Abstract
We have developed an association-based approach using classical inbred strains of mice in which we correct for population structure, which is very extensive in mice, using an efficient mixed-model algorithm. Our approach includes inbred parental strains as well as recombinant inbred strains in order to capture loci with effect sizes typical of complex traits in mice (in the range of 5 % of total trait variance). Over the last few years, we have typed the hybrid mouse diversity panel (HMDP) strains for a variety of clinical traits as well as intermediate phenotypes and have shown that the HMDP has sufficient power to map genes for highly complex traits with resolution that is in most cases less than a megabase. In this essay, we review our experience with the HMDP, describe various ongoing projects, and discuss how the HMDP may fit into the larger picture of common diseases and different approaches.
Introduction
The human genome-wide association studies (GWAS) of the last several years have provided the first unbiased views of the genetics of common complex diseases, such as coronary artery disease, diabetes, and cancer. Many of the loci contain novel genes not previously connected to their respective disease, indicating that there is great potential to discover new pathways and new targets for therapeutic intervention. These GWAS do, however, have some important limitations. First, human GWAS are not well powered to study genetic interactions, such as gene-by-gene or gene-by-environment interactions (Zuk et al. 2012). Second, it will be difficult to move from locus to a disease pathway directly in humans (Altshuler et al. 2008). And third, for most diseases, GWAS have identified only a small fraction of the total genetic contributions and, thus, there is a great deal more to be discovered (Altshuler et al. 2008; Manolio et al. 2009).
To simplify genetic analysis, natural variations relevant to disease have been studied in mice and rats (Ahlqvist et al. 2011; Flint and Mackay 2009; Keane et al. 2011). This has generally involved traditional linkage mapping methods with crosses between different strains to identify quantitative trait loci (QTLs). An important problem with such analysis has been poor mapping resolution because the QTLs generally contain hundreds of genes, making the identification of the causal genes difficult.
To address these limitations, we have developed an association-based approach using classical inbred strains of mice (Bennett et al. 2010). We follow previous attempts to apply association in mice (Cervino et al. 2007; Grupe et al. 2001; Guo et al. 2007; Liao et al. 2004; Liu et al. 2007; Pletcher et al. 2004) with two differences. First, we correct for population structure, which is very extensive in mice, using an efficient mixed-model algorithm (EMMA) (Kang et al. 2008). Second, to capture loci with effect sizes typical of complex traits in mice (in the range of 5 % of total trait variance), we supplemented the population with recombinant inbred (RI) strains.
Over the last few years, we have typed the hybrid mouse diversity panel (HMDP) strains for a variety of clinical traits as well as intermediate phenotypes, and have shown that the HMDP has sufficient power to map genes for highly complex traits with resolution that is in most cases less than a megabase. In this essay, we review our experience with the HMDP, describe various ongoing projects, and discuss how the HMDP may fit into the larger picture of common diseases and different approaches.
Overview of the HMDP
The hybrid mouse diversity panel (HMDP) consists of a population of over 100 inbred mouse strains selected for usage in systematic genetic analyses of complex traits (Table 1). Our goals in selecting the strains were to (1) increase resolution of genetic mapping, (2) have a renewable resource that is available to all investigators worldwide, and (3) provide a shared data repository that would allow the integration of data across multiple scales, including genomic, transcriptomic, metabolomic, proteomic, and clinical phenotypes. The core of our panel for association mapping (Bennett et al. 2010; Cervino et al. 2007; Grupe et al. 2001) consists of 29 classic parental inbred strains which are a subset of a group of mice commonly called the mouse diversity panel. We settled on our strains by eliminating closely related strains and removing wild-derived strains. The decision to remove wild-derived stains is based on the tradeoff between statistical power and genetic diversity. While we were sacrificing the genetic diversity by leaving out wild-derived strains, our panel increased the statistical power (assuming the same number of animals) to identify genetic variants polymorphic among the classical inbred strains which affect traits, and these variants account for a tremendous amount of phenotypic diversity among the classical inbred strains.
Table 1.
The 114 strains typed within the HMDP for metabolic phenotypes
These strains are commercially available from The Jackson Laboratory (JAX) (Bar Harbor, ME). Additional strains will soon be entered into the HMDP data base (see Table 2). For entry of phenotypic measures into the HMDP database and for calculating associations using EMMA programming (see Table 2), the JAX strain names need to be converted to the EMMA strain names that we have provided here
In order to increase power, we included panels of RI mice, including the BXD, CXB, BXA/AXB, and BXH panels. Power calculations with the inclusion of these additional strains indicated that we have 70 % power to detect SNPs that contribute ~10 % of the overall variance of a complex trait (Bennett et al. 2010). We have recently shown that power can be further increased by performing meta-analysis in which data from the HMDP are combined with data from traditional crosses (Furlotte et al. 2012). Power can also be increased by typing additional commercially available RI panels, as discussed below.
A key feature of the panel is that genotyping is not necessary due to the wealth of single nucleotide polymorphism (SNP) genotypes known across the mouse strains (Keane et al. 2011; Kirby et al. 2010). The inbred strains used for the HMDP were previously genotyped by the Broad Institute and then combined with genotypes from the Wellcome Trust Center for Human Genetics (WTCHG) (Table 2). Genotypes of RI strains at the Broad Institute were inferred from WTCHG genotypes by interpolating alleles at polymorphic SNPs among parental strains, calling ambiguous genotypes missing. Of the 140,000 SNPs available, 107,145 were informative with an allele frequency >5 % and were used for GWAS in our publications (Bennett et al. 2010; Farber et al. 2011; Park et al. 2011). Additional genotyping classifications have been performed recently (Keane et al. 2011; Kirby et al. 2010), and the resulting 4 million SNP genotypes are freely available (Table 2).
Table 2.
URL sites useful for the HMDP as well as sites used to develop the HMDP, including sites at The Jackson Laboratory (JAX) (Bar Harbor, ME)
| Name | Description | URL |
|---|---|---|
| EMMA | Obtain EMMA in an R package | http://mouse.cs.ucla.edu/emma/ |
| EMMA server | Input data and obtain analysis | http://mouse.cs.ucla.edu/emmaserver/ |
| EMMA power simulator | R package allowing statistical power experiment via in silico mapping | http://mouse.cs.ucla.edu/power/ |
| EMMA study design Webserver | Provides power simulations with various background genetic effects and threshold estimations | http://mouse.cs.ucla.edu/emmaserver/powerSimulation/ |
| Mouse HapMap | Latest genetic maps that are regularly updated | http://mouse.cs.ucla.edu/mousehapmap/ |
| Mouse genome informatics (JAX) | Search tools for gene names and chromosomal positions, phenotypes, expression, orthology, and other features | http://www.informatics.jax.org/ |
| SGR HMDP database | Database for HMDP and several F2 studies with cQTL, eQTL, heatmap and correlation analyses | http://systems.genetics.ucla.edu |
| Genenetwork | Study relationships among phenotypes and genotypes | http://www.genenetwork.org/webqtl/main.py |
| Mouse phenome database (JAX) | Phenotypes across hundreds of strains contributed by over 200 investigators | http://phenome.jax.org/ |
| Wellcome trust center for human genetics | Center to understand the genetic foundations of human variation and disease | http://www.well.ox.ac.uk/home |
| Perlegen | Perlegen mouse SNP browser covering 12 classical inbred strains and 4 wild-derived strains | http://mouse.cs.ucla.edu/perlegen/ |
| Mouse genomes project | Nucleotide sequence database for many key mouse strains | http://sanger.ac.uk/resources/mouse/genomes/ |
In the current HMDP panel consisting of over 100 inbred and RI strains, we used ~860,000 SNPs to examine linkage disequilibrium (LD) blocks. These SNPs have greater than 5 % minor allele frequency (mean and median minor allele frequency of 28 and 31 %) and are polymorphic between the strains. Using 0.8 as the r2 cutoff to define LD, there are a total of 13,706 LD blocks with the median size of 42.8 kb per block (mean of 143.3 kb per block) scattered throughout the genome (Fig. 1). Since LD blocks are more likely to define the window in which a candidate gene for a locus resides, the presence of small LD in the HMDP suggests that the number of causal candidate genes will be on average fewer than five genes per locus. This is a considerable improvement over mapping resolution in traditional linkage studies and/or in outbred stock mice where, on average, the number of candidate genes for each locus ranges from 10 to 50. Large blocks (defined as LD blocks >1 Mb) are also present in the HMDP panel but not frequently. Only 1.5 % of the LD blocks (211 of 13,706 total) have a large size encompassing 12.5 % of the genome. The X chromosome is noted to contain multiple large LD blocks, suggesting that mapping resolution for this chromosome is reduced as compared to the autosomes. These large blocks reflect the regions in the genome that were inherited by all strains from the shared ancestors as described in (Frazer et al. 2007). The high-resolution mapping property of the HMDP becomes particularly important in systems genetics as one common goal is to identify causal genes that coordinately regulate network function. Such drivers have been reported in numerous studies as candidate genes for “QTL hot spots,” but the true identity of such drivers has been elusive mainly due to lack of resolution in mapping.
Fig. 1.
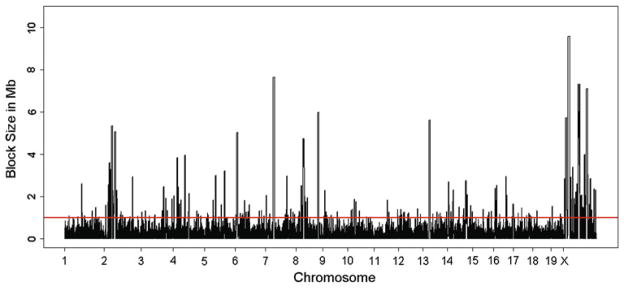
Map of linkage disequilibrium (LD) blocks along the genome in the hybrid mouse diversity panel (HMDP) population. The LD blocks were determined using ~860,000 SNPs that have >5 % minor allele frequency (mean and median minor allele frequency of 28 and 31 %) and are polymorphic between the strains. Using 0.8 as the r2 cutoff to define LD, there are a total of 13,706 LD blocks with a median size of 42.8 kb per block (mean of 143.3 kb per block) scattered throughout the genome. Only 1.5 % of the LD blocks (211 of 13,706 total) have a large size encompassing 12.5 % of the genome. The location of the large blocks in the genome can be identified by lines that cross the red bar. See text for more details
In addition to the excellent resolution, the HMDP has important advantages for systems genetics and for analysis of genetic interactions. The progeny from a genetic cross are unique and as such can be characterized for a limited number of phenotypes, whereas the inbred strains of the HMDP can be examined for an unlimited number of phenotypes since the data are cumulative. The same concept applies to interactions. Thus, mice of the same genotype can be examined under a variety of conditions to identify gene-by-environment interactions, and epistatic interactions can be tested using targeted perturbations on specific genetic backgrounds. This is also an important feature of other replicate mouse genetic systems such as consomic strains, collaborative cross strains, and wild-derived inbred strains.
Discoveries using the HMDP
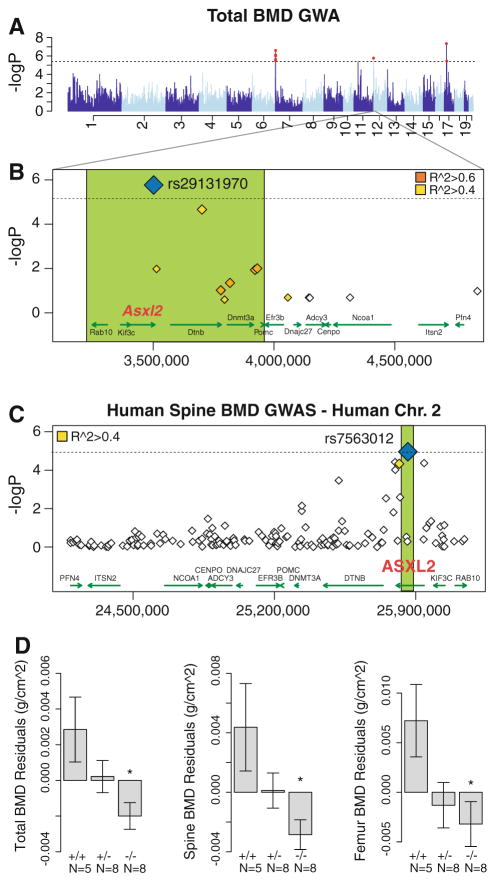
The successful use of the HMDP to identify complex trait genes was recently highlighted in a study of bone mineral density (BMD) (Farber et al. 2011). BMD is a polygenic phenotype that is commonly investigated in human and rodent genetic studies and is the single strongest predictor of osteoporotic fracture (Cummings et al. 2002; Farber and Rosen 2010). For this study, total body, spine, and femur areal BMD data were generated on 16 week-old male mice from 96 HMDP strains. The whole bone transcriptome was also profiled using Illumina gene expression microarrays. The authors used EMMA to perform genome-wide association for the three BMD measures. A total of four genome-wide significant associations were identified on Chromosomes (Chrs) 7, 11, 12, and 17, each affecting BMD at one or more sites. The Chr 12 association for total-body BMD was chosen for further analysis since the 3 Mb window surrounding the association contained only 14 candidate genes. Interestingly, the most significant association was with a nonsynonymous SNP (rs29131970) in the additional sex-combs like 2 (Asxl2) gene (Fig. 2). This polymorphism was predicted to have deleterious effects on ASXL2 protein function. To gain further support for Asxl2 being the causal gene, existing human genome-wide association data (generated in ~6,000 Icelandic subjects) was used to evaluate SNPs within the human syntenic region for association with BMD (Styrkarsdottir et al. 2008). One SNP (rs7563012) was significant after Bonferroni correction and was located in intron 3 of the human ASXL2 gene (Fig. 2). Together these data suggested that Asxl2 influenced BMD in both humans and mice. Consistent with this hypothesis, BMD was found to be lower in Asxl2−/− knockout mice.
Fig. 2.
Variation in Asxl2 in mice and humans is associated with bone mineral density (BMD). a Genome-wide association in the HMDP for total BMD identifies an association on chromosome (Chr) 12. b A nonsynonymous SNP (rs29131970) in Asxl2 that was predicted to alter protein function was the most significantly associated Chr 12 SNP in the HMDP. c Human SNPs within ASXL2 were also associated with BMD in ~6,000 Icelandic individuals. d Male mice deficient in Asxl2 (−/−) display significant decreases relative to wild-type controls (+/+) in total BMD, spine BMD, and femur BMD residuals after adjustments for age and body weight. Data shown in d are residual mean ± SEM, *P < 0.05
Network analysis in the HMDP is another powerful approach for investigating complex traits from a systems-level perspective. Coexpression networks can be used to annotate genes of unknown function based on the known functions of the genes to which they are most closely connected. This “guilt by association” approach has been shown to be a robust gene annotation tool (Wolfe et al. 2005) and was used to determine the mechanism through which Asxl2 influenced BMD. Weighted Gene Co-expression Network Analysis (WGNCA) was first used to generate a coexpression network using the bone microarray data, which identified Asxl2 as being connected to genes involved in myeloid cell differentiation. In bone, osteoclasts are bone-resorbing cells of myeloid origin (Teitelbaum and Ross 2003). Additionally, in a human protein-protein interaction network, ASXL2 interacts with TRAF6, a key component of the major signaling pathway regulating osteoclastogenesis (Teitelbaum and Ross 2003). Thus, based on network inferences, Asxl2 was predicted to be involved in the differentiation of osteoclasts. To test this prediction, expression of Asxl2 was knocked down in osteoclast precursors. An ~50 % reduction in Asxl2 transcript levels inhibited the formation of TRAP+ (a marker of mature osteoclasts) multinuclear cells. These data suggested that Asxl2 influences BMD, at least in part, through its regulation of osteoclastogenesis. This work highlights the ability of using the HMDP and systems genetics to move from association to gene to mechanism in a single step.
In another study, (Park et al. 2011) utilized the HMDP resource to report on gene networks associated with conditional fear. In this study, the authors combined behavioral phenotypes with gene expression data in two regions of the brain (striatum and hippocampus) to identify groups of genes that coordinately regulate the behavior of the animals. Overall, they observed significant overlap between the local QTLs and QTL hotspots in the two tissues, as well as module conservation and preservation of highly connected genes (also known as “hubs”) in the striatum and hippocampus networks. The authors were also able to identify tissue-specific network modules between the striatum and the hippocampus, and after performing functional enrichment analysis of the modules, they arrived at pathways likely to contribute to the differences in hippocampus and striatum function. Finally, using modules as functional units, the authors were able to demonstrate correlations with behavioral traits, thus helping to prioritize candidate genes and pathways for behavioral traits.
In addition to identifying cellular mechanisms and genes underlying physiological traits, the HMDP has been used to investigate the relationships across various biological scales at the global level (Ghazalpour et al. 2011). For example, among the intermediate phenotypes that have been examined in liver are transcript levels (in triplicate, using the Affymetrix platform) and a set of peptides corresponding to about 1,000 proteins (using quantitative mass spectrometry analysis) (Ghazalpour et al. 2011). The correlation between protein and transcript levels was quite weak, with a correlation coefficient of less than 0.4 in most cases, similar to what has been observed in previous studies with yeast and worms. More surprising was the finding that transcript levels were much more strongly correlated with clinical traits (primarily metabolic) than were protein levels. One possible explanation is that transcript levels may be reactive rather than causal with respect to physiologic traits (Ghazalpour et al. 2011).
Overall, the HMDP is being used to develop a multi-scale understanding of a number of complex traits, including a recent report on elevated heart rate (Smolock et al. 2012). There are already over 70 traditional clinical traits reported and there are ongoing studies related to diet-induced obesity, hearing loss, heart failure, atherosclerosis, lipoprotein metabolism, bone metabolism, vascular injury, hematopoietic stem cells, air pollution, gut flora, addictive behavior, hepatotoxicity, and diabetic complications. In addition, gene expression microarrays have been used to quantify mRNA levels in liver, bone, adipose, brain, peritoneal macrophages, aorta, and heart, and proteomic and metabolomic profiling has been performed in liver.
Integration of the HMDP with other resources
The HMDP is just one of several recently proposed approaches to improve the resolution of mouse genetic studies. Other approaches include the Collaborative Cross (CC) (Churchill et al. 2004), outbred designs (Valdar et al. 2006; Yalcin et al. 2010), and the use of consomic strains (Gregorova et al. 2008; Singer et al. 2004; Takada et al. 2008). Each approach has advantages and disadvantages relative to the HMDP. The CC is a recently developed panel of RI strains that are descendants from eight founder strains. A key difference between the HMDP and the CC is that three of the CC founders are wild-derived strains. Wild-derived strains introduce a significantly larger amount of genetic variation and corresponding phenotypic variation compared to the HMDP. For this reason, it is likely that more genes in the CC will have effects on traits than in the HMDP. However, the additional variation may make it relatively more difficult to map quantitative loci polymorphic in both panels since the increased variation of the CC and outbred panels will reduce the relative effect size of the same variant compared to the HMDP (Kang et al. 2008). The CC will ultimately contain approximately 300 strains compared to the current 100 strains of the HMDP. However, the HMDP may be enlarged to 260 strains (see Future directions and conclusions section) (Collaborative Cross Consortium 2012). Both the CC and the HMDP use inbred strains so they share the advantage of accumulation of data on each strain over time as more and more studies are performed. Utilizing inbred strains also facilitates performing studies with perturbations because identical animals can be phenotyped both with and without a perturbation.
An advantage of outbred designs is that they have higher resolution than the HMDP (up to 100 kb). On the other hand, a disadvantage in outbred designs, either utilizing a specially designed Heterogeneous Stock (Valdar et al. 2006) or commercially available outbred mice (Yalcin et al. 2010), is that each animal is unique. Overall, several mapping strategies are, or will be, available to tackle the genetics of complex diseases.
Consomic strains are also proving valuable for reducing genetic intervals containing candidate genes (Hoover-Plow et al. 2006; Prows et al. 2008) and for identifying causal genes (Burrage et al. 2010). However, consomic strains have limited uses for initial genomic studies, as only two alleles are sampled and consomic strains by definition consist of large areas of LD which were captured from the donor strain during breeding. Nonetheless, consomic strains have enriched the ability to test for epistasis and to reduce genetic intervals by the generation of congenic strains.
Resources for design and analysis of HMDP
UCLA maintains several resources for the design and analysis of HMDP studies (Table 2). The two most relevant to investigators are the EMMA association webserver and EMMA design webserver. Investigators utilizing the HMDP can upload their collected phenotypes to the association webserver which will perform association mapping using EMMA and return population structure-corrected P values for each SNP. The analysis is performed on a high-performance computing infrastructure at UCLA, eliminating the need for investigators applying the HMDP to invest in computational resources to perform the analysis. The EMMA design webserver allows an investigator to estimate the power of a proposed study design through simulations (Kirby et al. 2010) that can help guide an investigator in the design of HMDP studies. In addition, curated genotypes of the HMDP strains are also available.
A systems genetics database
Because the HMDP mice are inbred, with fixed genotypes, the data generated from their study are cumulative. To facilitate the integration and analysis of such data, we also developed a database called the Systems Genetics Resource (SGR). The database comprises mouse genomic, transcriptomic, metabolomic, proteomic, and clinical trait data from the HMDP as well as selected traditional mouse crosses and several human studies. The data are accompanied by detailed descriptions of how the data were acquired, with protocols and links to related published papers. A summary of current data sets contained in the database is presented in Table 3.
Table 3.
Data sets currently on line on the systems genetics resource (SGR) database
|
This resource is updated regularly with submissions from the HMDP user groups
We developed a web-based interface where data can be queried for information on specific gene and trait correlations, gene expression in various tissues, or quantitative trait loci, as well as be downloaded for other types of analyses. Such information can be used, for example, to prioritize candidate genes in genetic studies and transacting loci can be used to generate hypotheses about regulatory pathways (Fig. 3). The intermediate phenotypes can also be used to model gene networks and causal interactions. The power of the SGR is expected to expand as more data are added. Some of the data is also available in the Genenetwork and the Mouse Genome Informatics databases (Table 2).
Fig. 3.
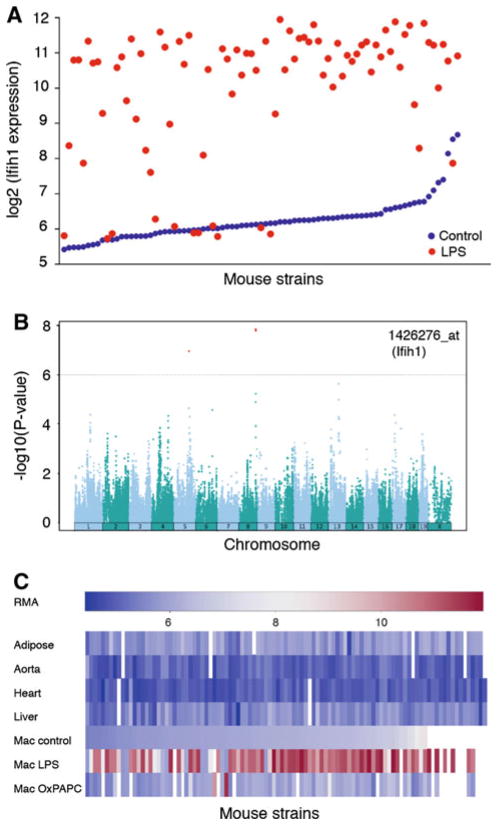
Database plots for interferon-inducible helicase 1 (Ifih1). Sample plots for a given gene of interest that can be obtained from our online database. a Lipopolysaccharide (LPS) response of Ifih1 in macrophages of the HMDP. b Genome-wide association for the expression of Ifih1 in LPS-treated macrophages. c Relative expression levels among mouse strains of the HMDP in adipose, aorta, heart, and liver, and for macrophages treated with control, LPS or oxidation products of 1-palmitoyl-2-arachidonyl-sn-glycero-3-phosphocholine (OxPAPC) media. Robust microarray average (RMA) refers to an algorithm for gene expression microarray background corrections. We used the Affymetrix GCOS RMA algorithm
The SGR is a resource that can be used to answer specific questions and to understand the relationships among different genes. For example, data generated from primary macrophages of the HMDP helped us to determine that a gene of interest, the interferon inducible helicase 1 (Ifih1), shows gene-by-environment interactions and that it is under the control of the inflammatory stimulus bacterial lipopolysaccharide (LPS) (Fig. 3a). Using eQTL, we also found that the expression of Ifih1 is controlled by three loci on Chrs 5, 8, and 13 (Fig. 3b). We can also compare the expression patterns of Ifih1 among the different tissues available on the database, which include adipose, aorta, heart, liver, and macrophages treated in three different conditions (Fig. 3c). Similarly, we found that the three trans-eQTL on Chrs 5, 8, and 13 are specific to macrophages treated with LPS, but there is also a strong cis-eQTL in the liver and other regulatory loci in Chr 2. Such information can allow us to identify candidate regulators, to examine gene-by-environment interactions and tissue specificities, and to prioritize candidate genes for clinical QTL.
Future directions and conclusions
While the resolution of the HMDP is excellent at most loci, the power is marginal. As judged by QTL studies, few loci contributing to complex clinical traits have effect sizes as large as 10 % and most are below 5 % (Flint and Mott 2008). Thus, using the panel of 100 strains (Bennett et al. 2010), only a subset of the loci contributing to complex traits are likely to be identified. As mentioned above, the power can be enhanced by integrating the results from traditional crosses or by expanding the number of inbred and recombinant inbred strains. Recently, two panels of advanced intercross RI lines have become available from The Jackson Laboratory: The LXS RI panel includes 62 strains and 50 more BXD strains have recently been developed. Also available from The Jackson Laboratory are several sets of cryopreserved strains: 19 strains from the AKXD RI panel, 13 from the AKXL panel, and 15 from the NXSM panel. Thus, it is possible to increase the size of the HMDP to over 260 strains. Recombinant Inbred Congenic Lines, Chromosome Substitution Strains, and Genome Tagged Mice (Peters et al. 2007) could also be employed to increase both power and resolution. A particularly useful complement to the HMDP will be the CC strain set now being generated (Casci 2012; Threadgill and Churchill 2012). Preliminary analyses of the partially inbred CC lines have been promising and substantial resources for their characterization, including complete genomic sequencing, have been planned (Collaborative Cross Consortium 2012).
As discussed above, human genetic studies of complex traits are limited in several respects, and the HMDP provides a partial solution to some of these limitations. First, human studies are poorly powered to identify genetic interactions, and these can be addressed more effectively in mice. As mentioned above, the HMDP is very convenient for genetic analysis of environmental interactions because mice of the same genotype can be examined under different conditions. Also, epistasis is likely to complicate studies of diseases such as diabetic complications and atherosclerosis, where one set of genes contributes to a predisposing factor (diabetes and elevated cholesterol, respectively), and another set of genes affects the response to these factors. In the HMDP, sensitizing genes can be introduced by breeding dominant mutations onto each of the HMDP strains and examining the F1 progeny. For example, we have bred a dominant hyperlipidemia-inducing gene, APOE-Leiden, onto a number of the HMDP strains and find atherosclerosis to be concordant with previous studies in which recessive mutations were transferred onto different backgrounds (B. Bennett and A. J. Lusis unpublished). Second, it will be difficult to identify, directly in humans, the pathways perturbed by novel GWAS genes. For example, the striking relationship between an allele of APOE and Alzheimer’s has been known for nearly 20 years and yet the mechanism remains uncertain. Clearly, studies in mice, where access to tissues and environmental conditions can be standardized, will simplify such analyses. Moreover, studying the genes in the context of natural variation, as opposed to transgenic or gene-targeted mice, may well offer important advantages. Third, for most common disease traits, human GWAS have been able to identify only a small fraction of the total heritability. There are undoubtedly many explanations but, clearly, much remains to be discovered. Studies in mice will most likely identify different, although overlapping, gene sets and perhaps different pathways. Traits that are substantially influenced by environmental factors will be addressed with greater power in mice because such factors can be controlled.
The HMDP panel should be useful as a tool for investigation of basic biological processes as well as complex clinical traits. An example is the study by (Ghazalpour et al. 2011) that examines the relationship between transcript levels, protein levels, and metabolic traits. By providing many thousands of genetic perturbations in various combinations, the HMDP enables global dissection of the relationships between biological scales such as DNA methylation, transcription factor binding, histone modification, and transcription.
Acknowledgments
Funding for this work was supported in part by the U.S. National Institutes of Health (NIH) grants NIAMS RO1 AR057759 (CRF) and NHLBI K99 HL102223 (BJB); the Ruth L. Kirschstein National Research Service Awards T32 HL69766 (CDR), T32-HL007895 (JW), NINDS R01 NS050148 and NIGMS R01 GM098273 (DJS), NHLBI RO1 HL098227 (RCL), NIDDK DP3 DK094311 (AJL, RCD, RCL), NIH RO1 GM095656 (MP), NIDDK RO1 DK072206 (TAD), RO1 DK080339 (STH), 5RO1 DC010856-02 (JDO, RAF), PO1 HL028481 (KR), and PO1 HL090553 (KR). EE was supported by National Science Foundation grants 0513612, 0731455, 0729049, 0916676, and 1065276, and NIH grants K25-HL080079, U01 DA024417, P01-HL30568, and PO1-HL28481. LDO was supported by USPHS National Research Service Award GM07104 and USPHS National Research Service Award T32-HG002536. This research was supported in part by the University of California, Los Angeles, CA, subcontract of contract N01-ES-45530 from the National Toxicology Program and National Institute of Environmental Health Sciences to Perlegen Sciences (EE) and from the Russian Program #2011-1.5-501-006-024 (VAK) and NIH RO1 HL105623 (VAK).
Contributor Information
Anatole Ghazalpour, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Christoph D. Rau, Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, CA, USA
Charles R. Farber, Departments of Medicine and Biochemistry and Molecular Genetics, and Center for Public Health Genomics, University of Virginia, Charlottesville, VA, USA
Brian J. Bennett, Department of Genetics, and Nutrition Research Institute, University of North Carolina, Chapel Hill, NC, USA
Luz D. Orozco, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Atila van Nas, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Calvin Pan, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Hooman Allayee, Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Simon W. Beaven, Division of Digestive Diseases, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Mete Civelek, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Richard C. Davis, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Thomas A. Drake, Department of Pathology and Laboratory Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Rick A. Friedman, Department of Otology/Skull Base Surgery, House Research Institute, Los Angeles, CA, USA
Nick Furlotte, Department of Computer Sciences, University of California, Los Angeles, CA, USA.
Simon T. Hui, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
J. David Jentsch, Department of Psychology & Behavioral Neuroscience and Psychiatry & Biobehavioral Sciences, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Emrah Kostem, Department of Computer Sciences, University of California, Los Angeles, CA, USA.
Hyun Min Kang, Department of Biostatistics, School of Public Health, University of Michigan, Ann Arbor, MI, USA.
Eun Yong Kang, Department of Computer Sciences, University of California, Los Angeles, CA, USA.
Jong Wha Joo, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA. Bioinformatics Program, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Vyacheslav A. Korshunov, Department of Medicine, Aab Cardiovascular Research Institute, University of Rochester School of Medicine and Dentistry, Rochester, NY, USA
Rick E. Laughlin, Semel Institute for Neuroscience and Human Behavior, University of California, Los Angeles, CA, USA
Lisa J. Martin, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Jeffrey D. Ohmen, Department of Cell Biology and Genetics, House Research Institute, Los Angeles, CA, USA
Brian W. Parks, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Matteo Pellegrini, Department of Molecular, Cell and Developmental Biology, University of California, Los Angeles, CA, USA.
Karen Reue, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Desmond J. Smith, Department of Molecular and Medical Pharmacology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA
Sotirios Tetradis, Molecular Biology Institute, David Geffen School of Medicine, University of California, Los Angeles, CA, USA. Division of Diagnostic and Surgical Science, School of Dentistry, University of California, Los Angeles, CA, USA.
Jessica Wang, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA. Department of Medicine/Cardiology, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
Yibin Wang, Division of Molecular Medicine, Department of Anesthesiology, Physiology and Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA.
James N. Weiss, Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, CA, USA
Todd Kirchgessner, Department of Cardiovascular Drug Discovery, Bristol-Myers Squibb Co, Pennington, NJ, USA.
Peter S. Gargalovic, Department of Cardiovascular Drug Discovery, Bristol-Myers Squibb Co, Pennington, NJ, USA
Eleazar Eskin, Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA. Department of Computer Sciences, University of California, Los Angeles, CA, USA.
Aldons J. Lusis, Department of Medicine, David Geffen School of Medicine, University of California, Los Angeles, CA, USA. Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, CA, USA. Department of Human Genetics, David Geffen School of Medicine, University of California, Los Angeles, CA, USA. Division of Diagnostic and Surgical Science, School of Dentistry, University of California, Los Angeles, CA, USA
Renée C. LeBoeuf, Email: leboeuf@u.washington.edu, Division of Metabolism, Endocrinology and Nutrition, Department of Medicine, University of Washington, 850 Republican Street, Seattle, WA 98109-4725, USA
References
- Ahlqvist E, Ekman D, Lindvall T, Popovic M, Forster M, Hultqvist M, Klaczkowska D, Teneva I, Johannesson M, Flint J, Valdar W, Nandakumar KS, Holmdahl R. High-resolution mapping of a complex disease, a model for rheumatoid arthritis, using heterogeneous stock mice. Hum Mol Genet. 2011;20:3031–3041. doi: 10.1093/hmg/ddr206. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang WP, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrage LC, Baskin-Hill AE, Sinasac DS, Singer JB, Croniger CM, Kirby A, Kulbokas EJ, Daly MJ, Lander ES, Broman KW, Nadeau JH. Genetic resistance to diet-induced obesity in chromosome substitution strains of mice. Mamm Genome. 2010;21:115–129. doi: 10.1007/s00335-010-9247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casci T. Mouse genetics: fruits of the Collaborative Cross. Nat Rev Genet. 2012;13:223. [Google Scholar]
- Cervino AC, Darvasi A, Fallahi M, Mader CC, Tsinoremas NF. An integrated in silico gene mapping strategy in inbred mice. Genetics. 2007;175:321–333. doi: 10.1534/genetics.106.065359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, Bleich A, Bogue M, Broman KW, Buck KJ, Buckler E, Burmeister M, Chesler EJ, Cheverud JM, Clapcote S, Cook MN, Cox RD, Crabbe JC, Crusio WE, Darvasi A, Deschepper CF, Doerge RW, Farber CR, Forejt J, Gaile D, Garlow SJ, Geiger H, Gershenfeld H, Gordon T, Gu J, Gu W, de Haan G, Hayes NL, Heller C, Himmelbauer H, Hitzemann R, Hunter K, Hsu HC, Iraqi FA, Ivandic B, Jacob HJ, Jansen RC, Jepsen KJ, Johnson DK, Johnson TE, Kempermann G, Kendziorski C, Kotb M, Kooy RF, Llamas B, Lammert F, Lassalle JM, Lowenstein PR, Lu L, Lusis A, Manly KF, Marcucio R, Matthews D, Medrano JF, Miller DR, Mittleman G, Mock BA, Mogil JS, Montagutelli X, Morahan G, Morris DG, Mott R, Nadeau JH, Nagase H, Nowakowski RS, O’Hara BF, Osadchuk AV, Page GP, Paigen B, Paigen K, Palmer AA, Pan HJ, Peltonen-Palotie L, Peirce J, Pomp D, Pravenec M, Prows DR, Qi Z, Reeves RH, Roder J, Rosen GD, Schadt EE, Schalkwyk LC, Seltzer Z, Shimomura K, Shou S, Sillanpaa MJ, Siracusa LD, Snoeck HW, Spearow JL, Svenson K, Tarantino LM, Threadgill D, Toth LA, Valdar W, de Villena FP, Warden C, Whatley S, Williams RW, Wiltshire T, Yi N, Zhang D, Zhang M, Zou F. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Collaborative Cross Consortium. The genome architecture of the collaborative cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings SR, Bates D, Black DM. Clinical use of bone densitometry: scientific review. JAMA. 2002;288:1889–1897. doi: 10.1001/jama.288.15.1889. [DOI] [PubMed] [Google Scholar]
- Farber CR, Rosen C. Genetics of osteoporosis. In: Robertson RP, editor. Translational endocrinology & metabolism. The Endocrine Society; Chevy Chase: 2010. pp. 87–116. [Google Scholar]
- Farber CR, Bennett BJ, Orozco L, Zou W, Lira A, Kostem E, Kang HM, Furlotte N, Berberyan A, Ghazalpour A, Suwanwela J, Drake TA, Eskin E, Wang QT, Teitelbaum SL, Lusis AJ. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclasto-genesis. PLoS Genet. 2011;7:e1002038. doi: 10.1371/journal.pgen.1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Mackay TF. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–733. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Mott R. Applying mouse complex-trait resources to behavioural genetics. Nature. 2008;456:724–727. doi: 10.1038/nature07630. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, Pethiyagoda CL, Stuve LL, Johnson FM, Daly MJ, Wade CM, Cox DR. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Furlotte NA, Kang EY, Van Nas A, Farber CR, Lusis AJ, Eskin E. Increasing association mapping power and resolution in mouse genetic studies through the use of meta-analysis for structured populations. Genetics. 2012;191(3):959–967. doi: 10.1534/genetics.112.140277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, Park CC, Wen PZ, Brewer H, Weitz K, Camp DG, 2nd, Pan C, Yordanova R, Neuhaus I, Tilford C, Siemers N, Gargalovic P, Eskin E, Kirchgessner T, Smith DJ, Smith RD, Lusis AJ. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet. 2011;7:e1001393. doi: 10.1371/journal.pgen.1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorova S, Divina P, Storchova R, Trachtulec Z, Fotopulosova V, Svenson KL, Donahue LR, Paigen B, Forejt J. Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 2008;18:509–515. doi: 10.1101/gr.7160508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, Klein RF, Ahluwalia MK, Higuchi R, Peltz G. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. doi: 10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- Guo Y, Lu P, Farrell E, Zhang X, Weller P, Monshouwer M, Wang J, Liao G, Zhang Z, Hu S, Allard J, Shafer S, Usuka J, Peltz G. In silico and in vitro pharmacogenetic analysis in mice. Proc Natl Acad Sci USA. 2007;104:17735–17740. doi: 10.1073/pnas.0700724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover-Plow J, Shchurin A, Hart E, Sha J, Hill AE, Singer JB, Nadeau JH. Genetic background determines response to hemostasis and thrombosis. BMC Blood Disord. 2006;6:6. doi: 10.1186/1471-2326-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A, Kang HM, Wade CM, Cotsapas C, Kostem E, Han B, Furlotte N, Kang EY, Rivas M, Bogue MA, Frazer KA, Johnson FM, Beilharz EJ, Cox DR, Eskin E, Daly MJ. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, Shafer S, Puech A, McPherson JD, Foernzler D, Peltz G, Usuka J. In silico genetics: identification of a functional element regulating H2-Ealpha gene expression. Science. 2004;306:690–695. doi: 10.1126/science.1100636. [DOI] [PubMed] [Google Scholar]
- Liu P, Vikis H, Lu Y, Wang D, You M. Large-scale in silico mapping of complex quantitative traits in inbred mice. PLoS One. 2007;2:e651. doi: 10.1371/journal.pone.0000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CC, Gale GD, de Jong S, Ghazalpour A, Bennett BJ, Farber CR, Langfelder P, Lin A, Khan AH, Eskin E, Horvath S, Lusis AJ, Ophoff RA, Smith DJ. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Syst Biol. 2011;5:43. doi: 10.1186/1752-0509-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, Mural RJ, Paigen B, Wiltshire T. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prows DR, Hafertepen AP, Winterberg AV, Gibbons WJ, Jr, Wesselkamper SC, Singer JB, Hill AE, Nadeau JH, Leikauf GD. Reciprocal congenic lines of mice capture the aliq1 effect on acute lung injury survival time. Am J Respir Cell Mol Biol. 2008;38:68–77. doi: 10.1165/rcmb.2006-0162OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer JB, Hill AE, Burrage LC, Olszens KR, Song J, Justice M, O’Brien WE, Conti DV, Witte JS, Lander ES, Nadeau JH. Genetic dissection of complex traits with chromosome substitution strains of mice. Science. 2004;304:445–448. doi: 10.1126/science.1093139. [DOI] [PubMed] [Google Scholar]
- Smolock EM, Ilyushkina IA, Ghazalpour A, Gerloff J, Murashev AN, Lusis AJ, Korshunov VA. A genetic locus on mouse chromosome 7 controls elevated heart rate. Physiol Genomics. 2012;44(13):689–698. doi: 10.1152/physiolgenomics.00041.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, Bagger Y, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- Takada T, Mita A, Maeno A, Sakai T, Shitara H, Kikkawa Y, Moriwaki K, Yonekawa H, Shiroishi T. Mouse inter-subspecific consomic strains for genetic dissection of quantitative complex traits. Genome Res. 2008;18:500–508. doi: 10.1101/gr.7175308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. Ten years of the collaborative cross. G3 (Bethesda) 2012;2:153–156. doi: 10.1534/g3.111.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B, Nicod J, Bhomra A, Davidson S, Cleak J, Farinelli L, Osteras M, Whitley A, Yuan W, Gan X, Goodson M, Klenerman P, Satpathy A, Mathis D, Benoist C, Adams DJ, Mott R, Flint J. Commercially available outbred mice for genome-wide association studies. PLoS Genet. 2010;6(9):e1001085. doi: 10.1371/journal.pgen.1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc Natl Acad Sci USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]