QTL) on chromosome 1.
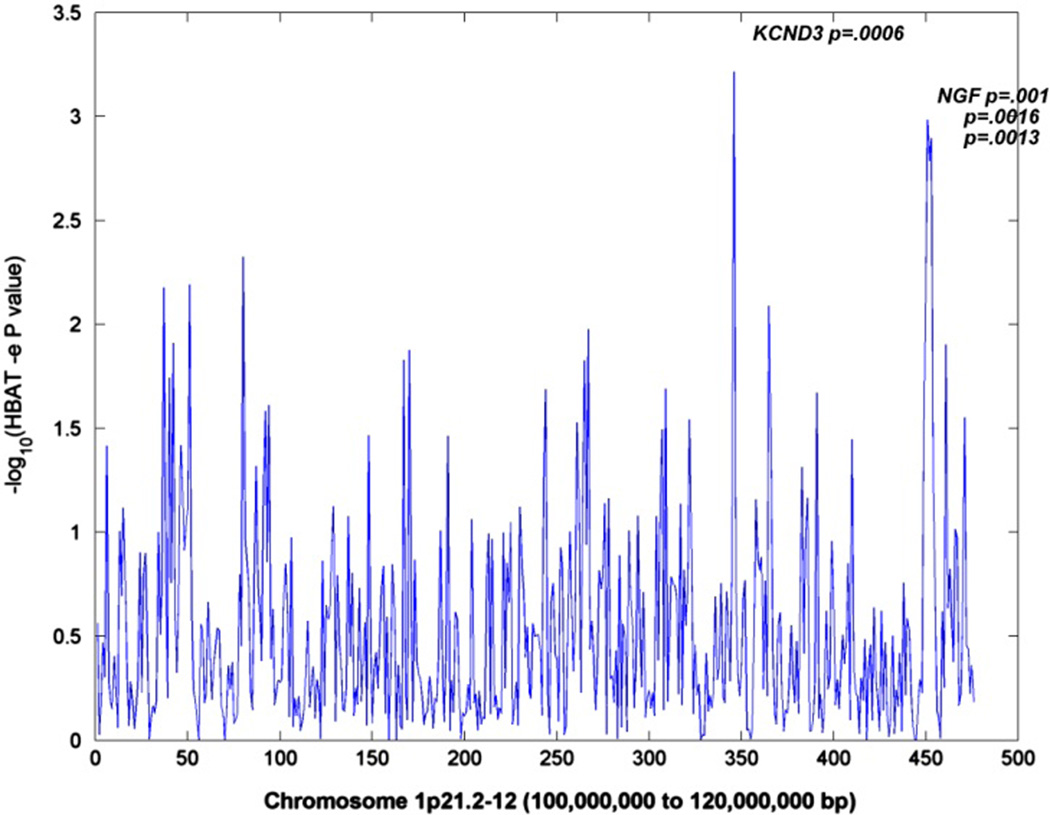
For the targeted-association analyses, a one Z-score drop interval restricted the replicated QTL to a 20-Mb region with 2518 SNPs genotyped on the Illumina 550k chip for which their tagging SNPs defined 476 haplotype blocks. The association results (P < 0.001) for these blocks are illustrated in Figure 3 and presented in greater detail in Table 2. Two QTL genes, KCND3 and nerve growth factor (NGF), are high-lighted by associated blocks in their introns. The table reports the information regarding these blocks. The gene name and study sample are given first, followed by the block number, the tagging SNPs that define it, their locations, the associated haplotypes within the block, the haplotype frequency, its individual P-value and the global P-value that highlighted the block and gene. Haplotypes in bold italics are associated with greater deficits in NVC, whereas the others may protect against such deficits. The Table also contains results from the study of the associated genes in the independent AGP sample.
Figure 3.
Nonverbal communcation (NVC) chromosome-1-targeted block association analysis in Autism Genetics Research Exchange (AGRE) sample.
Table 2.
Significantly associated blocks and haplotypes for genes in the NVC QTL
| Gene (study sample) |
Haplotype block |
Block tagging SNPs |
SNP base pair position |
Associated haplotypes |
Haplo freq |
Haplo P |
Block P |
|---|---|---|---|---|---|---|---|
| KCND3 (AGP) | 9th | rs11102353a | 112426542 | TTb,c | 0.20 | 0.008 | 0.02 |
| rs1373291 | 112426874 | ||||||
| KCND3 (AGRE) | 15th | rs1443928 | 112472773 | CC | 0.15 | 0.0001 | 0.0006 |
| rs12025303 | 112474929 | ||||||
| NGF (AGRE) | 2nd | rs2239622 | 115837709 | TAA | 0.27 | 0.003 | 0.001 |
| rs2856813 | 115837919 | CGC | 0.52 | 0.009 | |||
| rs2856811 | 115838282 | ||||||
| NGF (AGRE) | 3rd | rs6686615 | 115845046 | GT | 0.41 | 0.0003 | 0.002 |
| rs12058927 | 115846221 | AT | 0.42 | 0.03 | |||
| NGF (AGRE) | 4th | rs10776797 | 115847295 | TAGAG | 0.59 | 0.0002 | 0.001 |
| rs12760036 | 115848450 | GAGGA | 0.19 | 0.003 | |||
| rs4626883 | 115849938 | ||||||
| rs6537860 | 115856344 | ||||||
| rs12145726 | 115857251 | ||||||
| NGF (AGP) | 5th | rs10776798 | 115862254 | CGCT | 0.07 | 0.0005 | 0.02 |
| rs4565713 | 115869499 | ||||||
| rs17033692 | 115869850 | ||||||
| rs10858074 | 115870310 |
Abbreviations: AGP, Autism Genome Project; AGRE, Autism Genetics Research Exchange; NGF, nerve growth factor; NVC, nonverbal communication; SNP, single-nucleotide polymorphism.
Human genome assembly HG19.
Allele codes are based on the + strand.
Haplotypes in bold italics are associated with higher NVC scores in the NVC trait distribution, reflecting a more severe deficit. Haplotypes not in bold are associated with lower NVC scores.
Two genes are highlighted in the AGRE sample. The first, KCND3, has a single associated block, the 15th out of 22, with P = 0.0006 and an individual protective haplotype with P = 0.0001. KCND3 is a potassium-channel gene and plays a prominent role in the repolarization phase-1 of the cardiac action potential.31 Similar genes have been implicated in ASD. NGF with only seven blocks is highlighted by two blocks with a P-value of 0.001, and the block between them with a P-value of 0.002. Regarding the individual haplotypes in the three blocks, one is protective (0.003) and one provides risk (0.009) in block 2, one is protective (0.0003) and the other risk (0.03) in block 3, and one is risk (0.0002) and the other protective (0.003) in block 4. NGF is a member of the NGF-beta family and encodes a secreted protein whose function is nerve growth-stimulating activity. The complex is involved in the regulation of growth and the differentiation of sympathetic and certain sensory neurons.32
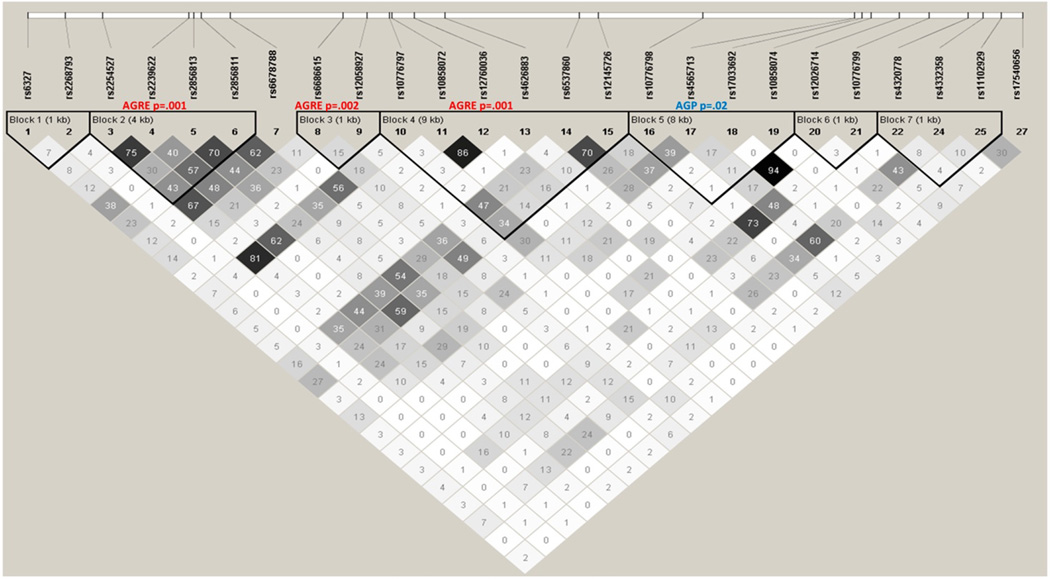
The same FBAT association analysis of 22 blocks in KCND3 and 7 blocks in NGF was conducted in the AGP sample. Results with P < 0.05 are shown in Table 2. The 9th block of KCND3, 46 kb away from the original signal, is associated with NVC (P = 0.02) and has an individual risk haplotype within it (P = 0.008). Block 5 of NGF, next to blocks 2, 3 and 4, is associated with NVC (P = 0.02) and has an individual protective haplotype within it (P = 0.0005). The same block has a P-value of 0.07 in the AGRE sample. To interpret the three individual NGF signals in the AGRE sample, we examined the pairwise linkage disequilibrium among the associated haplotype blocks and their tagging SNPs. We did this to help us decide whether we are finding a single association signal or several independent signals. Figure 4 gives the pairwise r2 estimates for each SNP pair used to define the haplotype blocks. It can be seen from the low r2 values between blocks that the associations are relatively independent, providing evidence to supporting NGF as a gene contributing to variation in NVC.
Figure 4.
Linkage disequilibrium and block structure for nerve growth factor (NGF) gene.
The multiple-association signals in NGF suggest that multiple NGF variants contribute to variation in NVC among those with ASD. It can be seen from Table 2 that some haplotypes tag variants that predispose to severe deficits, whereas others may be protective. To illustrate, the CGC haplotype in block 2 of NGF composed of three SNPs (rs2239622, rs2856813 and rs2856811) will contribute to severity in NVC deficits in those with ASD, as it is overtransmitted to those with higher NVC scores and undertransmitted to those with lower NVC scores. On the other hand, the pattern is reversed for those with the TAA haplotype in that block. This is inferred from an overall statistical analysis that does not allow for the unambiguous assignment of haplotypes to all of the individuals in the sample.
To follow-up, the question of whether the specific association signals could be attributed to specific subsets of the AGRE sample was explored among those with ethnic differences and differences in verbal ability. Eighty-five percent of the AGRE sample report themselves and their families as Caucasian, and multidimensional scaling on their SNP genotypes confirms this. Repeating the HBAT analyses for the associated blocks in the Caucasian subsample resulted in the same association signals with attenuated P-values. We attributed this to the reduced sample size. Differences in verbal ability, as assessed by the ADIR indicated that approximately 40% of the sample has the best verbal abilities, whereas the other 60% had more severe problems in their verbal abilities. HBAT analyses were conducted in both samples separately. The block associations were only observed in the sample of those who had substantial deficits in their language abilities. Thus, it does not appear that stratification by either of these two factors highlights particular block associations within the strata.
Discussion
The identification of predisposing genes has been a daunting endeavor for genetically complex neuro-psychiatric disorders such as ASD. This is in part due to extensive genetic heterogeneity that reduces statistical power for linkage and association studies. Hypothesizing that analyzing a quantitative trait that represents a single feature of ASD may reduce heterogeneity, we analyzed NVC to reveal potential contributory genes. Our earlier publication10 established that NVC can be measured using items from the ADI-R,11 a tool used extensively by researchers for the diagnosis of ASD. In those analyses, we established the necessary criteria that NVC is familial, varies among the affected and correlates with ASD. We also conducted analyses to identify QTL likely to harbor NVC genes, and among the five detected, a chromosome-1 locus was detected.
The work reported here builds sequentially on the findings of that previous study. The first analyses were conducted under the assumption that replication is an essential criterion for gene identification in complex disorders such as ASD. It requires a sufficiently large sample ascertained from an independent panel of families with a comparable diagnosis and should be based on the same quantitative trait measured with the same instrument. Those replication criteria were met by the chromosome-1 QTL, and given our confidence that this QTL harbors gene(s) contributing to deficits in NVC and the availability of GWAS data in this sample, we then conducted a targeted-association study. Those analyses operated under the principle that QTL identify the chromosome regions most likely to harbor trait genes with larger effects, which are easier to detect. One caveat, however, is that allelic heterogeneity in multiple-risk genes within a QTL will allow the region to be flagged by linkage, but may not provide sufficient effect sizes for gene detection with follow-up association studies. To capitalize on the likely heterogeneity within associated genes, we conducted association analyses of haplotype blocks rather than individual SNPs. Blocks divide the QTL into a smaller number of regions that require a less stringent correction for multiple testing than tests of individual SNPs. Causal variants within associated haplotype blocks are likely to reside on different individual haplotypes that can also be tested for association with NVC. To provide support for the associated genes, an additional analysis of their haplotype blocks was conducted in an independent study sample.
Two genes were flagged in these analyses, although they did not exhibit equivalent degrees of support. The first, KCND3, had the strongest association in a single block in the AGRE sample. Support in another block among 22 in KCND3 tested in the AGP sample was somewhat reassuring, but not overwhelming. On the other hand, the NGF gene showed association in three of seven blocks in the original sample and in an adjacent block in the AGP sample. These association signals are consistent with the model of multiple NGF variants contributing to variation in this trait, although the number of independent signals cannot be clearly discerned without sequencing for ‘causal variants’. The results do not preclude an important role for KCND3, as the presence of a replicated QTL reflects multiple signals that are likely to include the effects of more than one gene. Siblings with similar NVC scores are more likely to share such variants, consistent with the strong linkage signal observed and the weaker association findings at any one block of the NGF gene. Here, although the replication in NGF did not involve the same block or SNP, it does replicate at the gene level, with evidence for association in two cohorts in essentially the same region of the gene. These two adjacent haplotype blocks fall within an intronic region with low mammalian conservation, between exons 2 and 3 of this approximately 50 kb gene, so neither are likely to harbor the true functional variant.
NGF is the member of the neurotrophin family of genes and is characterized by its fundamental role in regulating nerve-cell growth, survival and differentiation during early brain development and in the adult both in the peripheral and the central nervous systems. NGF is expressed in the cerebral cortex, hippocampus and olfactory bulb in the central nervous systems,33 although its levels vary considerably by region. The majority of neuroscience research has focused on the neuroprotective role of NGF in neurodegenerative disease, especially as a potential therapy in Alzheimer Disease.34 Prior to this study, very little research has linked NGF to ASD. Riikonen and Vanhala35 showed that NGF levels were normal in children with infantile autism and low to negligible in children with Rett syndrome, however, they did not examine these levels during earlier or later time points. Nelson et al36 examined and compared archived newborn blood samples in children that developed ASD (n = 69), mental retardation without ASD (n = 60), cerebral palsy (n = 63) and control children (n = 54). They measured NGF, brain-derived neurotrophic factor, neurotrophin 3 (NT3) and neurotrophins 4/5 (NT4/5), finding that children with ASD and mental retardation without ASD showed higher levels of brain-derived neurotrophic factor, and NT4/5, but not NGF, when compared with control children. In a potentially related neurodevelopmental disorder, Schizophrenia, Parikh et al37 measured plasma NGF levels in 24 medicated first-episode psychotic patients and in 24 chronic medicated schizophrenia patients by measuring NGF levels. These investigators found that NGF levels were decreased in both groups when compared with the normal group, but NGF levels were significantly higher in chronic patients who were treated with antipsychotic medicine as compared with the first-episode psychosis patients.
With regards to previously published genetic evidence supporting a role for NGF in ASD, none of the published GWAS or studies of structural variation have identified clear pathogenic variants in NGF in patients with ASD. However, a hypothesis-driven candidate-gene association study focusing on a variety of neuronal signaling pathways did identify evidence for association in the gene NTRK1,38 which is the canonical receptor for NGF. Remarkably, NTRK1 was one of only 2 out of approximately 60 genes that survived significance thresholds in the two cohorts investigated in that study. Combined with the current study, these data suggest the involvement of the NGF signaling pathway in ASD pathogenesis.
It should also be noted that although NGF is well known for its neurotrophic activity, it has a significant role in immune modulation.39–41 Additionally, its receptor NTRK1 is expressed by several classes of immune cells.42 Given the emerging evidence for the role of immune or inflammatory activation in ASD,43 and the observation of an increase in autoimmune disorders in parents of autistic children,44 it is tempting to speculate a potential neuroimmune mechanism. Nevertheless, these results suggest that further in-depth sequence analysis of the NGF gene is warranted, so as to clarify its potential role in ASD.
Acknowledgments
These analyses were supported by NIH/NIMH Autism Center of Excellence network Grant MH081754 to Daniel Geschwind (PI). We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by Grant 1U24MH081810 from the National Institute of Mental Health to Clara M Lajonchere (PI). The AGRE Consortium: Dan Geschwind, MD, PhD, UCLA, Los Angeles, CA; Maja Bucan, PhD, University of Pennsylvania, Philadelphia, PA; W Ted Brown, MD, PhD, FACMG, NYS. Institute for Basic Research in Developmental Disabilities, Staten Island, NY; Rita M Cantor, PhD, UCLA School of Medicine, Los Angeles, CA; John N Constantino, MD, Washington University School of Medicine, St Louis, MO; T Conrad Gilliam, PhD, University of Chicago, Chicago, IL; Martha Herbert, MD, PhD, Harvard Medical School, Boston, MA Clara Lajonchere, PhD, Autism Speaks, Los Angeles, CA; David H Ledbetter, PhD, Emory University, Atlanta, GA; Christa Lese-Martin, PhD, Emory University, Atlanta, GA; Janet Miller, JD, PhD, Autism Speaks, Los Angeles, CA; Stanley F Nelson, MD, UCLA School of Medicine, Los Angeles, CA; Gerard D Schellenberg, PhD, University of Washington, Seattle, WA; Carol A Samango-Sprouse, EdD, George Washington University,Washington, DC; Sarah Spence, MD, PhD, UCLA, Los Angeles, CA; Matthew State, MD, PhD, Yale University, New Haven, CT Rudolph E Tanzi, PhD, Massachusetts General Hospital, Boston, MA. CIDR Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number N01-HG-65403. The authors gratefully acknowledge the families participating in the Autism Genome Project (AGP) and the main funders: Autism Speaks (USA), the Health Research Board (HRB; Ireland), the Medical Research Council (MRC; UK), Genome Canada/Ontario Genomics Institute and the Hilibrand Foundation (USA).
Footnotes
Web resources
AGRE, http://www.agre.org/index.cfm, AGP, http://www.autismgenome.org/, CDC, http://www.cdc.gov/ncbddd/autism/data.html, FBAT software, http://biosun1.harvard.edu/~fbat/fbat.htm, Haploview software, http://www.broad.mit.edu/mpg/haploview/, PLINK software http://pngu.mgh.harvard.edu/~purcell/plink/, Rutgers Map website, http://compgen.rutgers.edu/maps.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4th edn. Washington, DC: American Psychiatric Association; 2000. text revision. [Google Scholar]
- 2.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 3.Hallmayer JCS, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, et al. Genetic Heritability and Shared Environmental Factors Among Twin Pairs With Autism. Arch Gen Psychiatry. 2011 doi: 10.1001/archgenpsychiatry.2011.76. (e-pub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lainhart JE, Piven J. Diagnosis, treatment, and neurobiology of autism in children. Curr Opin Pediatr. 1995;7:392–400. doi: 10.1097/00008480-199508000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Constantino JN. The Social Responsiveness Scale. Los Angeles: Western Psychological Services; 2002. [Google Scholar]
- 6.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moldin SO, Rubenstein JLR, editors. Understanding Autism from Basic Neuroscience to Treatment. Boca Raton, FL: CRC Press; 2006. [Google Scholar]
- 8.Cantor RM. In: Autism Spectrum Disorders: Section 39 Autism Endophenotypes and Quantitative Trait Loci. Amaral DG, Dawson G, Geschwind DH, editors. New York, NY: Oxford University Press; 2011. [Google Scholar]
- 9.Dawson G, Webb S, Schellenberg GD, Dager S, Friedman S, Aylward E, et al. Defining the broader phenotype of autism: genetic, brain, and behavioral perspectives. Dev Psychopathol. 2002 Summer;14:581–611. doi: 10.1017/s0954579402003103. [DOI] [PubMed] [Google Scholar]
- 10.Chen GK, Kono N, Geschwind DH, Cantor RM. Quantitative trait locus analysis of nonverbal communication in autism spectrum disorder. Mol Psychiatry. 2006;11:214–220. doi: 10.1038/sj.mp.4001753. [DOI] [PubMed] [Google Scholar]
- 11.Rutter M, Couteur AL, Lord C. Autism Diagnostic Interview, Revised (ADI-R) 2003. [Google Scholar]
- 12.Kruglyak L, Lander ES. A nonparametric approach for mapping quantitative trait loci. Genetics. 1995;139:1421–1428. doi: 10.1093/genetics/139.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–466. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 15.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 17.W.H.O. International Statistical Classification of Diseases and Related Health Problems, 10th Revision. Geneva: World Health Organization; 1992. [Google Scholar]
- 18.Risi S, Lord C, Gotham K, Corsello C, Chrysler C, Szatmari P, et al. Combining information from multiple sources in the diagnosis of autism spectrum disorders. J Am Acad Child Adolesc Psychiatry. 2006;45:1094–1103. doi: 10.1097/01.chi.0000227880.42780.0e. [DOI] [PubMed] [Google Scholar]
- 19.Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 21.Samuels ML, Witmer JA. Statistics for the Life Sciences. 3rd edition. Upper Saddle River, NJ: Prentice Hall; 2002. [Google Scholar]
- 22.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 23.Kleensang A, Franke D, Alcais A, Abel L, Muller-Myhsok B, Ziegler A. An extensive comparison of quantitative trait Loci mapping methods. Hum Hered. 2010;69:202–211. doi: 10.1159/000289596. [DOI] [PubMed] [Google Scholar]
- 24.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.ip71. pdb ip71. [DOI] [PubMed] [Google Scholar]
- 26.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 27.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 28.Lake SL, Blacker D, Laird NM. Family-based tests of association in the presence of linkage. Am J Hum Genet. 2000;67:1515–1525. doi: 10.1086/316895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oudit GY, Kassiri Z, Sah R, Ramirez RJ, Zobel C, Backx PH. The molecular physiology of the cardiac transient outward potassium current (I(to)) in normal and diseased myocardium. J Mol Cell Cardiol. 2001;33:851–872. doi: 10.1006/jmcc.2001.1376. [DOI] [PubMed] [Google Scholar]
- 32.Freed WJ. The role of nerve-growth factor (NGF) in the central nervous system. Brain Res Bull. 1976;1:393–412. doi: 10.1016/0361-9230(76)90033-2. [DOI] [PubMed] [Google Scholar]
- 33.Korsching S, Auburger G, Heumann R, Scott J, Thoenen H. Levels of nerve growth factor and its mRNA in the central nervous system of the rat correlate with cholinergic innervation. EMBO J. 1985;4:1389–1393. doi: 10.1002/j.1460-2075.1985.tb03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Covaceuszach S, Capsoni S, Ugolini G, Spirito F, Vignone D, Cattaneo A. Development of a non invasive NGF-based therapy for Alzheimer0s disease. Curr Alzheimer Res. 2009;6:158–170. doi: 10.2174/156720509787602870. [DOI] [PubMed] [Google Scholar]
- 35.Riikonen R, Vanhala R. Levels of cerebrospinal fluid nerve-growth factor differ in infantile autism and Rett syndrome. Dev Med Child Neurol. 1999;41:148–152. doi: 10.1017/s0012162299000328. [DOI] [PubMed] [Google Scholar]
- 36.Nelson KB, Grether JK, Croen LA, Dambrosia JM, Dickens BF, Jelliffe LL, et al. Neuropeptides and neurotrophins in neonatal blood of children with autism or mental retardation. Ann Neurol. 2001;49:597–606. [PubMed] [Google Scholar]
- 37.Parikh V, Evans DR, Khan MM, Mahadik SP. Nerve growth factor in never-medicated first-episode psychotic and medicated chronic schizophrenic patients: possible implications for treatment outcome. Schizophr Res. 2003;60:117–123. doi: 10.1016/s0920-9964(02)00434-6. [DOI] [PubMed] [Google Scholar]
- 38.Chakrabarti B, Dudbridge F, Kent L, Wheelwright S, Hill-Cawthorne G, Allison C, et al. Genes related to sex steroids, neural growth, and social-emotional behavior are associated with autistic traits, empathy, and Asperger syndrome. Autism Res. 2009;2:157–177. doi: 10.1002/aur.80. [DOI] [PubMed] [Google Scholar]
- 39.Ralainirina N, Brons NH, Ammerlaan W, Hoffmann C, Hentges F, Zimmer J. Mouse natural killer (NK) cells express the nerve growth factor receptor TrkA, which is dynamically regulated. PLoS One. 2010;5:e15053. doi: 10.1371/journal.pone.0015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linker R, Gold R, Luhder F. Function of neurotrophic factors beyond the nervous system: inflammation and autoimmune demyelination. Crit Rev Immunol. 2009;29:43–68. doi: 10.1615/critrevimmunol.v29.i1.20. [DOI] [PubMed] [Google Scholar]
- 41.Fiore M, Chaldakov GN, Aloe L. Nerve growth factor as a signaling molecule for nerve cells and also for the neuroendocrine-immune systems. Rev Neurosci. 2009;20:133–145. doi: 10.1515/revneuro.2009.20.2.133. [DOI] [PubMed] [Google Scholar]
- 42.Ralainirina N, Brons NH, Ammerlaan W, Hoffmann C, Hentges F, Zimmer J. Mouse natural killer (NK) cells express the nerve growth factor receptor TrkA, which is dynamically regulated. PLoS One. 2010;5:e15053. doi: 10.1371/journal.pone.0015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashwood P, Krakowiak P, Hertz-Picciotto I, Hansen R, Pessah IN, Van de Water J. Altered T cell responses in children with autism. Brain Behav Immun. 2011;25:840–849. doi: 10.1016/j.bbi.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keil A, Daniels JL, Forssen U, Hultman C, Cnattingius S, Soderberg KC, et al. Parental autoimmune diseases associated with autism spectrum disorders in offspring. Epidemiology. 2010;21:805–808. doi: 10.1097/EDE.0b013e3181f26e3f. [DOI] [PMC free article] [PubMed] [Google Scholar]