Abstract
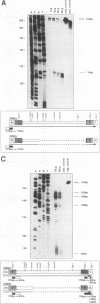
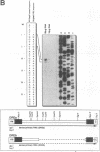
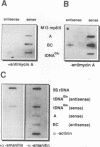
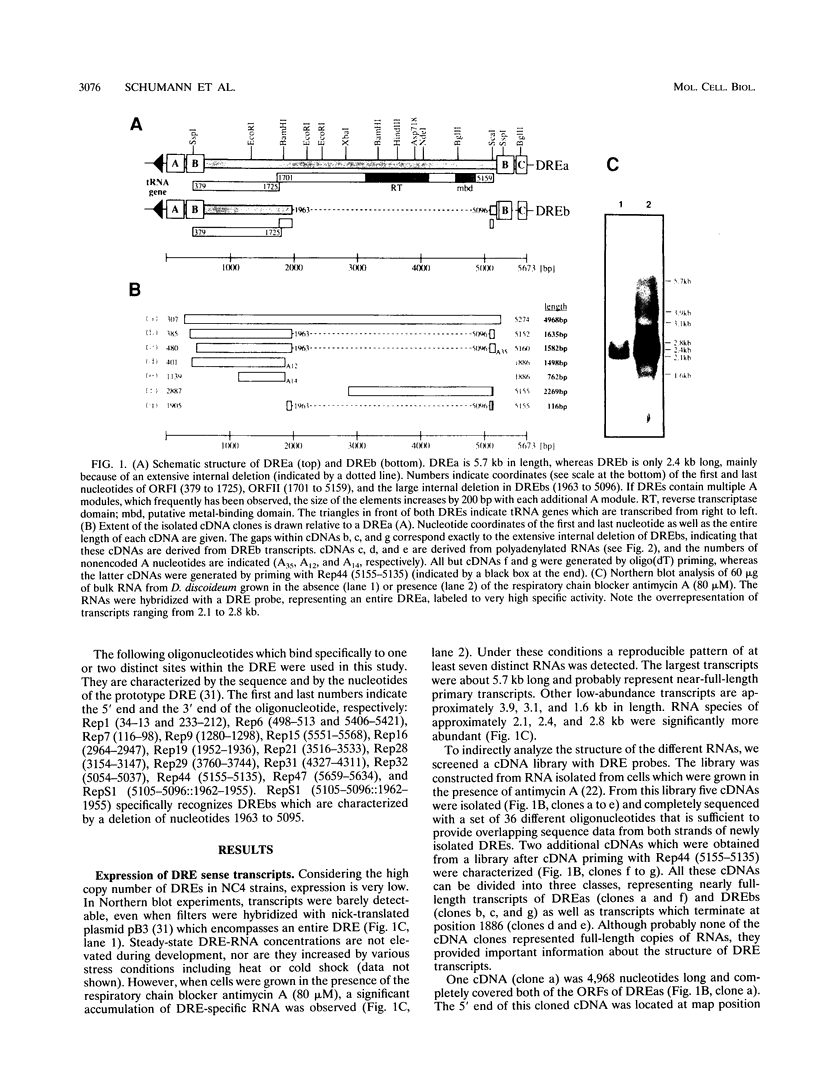
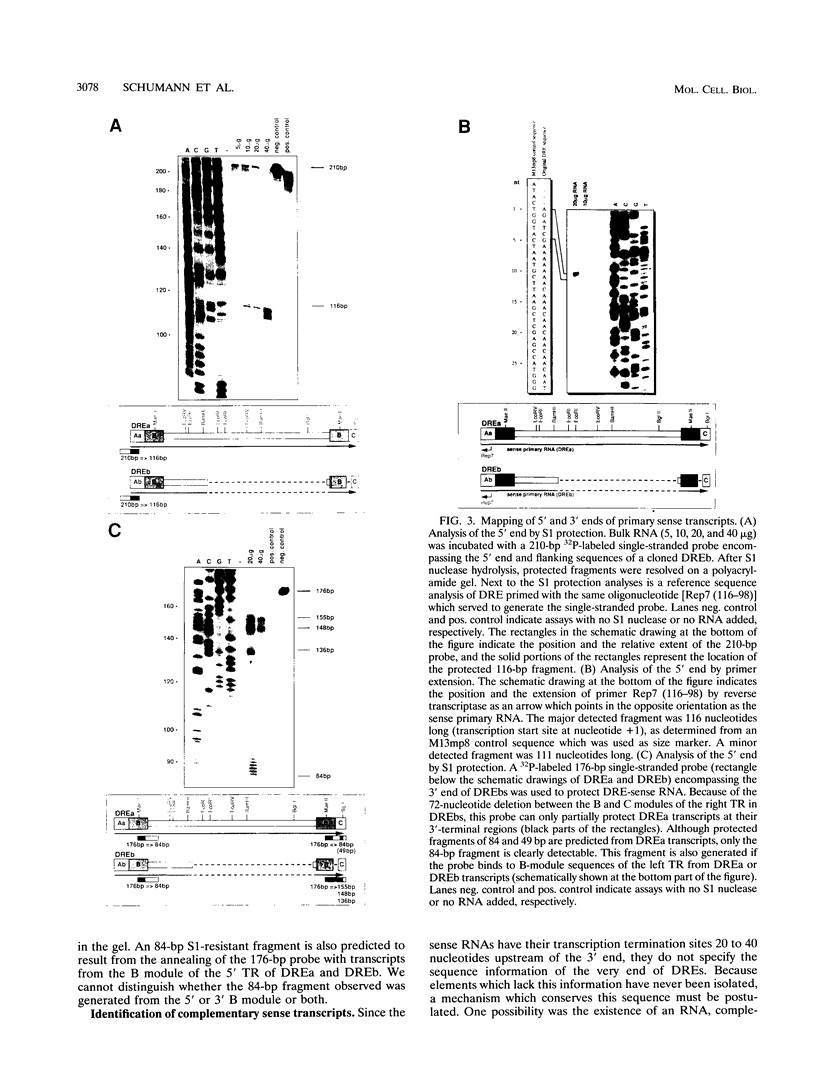
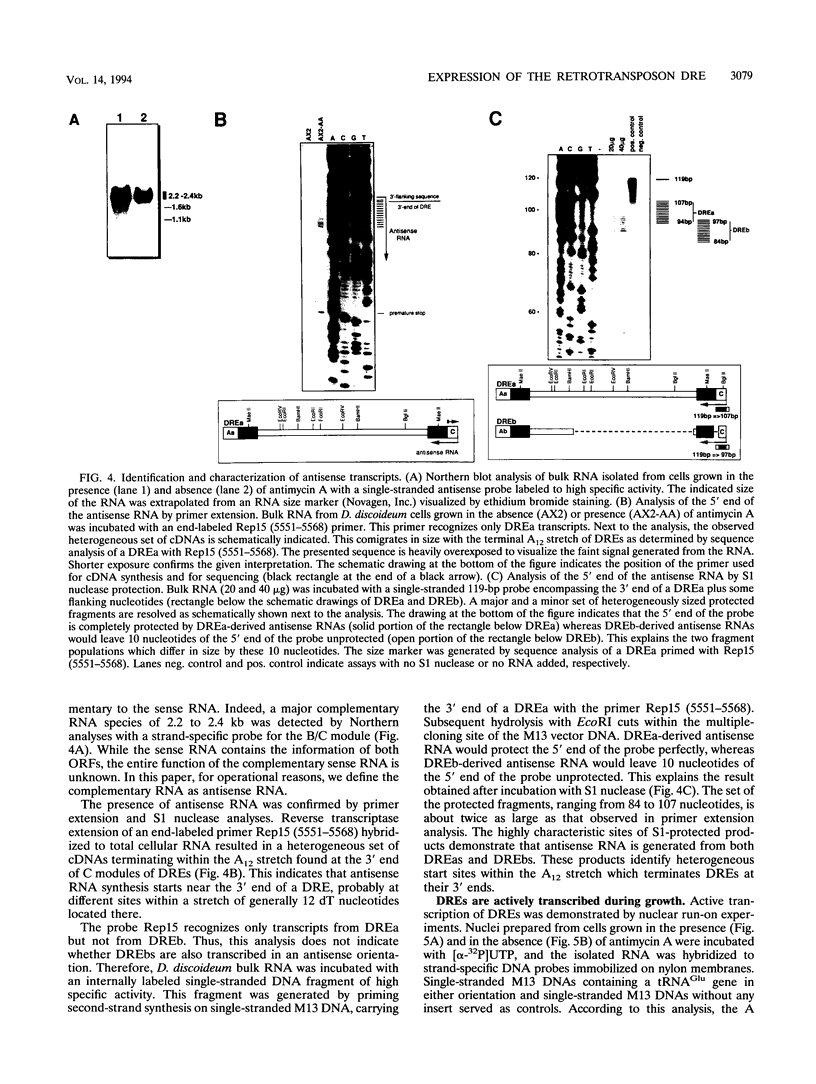
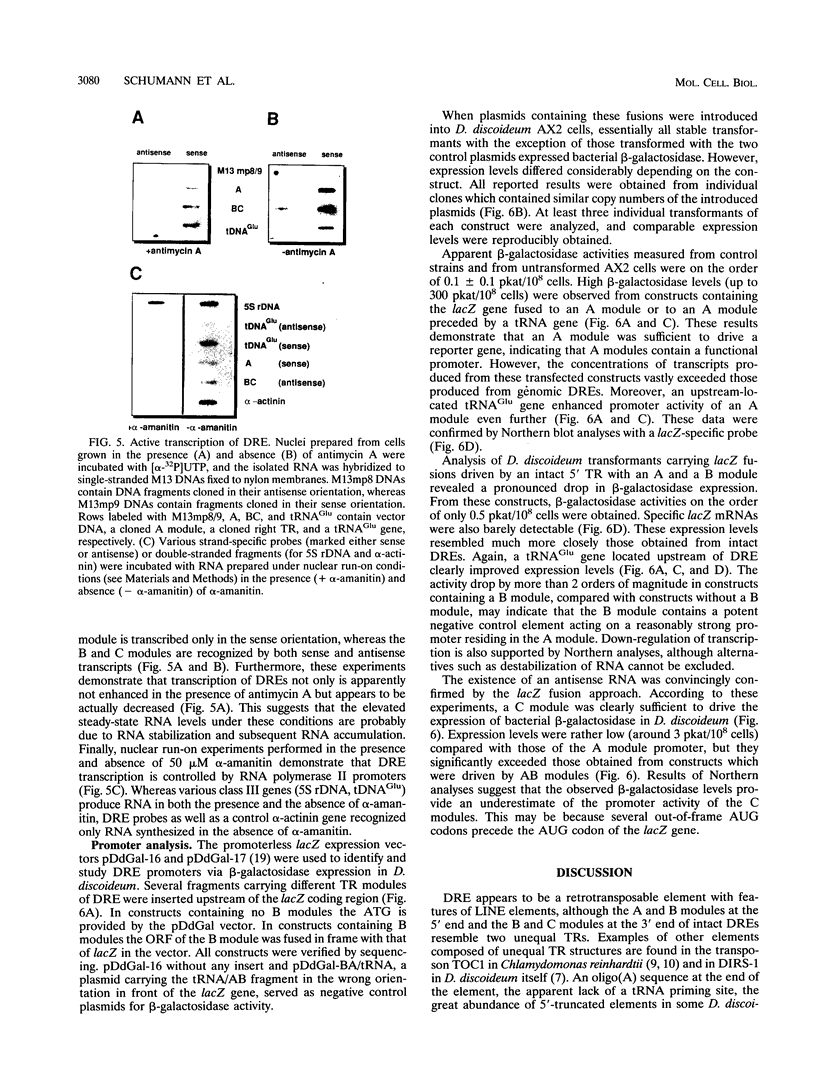
The Dictyostelium discoideum NC4 genome harbors approximately 150 individual copies of a retrotransposable element called the Dictyostelium repetitive element (DRE). This element contains nonidentical terminal repeats (TRs) consisting of conserved building blocks A and B in the left TR and B and C in the right TR. Seven different-sized classes of RNA transcripts from these elements were resolved by Northern (RNA) blot analysis, but their combined abundance was very low. When D. discoideum cells were grown in the presence of the respiratory chain blocker antimycin A, steady-state concentrations of these RNA species increased 10- to 20-fold. The D. discoideum genome contains two DRE subtypes, the full-length 5.7-kb DREa and the internally deleted 2.4-kb DREb. Both subtypes are transcribed, as confirmed by analysis of cloned cDNA. Primary transcripts from the sense strand originate at nucleotide +1 and terminate at two dominant sites, located 21 or 28 nucleotides upstream from the 3' end of the elements. The activity of a reasonably strong polymerase II promoter in the 5'-terminal A module is slightly upregulated by the tRNA gene located 50 +/- 4 nucleotides upstream and drastically reduced by the adjacent B module of the DRE. Transcripts from the opposite DNA strand (complementary-sense transcripts) were also detected, directed by an internally located polymerase II promoter residing within the C module. This latter transcription was initiated at multiple sites within the oligo(dA12) stretch which terminates DREs.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aksoy S., Williams S., Chang S., Richards F. F. SLACS retrotransposon from Trypanosoma brucei gambiense is similar to mammalian LINEs. Nucleic Acids Res. 1990 Feb 25;18(4):785–792. doi: 10.1093/nar/18.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilanchone V. W., Claypool J. A., Kinsey P. T., Sandmeyer S. B. Positive and negative regulatory elements control expression of the yeast retrotransposon Ty3. Genetics. 1993 Jul;134(3):685–700. doi: 10.1093/genetics/134.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Corces V. G. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- Busseau I., Pelisson A., Bucheton A. Characterization of 5' truncated transposed copies of the I factor in Drosophila melanogaster. Nucleic Acids Res. 1989 Sep 12;17(17):6939–6945. doi: 10.1093/nar/17.17.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboissier M. C., Busseau I., Prosser J., Finnegan D. J., Bucheton A. Identification of a potential RNA intermediate for transposition of the LINE-like element I factor in Drosophila melanogaster. EMBO J. 1990 Nov;9(11):3557–3563. doi: 10.1002/j.1460-2075.1990.tb07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Zuker C., Lodish H. F. A repetitive and apparently transposable DNA sequence in Dictyostelium discoideum associated with developmentally regulated RNAs. Nucleic Acids Res. 1983 Jul 25;11(14):4835–4852. doi: 10.1093/nar/11.14.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. M., Cappello J., Lodish H. F. Transcription of Dictyostelium discoideum transposable element DIRS-1. Mol Cell Biol. 1984 Nov;4(11):2332–2340. doi: 10.1128/mcb.4.11.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Errede B. Cell-type-dependent gene activation by yeast transposon Ty1 involves multiple regulatory determinants. Mol Cell Biol. 1987 Sep;7(9):3205–3211. doi: 10.1128/mcb.7.9.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A., Rochaix J. D. Structure and inheritance of sense and anti-sense transcripts from a transposon in the green alga Chlamydomonas reinhardtii. J Mol Biol. 1991 Mar 20;218(2):273–291. doi: 10.1016/0022-2836(91)90712-f. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Amon E., Williams K. L., Welker D. L. Chromosomal mapping of tRNA genes from Dictyostelium discoideum. Mol Gen Genet. 1987 Apr;207(1):176–187. doi: 10.1007/BF00331507. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Reindl N., Brechner T., Werner H., Nerke K. Nonsense suppression in Dictyostelium discoideum. Dev Genet. 1990;11(5-6):410–417. doi: 10.1002/dvg.1020110514. [DOI] [PubMed] [Google Scholar]
- Dingermann T., Reindl N., Werner H., Hildebrandt M., Nellen W., Harwood A., Williams J., Nerke K. Optimization and in situ detection of Escherichia coli beta-galactosidase gene expression in Dictyostelium discoideum. Gene. 1989 Dec 28;85(2):353–362. doi: 10.1016/0378-1119(89)90428-9. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Vimaladithan A., Türkel S., Johnson R., Zhao H. Three downstream sites repress transcription of a Ty2 retrotransposon in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Apr;13(4):2081–2090. doi: 10.1128/mcb.13.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P., Liao X. B., Belcourt M., Zhao H., Kapakos J., Clare J. Enhancer and silencerlike sites within the transcribed portion of a Ty2 transposable element of Saccharomyces cerevisiae. Mol Cell Biol. 1989 Nov;9(11):4824–4834. doi: 10.1128/mcb.9.11.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett D. H., Lister C. K., Kellett E., Finnegan D. J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986 Dec 26;47(6):1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Harwood A. J., Drury L. New vectors for expression of the E.coli lacZ gene in Dictyostelium. Nucleic Acids Res. 1990 Jul 25;18(14):4292–4292. doi: 10.1093/nar/18.14.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt M., Nellen W. Differential antisense transcription from the Dictyostelium EB4 gene locus: implications on antisense-mediated regulation of mRNA stability. Cell. 1992 Apr 3;69(1):197–204. doi: 10.1016/0092-8674(92)90130-5. [DOI] [PubMed] [Google Scholar]
- Hofmann J., Schumann G., Borschet G., Gösseringer R., Bach M., Bertling W. M., Marschalek R., Dingermann T. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5' and 3'-flanking regions. J Mol Biol. 1991 Dec 5;222(3):537–552. doi: 10.1016/0022-2836(91)90495-r. [DOI] [PubMed] [Google Scholar]
- Hoja U., Hofmann J., Marschalek R., Dingermann T. Nucleotide sequence of a Dictyostelium discoideum gene encoding a protein homologous to the yeast ribosomal protein S31. Biochem Biophys Res Commun. 1993 Jan 15;190(1):134–139. doi: 10.1006/bbrc.1993.1021. [DOI] [PubMed] [Google Scholar]
- Khan E., Mack J. P., Katz R. A., Kulkosky J., Skalka A. M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991 Feb 25;19(4):851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A. R., Firtel R. A. Sequence organization in Dictyostelium: unique structure at the 5'-ends of protein coding genes. Nucleic Acids Res. 1983 Jan 25;11(2):541–552. doi: 10.1093/nar/11.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey P. T., Sandmeyer S. B. Adjacent pol II and pol III promoters: transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991 Mar 25;19(6):1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993 Feb 26;72(4):595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Brechner T., Amon-Böhm E., Dingermann T. Transfer RNA genes: landmarks for integration of mobile genetic elements in Dictyostelium discoideum. Science. 1989 Jun 23;244(4911):1493–1496. doi: 10.1126/science.2567533. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Hofmann J., Schumann G., Bach M., Dingermann T. Different organization of the tRNA-gene-associated repetitive element, DRE, in NC4-derived strains and in other wild-type Dictyostelium discoideum strains. Eur J Biochem. 1993 Oct 15;217(2):627–631. doi: 10.1111/j.1432-1033.1993.tb18285.x. [DOI] [PubMed] [Google Scholar]
- Marschalek R., Hofmann J., Schumann G., Dingermann T. Two distinct subforms of the retrotransposable DRE element in NC4 strains of Dictyostelium discoideum. Nucleic Acids Res. 1992 Dec 11;20(23):6247–6252. doi: 10.1093/nar/20.23.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschalek R., Hofmann J., Schumann G., Gösseringer R., Dingermann T. Structure of DRE, a retrotransposable element which integrates with position specificity upstream of Dictyostelium discoideum tRNA genes. Mol Cell Biol. 1992 Jan;12(1):229–239. doi: 10.1128/mcb.12.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C., Bucheton A., Finnegan D. J. The 5' untranslated region of the I factor, a long interspersed nuclear element-like retrotransposon of Drosophila melanogaster, contains an internal promoter and sequences that regulate expression. Mol Cell Biol. 1993 Feb;13(2):1042–1050. doi: 10.1128/mcb.13.2.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchiotti G., Di Nocera P. P. Convergent transcription initiates from oppositely oriented promoters within the 5' end regions of Drosophila melanogaster F elements. Mol Cell Biol. 1991 Oct;11(10):5171–5180. doi: 10.1128/mcb.11.10.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrokhi L. J., Georgieva S. G., Ilyin Y. V. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988 Aug 26;54(5):685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- Mizrokhi L. J., Mazo A. M. Evidence for horizontal transmission of the mobile element jockey between distant Drosophila species. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9216–9220. doi: 10.1073/pnas.87.23.9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchès C., Agarwal M., Campbell K., Lemieux L., Abadon M. Sequence of a truncated LINE-like retroposon dispersed in the genome of Culex mosquitoes. Gene. 1991 Oct 15;106(2):279–280. doi: 10.1016/0378-1119(91)90211-s. [DOI] [PubMed] [Google Scholar]
- Murphy N. B., Pays A., Tebabi P., Coquelet H., Guyaux M., Steinert M., Pays E. Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J Mol Biol. 1987 Jun 20;195(4):855–871. doi: 10.1016/0022-2836(87)90490-6. [DOI] [PubMed] [Google Scholar]
- Priimägi A. F., Mizrokhi L. J., Ilyin Y. V. The Drosophila mobile element jockey belongs to LINEs and contains coding sequences homologous to some retroviral proteins. Gene. 1988 Oct 30;70(2):253–262. doi: 10.1016/0378-1119(88)90197-7. [DOI] [PubMed] [Google Scholar]
- Pritchard M. A., Dura J. M., Pélisson A., Bucheton A., Finnegan D. J. A cloned I-factor is fully functional in Drosophila melanogaster. Mol Gen Genet. 1988 Nov;214(3):533–540. doi: 10.1007/BF00330491. [DOI] [PubMed] [Google Scholar]
- Rae P. M., Kohorn B. D., Wade R. P. The 10 kb Drosophila virilis 28S rDNA intervening sequence is flanked by a direct repeat of 14 base pairs of coding sequence. Nucleic Acids Res. 1980 Aug 25;8(16):3491–3504. doi: 10.1093/nar/8.16.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Leclercq L., Göbel E., Saedler H. Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 1987 Dec 20;6(13):3873–3880. doi: 10.1002/j.1460-2075.1987.tb02727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Purification and characterization of a heat-shock element binding protein from yeast. EMBO J. 1987 Oct;6(10):3035–3041. doi: 10.1002/j.1460-2075.1987.tb02609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold G. D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990 Dec;10(12):6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M. F., Bossier P., Fitch I. T. A highly conserved sequence in yeast heat shock gene promoters. Nucleic Acids Res. 1988 Dec 23;16(24):11845–11845. doi: 10.1093/nar/16.24.11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D. J., Ashworth J. M. Growth of myxameobae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem J. 1970 Sep;119(2):171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]