Abstract
Background
Supravalvular aortic stenosis (SVAS) is caused by mutations in the elastin (ELN) gene and is characterized by abnormal proliferation of vascular smooth muscle cells (SMCs) that can lead to narrowing or blockage of the ascending aorta and other arterial vessels. Availability of patient-specific SMCs may facilitate studying disease mechanisms and developing novel therapeutic interventions.
Methods and Results
Here, we report the development of a human induced pluripotent stem cell (iPSC) line from a patient with SVAS caused by the premature termination in exon 10 of the ELN gene due to an exon 9 4-nucleotide insertion. We showed that SVAS iPSC-derived SMCs (iPSC-SMCs) had significantly fewer organized networks of smooth muscle alpha actin (SM α-actin) filament bundles, a hallmark of mature contractile SMCs, compared to control iPSC-SMCs. Addition of elastin recombinant protein or enhancement of small GTPase RhoA signaling was able to rescue the formation of SM α-actin filament bundles in SVAS iPSC-SMCs. Cell counts and BrdU analysis revealed a significantly higher proliferation rate in SVAS iPSC-SMCs than control iPSC-SMCs. Furthermore, SVAS iPSC-SMCs migrated at a markedly higher rate to the chemotactic agent platelet-derived growth factor (PDGF) in comparison with the control iPSC-SMCs. We also provided evidence that elevated activity of extracellular signal-regulated kinase 1/2 (ERK1/2) is required for hyper-proliferation of SVAS iPSC-SMCs. The phenotype was confirmed in iPSC-SMCs generated from a patient with deletion of elastin due to Williams-Beuren syndrome (WBS).
Conclusions
Thus, SVAS iPSC-SMCs recapitulate key pathological features of patients with SVAS and may provide a promising strategy to study disease mechanisms and to develop novel therapies.
Keywords: elastin, induced pluripotent stem cells, smooth muscle alpha actin filament bundle, smooth muscle cells, supravalvular aortic stenosis
Introduction
Supravalvular aortic stenosis (SVAS) is an autosomal dominant disease characterized by abnormal proliferation of vascular smooth muscle cells (SMCs) that can lead to narrowing or blockage of the ascending aorta and other arterial vessels and a propensity toward sudden cardiac death 1. The most common underlying causes for SVAS are heterozygous, loss-of-function mutations in the elastin (ELN) gene that produce haploinsufficiency 2, 3. Patients with Williams-Beuren syndrome (WBS) also display SVAS; these patients are hemizygous for 26–28 contiguous genes, including the ELN gene, due to a 1.5 – 1.8 Mb microdeletion on chromosome 7q11.23. While WBS patients display a more complex phenotype including craniofacial and other neurobehavioral defects, the spectrum and pathological characteristics of cardiovascular lesions in patients with SVAS and WBS are virtually identical and have been denoted as ELN arteriopathy1, 4.
The encoded product of the ELN gene is the monomeric precursor protein, tropoelastin, which is secreted, crosslinked and organized into an ELN polymer by vascular SMCs 5. ELN polymers are the main extracellular matrix components deposited in the arterial wall where they endow elastic resilience. Aside from its essential role in providing biomechanical support for blood vessels, ELN plays a critical role in inducing a quiescent contractile state in vascular SMCs by inhibiting cellular proliferation and promoting the organization of actin filament bundles, the scaffold for the contractile apparatus in SMCs 3, 6.
The use of genetic animal models and primary vascular SMCs to study the mechanisms underlying SVAS has been very informative 3, 6. However, the study of the disease has been significantly hampered by functional differences in SMCs between species 7, limited accessibility to patient vascular SMCs, rapid loss of SMC properties in primary cell culture 8 and an inability to model patient-specific disease variations. Thus, it would be very useful to establish a human cell-based model to obtain an abundant and renewable source of functional SMCs for studying the pathogenesis of this disease and for developing patient-specific therapeutic interventions.
The generation of induced pluripotent stem cells (iPSCs) from human adult somatic cells has opened an exciting avenue for disease modeling and regenerative medicine 9–11. Recently, several human cardiovascular disease models have been generated from patients with the Long-QT 12, 13, LEOPARD 14 and Hutchinson-Gilford Progeria syndromes 15, 16. In these studies, cardiovascular cells derived from patient-specific iPSCs have recapitulated the pathological features of each disorder and have provided unique human models to study disease mechanisms.
In this study, we report the generation of an iPSC model of SVAS using two different ELN mutations. We have found that SVAS iPSC-derived SMCs (iPSC-SMCs) exhibit a lower degree of organized smooth muscle alpha actin (SM α-actin) filament bundles, proliferate at a higher rate and migrate significantly faster in response to the chemotactic cytokine platelet-derived growth factor (PDGF) than control iPSC-SMCs, recapitulating key pathologic features of the human disease. Our results further show that recombinant ELN or enhancement of small GTPase RhoA signaling rescues SM α-actin filament bundle formation and that attenuation of extracellular signal-regulated kinase 1/2 (ERK1/2) activity inhibits hyper-proliferation of SVAS iPSC-SMCs, providing a promising paradigm to study disease mechanisms and to develop novel personalized therapies.
Methods
Establishment of patient-derived iPSCs
Human iPSC clones were established from vascular SMCs derived by explant outgrowth from excised epicardial coronary arteries of a patient with SVAS and from foreskin fibroblasts from a patient with WBS collected under an Institutional Review Board-approved protocol by transduction with the hSTEMCCA polycistronic lentiviral vector (encoding OCT4, KLF4, SOX2, and C-MYC), as previously published 17. The human iPSC clones were maintained and propagated on mitotically arrested mouse embryonic fibroblast (MEF) feeder layers. Detailed information of human iPSC generation can be found in Supplemental Methods.
Genomic sequencing
See Supplemental Methods for details.
Detection of mutant ELN mRNA by inhibiting nonsense-mediated decay (NMD)
Primary SMCs from a control donor or a SVAS patient were plated and treated with 100μg/ml of cycloheximide (Sigma) or DMSO (control) for 4 hours, as previously described 3, 18. RNA was extracted with Trizol (Life Technologies) following manufacturer’s instructions. Mutant ELN mRNA was detected by the RT-PCR assay using an allele-specific forward primer that contained a 4-nucleotide GTAT insertion at its 3′ end and an ELN downstream reverse primer. This results in the detection of a PCR product only in the SVAS cells but not in the healthy control cells. GAPDH was used as an internal control. The fragment amplified by PCR was gel extracted (Qiagen) and sequenced to confirm detection of the ELN gene mutation.
Immunofluorescence and alkaline phosphatase staining
See Supplemental Methods for details.
Chromosome integrity (karyotype) and fluorescence in situ hybridization (FISH) analyses
See Supplemental Methods for details.
Bisulphite sequencing
See Supplemental Methods for details.
Gene expression analysis
See Supplemental Methods for details.
Teratoma formation assay
See Supplemental Methods for details.
SMC differentiation
SMC differentiation was induced using an embryoid body differentiation system as previously described 19. Briefly, undifferentiated human iPSCs were dispersed into small cell clumps using 1 mg/ml of collagenase IV (Invitrogen) at 37° for 5 min. These cells were then cultured in suspension for 6 days in differentiation medium (DMEM supplemented with 10% FBS from Hyclone, 1% nonessential amino acids, 0.1 mM β-mercaptoethanol and 1% L-glutamine) to form embryoid bodies. The embryoid bodies were then plated on 0.1% gelatin-coated culture dishes and cultured with fresh differentiation medium. At 6 days after plating, small clusters were dissociated and transferred to matrigel-coated plate in SmGM-2 media (Lonza). After one week of culture on matrigel-coated plates, cells were transferred to 0.1% gelatin-coated culture dishes again and cultured with the 5% FBS differentiation medium for at least 5 days to complete differentiation.
Western blot analysis
See Supplemental Methods for details.
Fluorescence-activated cell sorting (FACS) analysis
See Supplemental Methods for details.
Rescue of SM α-actin filament bundle formation by ELN treatment
These procedures were described in a previous study 6. Briefly, SVAS iPSC-SMCs, WBS iPSC-SMCs and control iPSC-SMCs were seeded at a density of 50,000 cells/well on a 12-well plate. After attachment, cells were starved in 0.1% BSA (Fisher) in Amniomax Basal Medium (Invitrogen) for 24 hours. Cells were then grown in whole Amniomax medium in the presence or absence of 50 μg/ml tropoelastin (Advanced BioMatrix) for 12–14 hrs, followed by immunofluorescent staining for SM α-actin (Sigma) and nuclear counterstaining with Hoechst dye.
Migration of SVAS and WBS iPSC-SMCs
Cell migration was assayed in a modified Boyden chamber system using 6.5 mm transwell polycarbonate chemotaxis filter inserts in a plastic 24-well tissue culture plate (Corning), as previously described 6. The inserts were covered with a polycarbonate membrane filter (8 μm porosity). PDGF-BB (100 ng/ml; R&D Systems) was added to the designated lower chambers. After serum starvation for 24 hrs, 2×104 iPSC-SMCs grown in SmGM2 media (Lonza) were placed in the upper wells of the chamber and incubated at 37°C/5% CO2 overnight in the media containing high glucose DMEM (Invitrogen), 1% BSA (Sigma) and 0.5% FBS (Sigma). The cells adhering to the upper surface of the filter were scraped off with Q-tips, and the cells that had migrated to the lower surface were fixed with 4% paraformaldehyde, stained with Hoechst 33258 (Sigma) and viewed under a fluorescent microscope (Leica). Five to seven randomly selected fields were counted on each filter. The chemotactic response was calculated as the number of cells that had traversed the filter in response to PDGF and expressed as fold increase over baseline. Each experiment was repeated at least three times.
Blockage of ERK signaling pathway and analysis of cell proliferation
See Supplemental Methods for details.
Calcium imaging
See Supplemental Methods for details.
Statistical analysis
Results were reported as mean ± standard error of the mean (SEM). Wilcoxon rank-sum test (Mann-Whitney U test) was used to compare two groups. Kruskal-Wallis test was used for one-factor design with three or more groups, and Scheirer-Ray-Hare test was used for the two-way design experiments. Both tests were followed by two-group pairwise comparison using Wilcoxon rank-sum test. A probability value <0.05 was considered statistically significant.
Results
Generate and characterize SVAS-iPSCs
Coronary vascular SMCs were isolated from a 39-year-old Caucasian male with SVAS who had undergone heart transplantation. The patient is heterozygous for a 4-base pair nucleotide (GTAT) insertion in exon 9 of ELN that was predicted to result in a frameshift and a premature termination codon in exon 10 (Figure 1A). Premature termination mutations frequently result in the specific degradation of mutant mRNA through nonsense-mediated decay (NMD) that can be blocked by protein synthesis inhibitor cycloheximide (CHX) 3, 18. The expression of the mutant mRNA was restored by CHX treatment (Supplemental Figure 1A), indicating that the mutant mRNA undergoes NMD. The cultured vascular SMCs were reprogrammed with a polycistronic lentiviral vector [hSTEMCCA; 17] expressing OCT4, KLF4, SOX2 and c-MYC to generate SVAS patient-specific iPSC clones (Figure 1B). Control human iPSC clones were also created from vascular SMCs from a 12-year-old healthy Caucasian male using the same reprogramming system (Supplemental Figure 1B). Multiple iPSC clones were generated, and two SVAS clones and two control clones were continuously propagated and used for SMC differentiation and characterization.
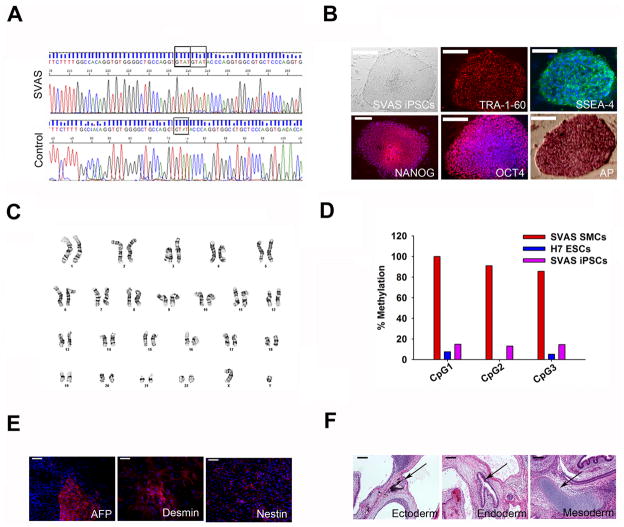
Figure 1.
Establishment and characterization of SVAS iPSCs (clone 1). (A) Sequencing of the ELN gene identifying a heterozygous 4-nucleotide insertion mutation in SMCs from the patient with SVAS. (B) Reprogramming of the patient’s SMCs into SVAS iPSCs. Shown from left to right are a typical human iPSC colony, positive immunostaining for pluripotency markers (TRA-1-60, SSEA-4, NANOG, and OCT4), and positive staining for alkaline phosphatase (AP). Scale bars, 100 μm. (C) Karyotype analysis of SVAS iPSCs. (D) Bisulphite sequencing analysis of the NANOG promoter in primary SMCs, SVAS iPSCs and H7 human embryonic stem cells (ESCs). (E) Immunostaining of differentiated embryoid bodies for nestin (ectoderm), α-fetoprotein (AFP, endoderm) and desmin (mesoderm). Scale bars, 50 μm. (F) Teratoma formation following injection of undifferentiated SVAS iPSCs in NOD/SCID mice. Note the formation of pigmented epithelium (ectoderm, left panel), gastrointestinal epithelium (endoderm, center panel) and hyaline cartilage (mesoderm, right panel), as identified by the arrows. Scale bars, 200 μm.
The established iPSC clones showed typical, compact human embryonic stem cell (ESC) morphology (Figure 1B and Supplemental Figure 1B), expressed the pluripotency markers TRA-1-60, SSEA-4, NANOG and OCT4 (Figure 1B and Supplemental Figure 1B), exhibited alkaline phosphatase activity (Figure 1B and Supplemental Figure 1B), and maintained a normal karyotype of 46 XY (Figure 1C and Supplemental Figure 1C). Silencing of the four lentiviral transgenes (Supplemental Figure 2A) and reactivation of endogenous pluripotency genes (OCT4, SOX2, NANOG, FOXD3 and REX1) (Supplemental Figure 2B) in these iPSC clones were confirmed by qRT-PCR. Bisulphite sequencing revealed marked demethylation of the NANOG promoter in each iPSC clone compared to their parental somatic cells (Figure 1D and Supplemental Figure 1D). Pluripotency of human iPSC clones was confirmed by spontaneous differentiation into the three embryonic germ layers in vitro using embryoid body (EB) formation (Figure 1E and Supplemental Figure 3A) and by in vivo teratoma formation after injection of undifferentiated iPSCs into immunocompromised NOD/SCID mice (Figure 1F and Supplemental Figure 3B).
To model SVAS syndrome in a second patient with ELN deficiency, and to confirm that the observed disease phenotypes were the result of an ELN defect, we generated and characterized a second iPSC line using the foreskin fibroblasts from a 1-year-old Caucasian male with Williams-Beuren syndrome (WBS) who displays SVAS and has a heterozygous, 1.4-Mb microdeletion at chromosome 7q11.23, including the ELN gene20. The control human iPSC line was created using the foreskin fibroblasts from a 1-year-old healthy Caucasian male. Multiple iPSC clones were generated, and two clones were expanded and analyzed showing no morphological or phenotypical differences between them; we focused primarily on WBS iPSC clone2 and control iPSC clone2 for our studies (hereafter referred to as WBS iPSCs or control iPSCs). Fluorescence in situ hybridization (FISH) analysis confirmed the deletion of one copy of the ELN gene (Supplemental Figure 4A) in WBS iPSCs. Control and WBS iPSC lines demonstrated pluripotency gene expression (Supplemental Figure 4B), demethylation of the NANOG promoter (Supplemental Figure 4D), silencing of transgenes (Supplemental Figure 5A) and reactivation of endogenous pluripotency genes (Supplemental Figure 5B). The pluripotency of each iPSC line was further confirmed by differentiation into the three embryonic germ layers in vitro and by in vivo teratoma formation (Supplemental Figure 4E–F).
SVAS human iPSC-SMCs have defective actin filament bundle formation
We used an embryoid-body differentiation system to induce human iPSC differentiation toward a SMC lineage 19. This resulted in the production of highly homogenous SMC-like cells identified by the SMC marker calponin 21 in both control (96.6±0.8%) and SVAS iPSC cultures (96.1±1.5%) upon differentiation (Figure 2A–B and Supplemental Figure 6A–B). This result was confirmed by flow cytometry analysis using a calponin antibody (Supplemental Figure 6C). Loss of SMC contractile phenotype, such as actin filament bundles, has been implicated as a causative factor in the occlusive vascular pathology observed in patients with SVAS and elastin-deficient mice 6, 22. We evaluated actin filament bundle formation in control and SVAS iPSC-SMCs by immunofluorescence analysis of smooth muscle alpha actin (SM α-actin). Well-defined actin filament bundles were evident in 91.8±1.1% of control iPSC-SMCs (Figure 2A–B and Supplemental Figure 6A–B). However, only 17.4±2.3% of SVAS iPSC-SMCs exhibited detectable actin filament bundle formation (Figure 2A–B and Supplemental Figure 6A–B). Western blot analysis revealed that SM α-actin protein levels were comparable in control and SVAS iPSC-SMCs (Figure 2C and Supplemental Figure 6D), suggesting that defective actin filament bundle formation in SVAS iPSC-SMCs is not caused by reduced SM α-actin protein expression but is due to a defect in contractile apparatus organization. Additionally, flow cytometry analysis revealed a reduction in the percentage of cells expressing SM α-actin in SVAS iPSC-SMCs (Supplemental Figure 6C). Notably, human embryonic stem cell (ESC)-derived SMCs also exhibited well-defined actin filament bundles (Supplemental Figure 6E), suggesting that the tissue origin of iPSC (vascular SMC in this study) and ESC (blastocyst) plays a minimal role in contributing to the contractile phenotype of SMCs derived from pluripotent stem cell lines.
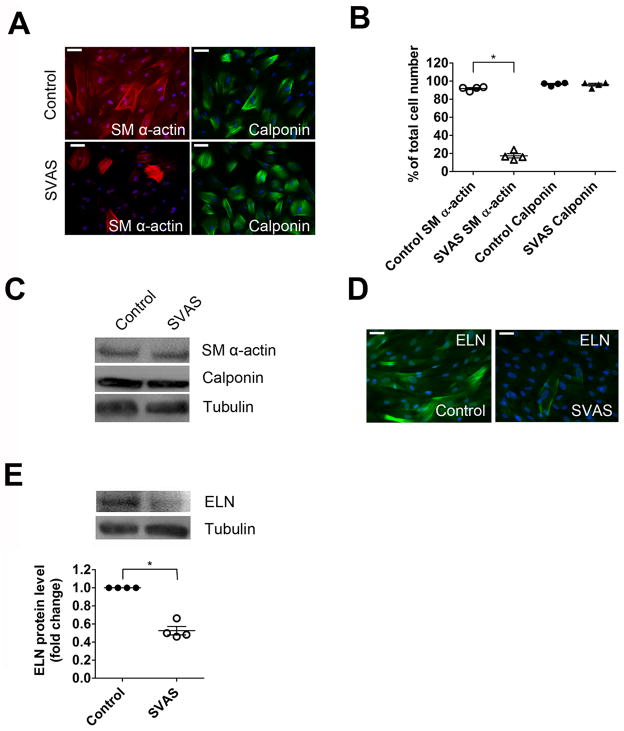
Figure 2.
Phenotypic characterization of SVAS iPSC-SMCs. (A) SVAS iPSC-SMCs and control iPSC-SMCs were immunostained with antibodies for SM α-actin and calponin. Nuclei: Hoechst 33258. Scale bar, 50 μm. (B) SM α-actin filament bundle and calponin positive cells from each group in (A) were quantified as number of cells stained positively divided by total number of cells (mean ± s.e.m., n=4; *p<0.05). (C) SVAS iPSC-SMCs and control iPSC-SMCs were lysed and immunoblotted with anti-SM α-actin, anti-calponin and anti-tubulin antibodies. (D) SVAS iPSC-SMCs and control iPSC-SMCs were immunostained with antibodies for ELN. Nuclei: Hoechst 33258. Scale bar, 50 μm. (E) SVAS iPSC-SMCs and control iPSC-SMCs were lysed and immunoblotted with anti-ELN and anti-tubulin antibodies. Mean± s.e.m. (n=4). *p<0.05.
To examine whether the defective formation of actin filament bundles in SVAS iPSC-SMCs correlates with an inability to produce sufficient quantities of ELN, we performed immunofluorescence (Figure 2D and Supplemental Figure 7A) and western blot analyses (Figure 2E and Supplemental Figure 7B) using an ELN antibody. There was a significantly lower level of ELN protein in SVAS iPSC-SMCs than that in control iPSC-SMCs, consistent with haploinsufficiency resulting from the ELN mutation in our patient with SVAS. Similar to the previous studies using cultured primary SMCs 3, 6, our iPSC-SMCs showed a pattern of both intracellular and extracellular ELN staining (Figure 2D and Supplemental Figure 7A), possibly reflecting the pattern of ELN expression during in vitro tissue culture. Our findings provide important evidence for decreased ELN expression in the SVAS cells.
Elastin and small GTPase RhoA rescue defective actin filament bundle formation in SVAS iPSC-SMCs
To investigate whether exogenous ELN is able to rescue the failed formation of actin filament bundles in SVAS iPSC-SMCs, we treated these cells with recombinant tropoelastin protein (ELN monomeric form) for 12 hours, and observed that the percentage of SVAS iPSC-SMCs with organized actin filament bundles increased 2.3-fold to 63.6±4.8% (Figure 3A–B and Supplemental Figure 8A–B). In contrast, tropoelastin treatment had no effect on the actin filament bundle formation in control iPSC-SMCs (Supplemental Figure 8A–B).
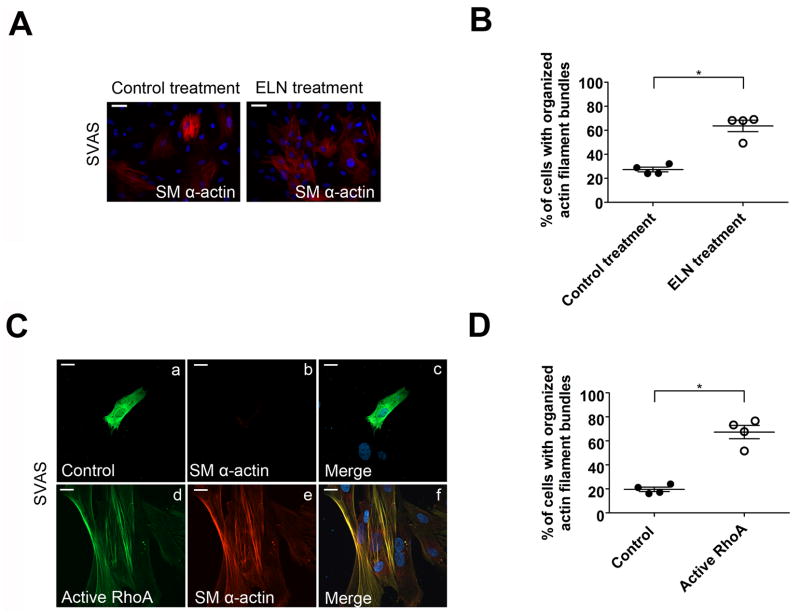
Figure 3.
Elastin and small GTPase RhoA rescue defective actin filament bundle formation in SVAS iPSC-SMCs. (A) Immunostaining of SM α-actin in SVAS iPSC-SMCs with or without 50 μg/ml ELN pretreatment. Nuclei: Hoechst 33258. (B) Quantification of cells with organized SMα-actin filament bundles in (A) (mean ± s.e.m., n=4). *p<0.05. (C) Immunostaining of SVAS iPSC-SMCs transfected with GFP-tagged vector (control) or cMyc-tagged RhoA constitutively active construct (active RhoA) for SM α-actin. Rabbit anti-SM α-actin conjugated with Alexa 565 and mouse anti-cMyc conjugated with Alexa 488 were used to detect SM α-actin and active RhoA. Nuclei: Hoechst 33258. Scale bars, 15 μm. (D) Quantification of SVAS iPSC-SMCs exhibiting organized SM α-actin filament bundles. Mean ± s.e.m. (n=4, minimum 100 cells/experiment). *p<0.05.
The RhoA signaling pathway is known to regulate actin filament bundle formation 6, 23. Furthermore, activation of RhoA signaling by tropoelastin has been implicated in inducing actin filament bundle formation in murine Eln null SMCs 6. We investigated whether activation of RhoA signaling would mimic tropoelastin treatment and rescue the defective actin filament bundle formation in SVAS iPSC-SMCs. Transient transfection of SVAS iPSC-SMCs with constitutively active RhoA (G14V) resulted in a 3.4-fold increase of the cells with organized actin filament bundles (67.3±5.5%) (Figure 3C–D). In contrast, constitutively active RhoA had no effect on the actin filament bundle formation in control iPSC-SMCs (Supplemental Figure 8C–D). Furthermore, inhibition of endogenous RhoA in control iPSC-SMCs by dominant negative RhoA (T19N) resulted in a significant loss of organized actin filament bundles (Supplemental Figure 8C–D). These results suggest that RhoA activity plays a critical role in inducing actin filament bundle formation in SVAS iPSC-SMCs.
SVAS iPSC-SMCs proliferate and migrate at higher rates than control iPSC-SMCs
It has been reported previously that vascular SMCs derived from ELN deficient patients or mouse tissue have abnormally high proliferation rates compared to cells derived from healthy controls 3, 6. We investigated whether SVAS iPSC-SMCs recapitulate the pathology of vascular SMCs derived from primary tissues of patients with SVAS and whether they proliferate at a higher rate than control iPSC-SMCs. Cells were plated at the same density and growth rates were evaluated by counting cells on days 1, 4 and 7. The number of SVAS iPSC-SMCs was 2.0-fold and 2.9-fold higher than the number of control iPSC-SMCs 4 and 7 days after seeding, respectively (Figure 4A and Supplemental Figure 9A). BrdU incorporation in SVAS iPSC-SMCs was 2.3-fold higher than that in control iPSC-SMCs 7 days after seeding (Figure 4B–C and Supplemental Figure 9B–C). These results suggest that SVAS iPSC-SMCs mimic the pathology of vascular SMCs derived from primary tissues of the patient with SVAS and have abnormally high proliferation rates compared to control iPSC-SMCs.
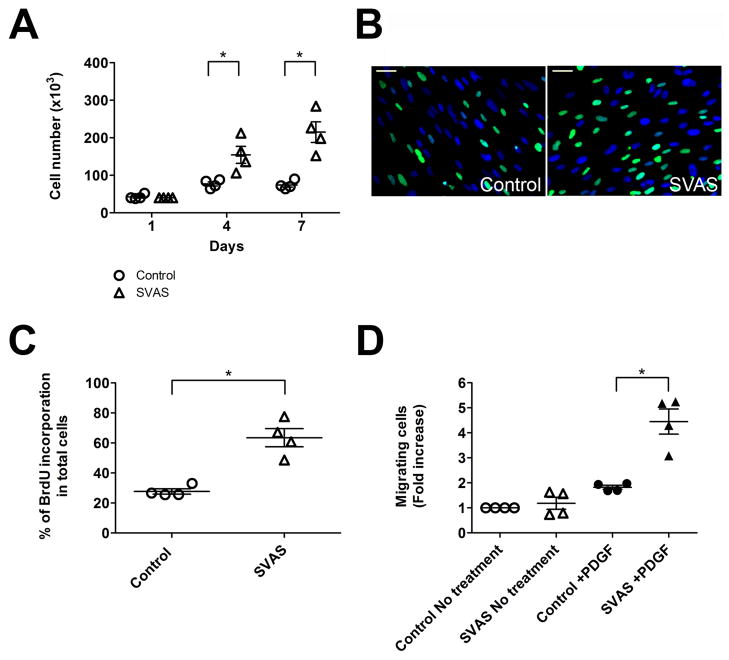
Figure 4.
SVAS iPSC-SMCs proliferate and migrate at higher rates than control iPSC-SMCs. (A) Cellular proliferation of SVAS iPSC-SMCs and control iPSC-SMCs. Mean± s.e.m. (n=4). *p<0.05. (B–C) 7-day old SVAS iPSC-SMC and control iPSC-SMC culture were labeled with BrdU, immunostained with antibodies for BrdU (B), and quantified (C). Nuclei: Hoechst 33258. Scale bar, 50 μm. Mean± s.e.m. (n=4). *p<0.05. (D) A modified Boyden chamber assay was used to determine the total number of migrated cells in five to seven randomly selected fields acquired by the fluorescence microscopy. Data shown are the mean±s.e.m. from four independent experiments. *p<0.05.
Cytokines and growth factors, such as PDGF, have been implicated in mediating the subendothelial migration of vascular SMCs in occlusive vascular lesions in Eln null mice or in patients with vascular diseases6, 24. To test if SVAS iPSC-SMCs migrate at a higher rate than control iPSC-SMCs, we used a modified Boyden chamber chemotaxis assay and observed that SVAS iPSC-SMCs migrated to PDGF at a 2.4-fold higher rate than control iPSC-SMCs did (Figure 4D and Supplemental Figure 9D).
WBS iPSC-SMCs have defective actin filament bundle formation, and proliferate and migrate at higher rates compared to control iPSC-SMCs
To determine whether the disease phenotypes definitively correspond with the ELN deficiency, we used iPSC to model SVAS in a second patient. We generated a second iPSC line from a patient with WBS who displays SVAS and has the ELN deletion (Supplemental Figure 4). Using the same approach employed to induce SVAS iPSC-SMCs, we derived WBS iPSC-SMCs and observed significantly fewer organized networks of SM α-actin filament bundles in WBS iPSC-SMCs compared to control iPSC-SMCs (Supplemental Figure 10A–B). Western blot analysis revealed a comparable expression of SM α-actin between control and WBS iPSC-SMCs (Supplemental Figure 10C). In contrast, there was a 3.6-fold lower ELN expression in WBS iPSC-SMCs compared to that of control iPSC-SMCs (Supplemental Figure 10C), consistent with ELN deficiency in our patient with WBS. Importantly, recombinant tropoelastin protein was able to significantly rescue the defective actin filament bundle formation in WBS iPSC-SMCs (Supplemental Figure 10D–E). In addition, cell counts and BrdU analysis revealed a significantly higher proliferation rate in WBS iPSC-SMCs than in control iPSC-SMCs (Supplemental Figure 11A–C). Furthermore, the Boyden chamber chemotaxis assay showed that WBS iPSC-SMCs migrated at a markedly higher rate to PDGF than that of control iPSC-SMCs (Supplemental Figure 11D). In summary, both WBS and SVAS iPSC-SMCs recapitulate key cardiovascular lesions in WBS and SVAS patients, suggesting that the ELN deficiency tracks with the disease phenotypes.
Elevated ERK1/2 activity is implicated in the hyper-proliferation of SVAS iPSC-SMC
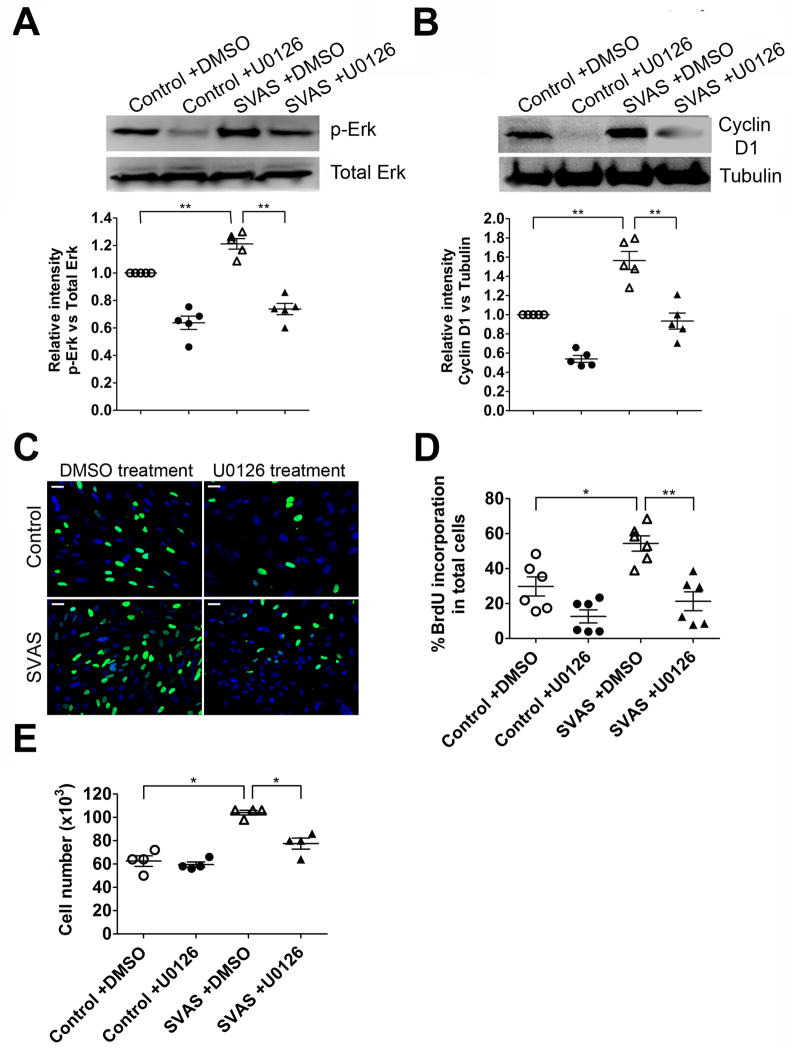
ERK1/2 signaling has been reported to play an important role in the regulation of vascular SMC proliferation25, 26, but has not been investigated in vascular SMCs from SVAS. We investigated whether elevated ERK1/2 activity correlates with hyper-proliferation of SVAS iPSC-SMCs. Analyses of ERK1/2 activity revealed significantly higher ERK phosphorylation in SVAS iPSC-SMCs than in control iPSC-SMCs (Figure 5A). Since cyclin D1 is a downstream mediator of ERK signaling during cell cycle control27, 28, we measured cyclin D1 expression and observed a significantly higher level of cyclin D1 in SVAS iPSC-SMCs than in control iPSC-SMCs (Figure 5B).
Figure 5.
Elevated activity of ERK1/2 is implicated in the hyper-proliferation of SVAS iPSC-SMCs. (A–B) SVAS iPSC-SMCs and control iPSC-SMCs were treated with vehicle control (DMSO) or a specific MEK1/2 inhibitor U0126 (10 μM) for 72 hrs. Cells were then lysed, immunoblotted with anti-total Erk1/2 (Total Erk ), anti-phosphorylated Erk1/2 (p-Erk) (A) or anti-cyclin D1 and anti-tubulin antibodies (B), and quantified. Mean ± s.e.m. (n=5). **p<0.01. (C) SVAS iPSC-SMCs and control iPSC-SMCs were cultured in the presence of DMSO or U0126 (10 μM) for 72 hrs, labeled with BrdU, immunostained with antibody for BrdU (C), and quantified (D). Nuclei: Hoechst 33258. Scale bar, 50 μm. Mean± s.e.m. (n=6). *p<0.05, **p<0.01. (E) Cellular proliferation of SVAS iPSC-SMCs and control iPSC-SMCs in the presence of DMSO or U0126 (10 μM) for 72 hrs. Mean± s.e.m. (n=4). *p<0.05.
To investigate if elevated activity of ERK1/2 and up-regulation of cyclin D1 are required for hyper-proliferation of SVAS iPSC-SMCs, we inhibited ERK1/2 activity by treating cells with U0126, a specific inhibitor for MEK 1/2, which activates ERK1/2 through phosphorylation27, 28. Addition of U0126 markedly decreased ERK1/2 phosphorylation and cyclin D1 expression in SVAS iPSC-SMCs (Figure 5A–B). Furthermore, U0126 significantly inhibited the hyper-proliferation of SVAS iPSC-SMCs, shown by BrdU incorporation (Figure 5C–D) and direct cell counting (Figure 5E). Taken together, these results suggest that elevated activity of ERK signaling is necessary for the hyper-proliferation of SVAS iPSC-SMCs.
Discussion
The current study demonstrates the feasibility of using iPSCs to model severe human vascular disease phenotypes caused by genetic defects. We have validated that SMCs derived from SVAS iPSCs have the prototypical hyper-proliferation response seen in primary SMCs from SVAS patients (Figure 4A-C) 3. More importantly, our iPSC model of SVAS has led to the novel finding that SVAS iPSC-SMCs had significantly fewer organized networks of SM α-actin filament bundles, compared to control iPSC-SMCs (Figure 2A–B). Furthermore, we showed for the first time that either recombinant elastin or small GTPase RhoA is able to rescue defective SM α-actin filament bundles in SVAS (Figure 3). We further made a new discovery that SVAS iPSC-SMCs migrated at a markedly higher rate in response to the chemotactic agent PDGF in comparison with the control iPSC-SMCs (Figure 4D). We also provided novel evidence that ERK1/2 signaling is activated and its activity is required for hyper-proliferation of SVAS iPSC-SMCs (Figure 5). Thus, we not only validate the iPSC approach and the vascular SMC defect from prior studies, but also provide new mechanistic insights in SVAS. Our studies specifically suggest that enhancement of SMC actin filament bundle formation by elastin or RhoA as well as inhibition of cellular proliferation by decreasing ERK1/2 signaling, might be potential therapeutic strategies in SVAS patients.
To model SVAS in a second patient and to ensure that the disease phenotypes corresponded with ELN deficiency, we generated a second iPSC line from a patient with WBS who displays SVAS and has a typical WBS microdeletion on chromosome 7q11.23, including ELN (Supplemental Figure 4). Although WBS patients display a more complex phenotype, the spectrum and pathological characteristics of cardiovascular lesions in patients with SVAS and WBS are virtually identical and have been denoted as ELN arteriopathy1, 4. Importantly, WBS iPSC-SMCs recapitulate all three defects observed in SVAS iPSC-SMCs (Supplemental Figure 10 and Supplemental Figure 11), suggesting that these disease phenotypes correspond well with ELN deficiency in SVAS and WBS patients.
Differences in ELN gene structure and alternative splicing variants between animal models and humans confounds the understanding of ELN biology 29, highlighting the importance of human models to understand SVAS. Previous studies using elastin deficiency mice to study the mechanisms underlying SVAS have also been informative. However, unlike humans with ELN haploinsufficiencies such as SVAS or WBS, Eln+/− or WBS haploinsufficent mice do not develop aortic occlusive defects 30, 31. Eln−/− null mice develop fulminant aortic occlusive defects, but die around postnatal day 4.5 30, preventing the study of ELN deficiency in adult animals. Primary Eln−/− vascular SMCs from postnatal day 0.5 pups have defective actin filament bundle formation and proliferate and migrate at a higher rate, compared to control SMCs 6, suggesting a requirement for elastin in vascular morphogenesis. However, while the Eln−/− null mouse cells have provided very useful information, they are not SVAS cells and do not replicate the ELN gene dosage defect in human patients. Thus, the mouse model has limitations to study the pathophysiology of human disease. It is therefore of great importance to establish a human model for studying the pathogenesis of this disease. Because SVAS and WBS iPSCs can self-perpetuate, they are able to produce unlimited quantities of SMCs and thereby, provide a promising model both to study human SVAS and WBS disease mechanisms and to develop new therapies. To our knowledge, this is the first report of a human iPSC-based model of SVAS.
SVAS and WBS iPSC isolates showed similar characteristics of pluripotency that was confirmed by differentiation into the three embryonic germ layers in vitro and by in vivo teratoma formation (Figure 1; Supplemental Figure 1–5). In this study, SVAS iPSC-SMCs expressed around 50% of the elastin made by control iPSC-SMCs (Figure 2E and Supplemental Figure 7B–C), while WBS iPSC-SMCs expressed around 30% of the elastin made by control iPSC-SMCs (Supplemental Figure 10C). We did not observe a significant difference in the migration (Figure 4D; Supplemental Figure 9D; Supplemental Figure 11D), proliferation (Figure 4A–C; Supplemental Figure 9A–C; Supplemental Figure 11A–C), and formation of SM α-actin filament bundles (Figure 2A–B; Supplemental Figure 6A–C; Supplemental Figure 10A–B) between SMCs derived from SVAS iPSCs and WBS iPSCs. This might be due to the modest difference in elastin expression between the two lines, or due to the loss of additional gene(s) in the WBS patient that may modify the phenotype. Future efforts will be made to generate additional iPSC lines from multiple SVAS and WBS patients in order to better assess a correlation between elastin gene dosage and disease phenotype.
Current therapies targeting vascular SMC proliferation, such as rapamycin (a protein synthesis and cell cycle inhibitor), tend to have off-target effects, such as endothelial cell function inhibition 32. Previous studies indicate that the rescue of defective actin filament bundle formation via ELN protein inhibits the abnormally high proliferation rate in primary vascular SMCs from Eln null mice or patients with SVAS 3, 6. This raises the possibility that inducing assembly of the contractile apparatus, such as actin filament bundles, may be a more specific and efficacious target for therapy for vascular diseases caused by hyper-proliferation of SMCs. The defective actin filament bundle formation (Figure 2A–B) and the ERK-mediated hyper-proliferation observed in SVAS iPSC-SMCs (Figure 5) support the notion that enhancing the formation of actin filament bundles might be a preferred target for ameliorating the vascular diseases caused by ERK-mediated hyper-proliferation of SMCs. Further studies are warranted to investigate the potential link of defective actin filament bundle formation and hyper-activity of ERK in SVAS iPSC-SMCs. Actin filament bundle formation has been reported to induce nuclear localization of myocardin-related transcriptional factor (MRTF) co-factors, which inhibit cellular proliferation mediated by serum response factor (SRF) and the ERK signaling 33, 34, thereby linking actin dynamics with cellular proliferation.
We also provided novel evidence that SVAS and WBS iPSC-SMCs migrated to PDGF at a markedly higher rate than control iPSC-SMCs (Figure 4D, Supplemental Figure 9D, Supplemental Figure 11D). Formation of lamellipodia at the leading edge of cell is one of the key events regulating directional cell migration. This process requires the assembly of actin subunits into linear polymers and the cross-linking of the filaments 35. Activation of PI3K/Akt and/or PLCγ signaling by PDGF has been implicated in enhancing migration by promoting actin polymerization and the formation of actin networks in lamellipodia 36, 37. The defective actin filament bundle formation in SVAS and WBS SMCs may result in a larger pool of short fragments or monomeric subunits which are key substrates for the actin assembly reactions that drive the formation of lamellipodia and promote cell migration. Further studies investigating the mechanism of migration defect in SVAS SMCs may help to unravel the pathological mechanisms of vascular proliferative diseases.
The production of SVAS and WBS iPSC-SMCs also offers a patient-specific platform for small molecule screening. The effects of small molecules on ELN expression, RhoA activity or novel signaling pathways could be assessed, or perhaps more directly their efficacy to increase actin filament bundle formation or decrease cellular proliferation might prove useful. Further studies to elucidate the intracellular signaling events consequent to the exogenous addition of ELN or activation of RhoA, as well as to determine whether those effects are sufficient to alleviate in vivo arterial narrowing, represent important future steps for the understanding and treatment of SVAS. Because this SVAS iPSC model recapitulates key pathological features of SVAS due to elastin haploinsufficiency, it should facilitate future translational research into SVAS disease mechanisms and novel therapeutic interventions.
Supplementary Material
Acknowledgments
We thank Drs. Lawrence H. Young, Jeffrey R. Bender, Michael Simons, William Sessa, Diane Krause and Marsha W. Rolle for reviewing this manuscript; Drs. Yong Deng, Min Ding, Rong Ju, Yi Xie and Elenoe Smith’s assistance on our experimental design; and Caroline Greenberg, Carol Suh, Amogh Sivarapatna, Jessica Tuan’s assistance on the preparation of the manuscript. Yongming Ren and Xin Ge are listed interchangeably in this manuscript.
Funding Sources: This work was supported by the Connecticut Stem Cell (09SCAYALE10, 10SCA35, 11SCB18), NIH 1K02HL101990-01 and UL1 RR024139 (Y.Q.), NIH 5T32 HL007950 (P.J.A.), NIH R01HL091013 (K.A.M.), NIH R01HL078960 (L.Y.), NIH 1RC2HL101535 (D.N.K.) and Charles Hood Foundation (I.H.P.).
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Pober BR, Johnson M, Urban Z. Mechanisms and treatment of cardiovascular disease in Williams-Beuren syndrome. J Clin Invest. 2008;118:1606–1615. doi: 10.1172/JCI35309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li DY, Toland AE, Boak BB, Atkinson DL, Ensing GJ, Morris CA, Keating MT. Elastin point mutations cause an obstructive vascular disease, supravalvular aortic stenosis. Hum Mol Genet. 1997;6:1021–1028. doi: 10.1093/hmg/6.7.1021. [DOI] [PubMed] [Google Scholar]
- 3.Urban Z, Riazi S, Seidl TL, Katahira J, Smoot LB, Chitayat D, Boyd CD, Hinek A. Connection between elastin haploinsufficiency and increased cell proliferation in patients with supravalvular aortic stenosis and Williams-Beuren syndrome. Am J Hum Genet. 2002;71:30–44. doi: 10.1086/341035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris CA, Mervis CB. Williams syndrome and related disorders. Annu Rev Genomics Hum Genet. 2000;1:461–484. doi: 10.1146/annurev.genom.1.1.461. [DOI] [PubMed] [Google Scholar]
- 5.Wagenseil JE, Mecham RP. Vascular extracellular matrix and arterial mechanics. Physiol Rev. 2009;89:957–989. doi: 10.1152/physrev.00041.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karnik SK, Brooke BS, Bayes-Genis A, Sorensen L, Wythe JD, Schwartz RS, Keating MT, Li DY. A critical role for elastin signaling in vascular morphogenesis and disease. Development. 2003;130:411–423. doi: 10.1242/dev.00223. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Torres A, Gimeno A, Melon J, Mendez L, Munoz FJ, Macia M. Age-related loss of proliferative activity of human vascular smooth muscle cells in culture. Mech Ageing Dev. 1999;110:49–55. doi: 10.1016/s0047-6374(99)00042-1. [DOI] [PubMed] [Google Scholar]
- 8.Thyberg J. Differentiated properties and proliferation of arterial smooth muscle cells in culture. Int Rev Cytol. 1996;169:183–265. doi: 10.1016/s0074-7696(08)61987-7. [DOI] [PubMed] [Google Scholar]
- 9.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 12.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 13.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, Seyfarth M, Sinnecker D, Schomig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 14.Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, Yang L, Kaplan AD, Adler ED, Rozov R, Ge Y, Cohen N, Edelmann LJ, Chang B, Waghray A, Su J, Pardo S, Lichtenbelt KD, Tartaglia M, Gelb BD, Lemischka IR. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Lian Q, Zhu G, Zhou F, Sui L, Tan C, Mutalif RA, Navasankari R, Zhang Y, Tse HF, Stewart CL, Colman A. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Liu GH, Barkho BZ, Ruiz S, Diep D, Qu J, Yang SL, Panopoulos AD, Suzuki K, Kurian L, Walsh C, Thompson J, Boue S, Fung HL, Sancho-Martinez I, Zhang K, Yates J, 3rd, Izpisua Belmonte JC. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW, Lafyatis R, Demierre MF, Weiss DJ, French DL, Gadue P, Murphy GJ, Mostoslavsky G, Kotton DN. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urban Z, Michels VV, Thibodeau SN, Davis EC, Bonnefont JP, Munnich A, Eyskens B, Gewillig M, Devriendt K, Boyd CD. Isolated supravalvular aortic stenosis: functional haploinsufficiency of the elastin gene as a result of nonsense-mediated decay. Hum Genet. 2000;106:577–588. doi: 10.1007/s004390000285. [DOI] [PubMed] [Google Scholar]
- 19.Xie CQ, Zhang J, Villacorta L, Cui T, Huang H, Chen YE. A highly efficient method to differentiate smooth muscle cells from human embryonic stem cells. Arterioscler Thromb Vasc Biol. 2007;27:e311–312. doi: 10.1161/ATVBAHA.107.154260. [DOI] [PubMed] [Google Scholar]
- 20.Zhang P, Huang A, Ferruzzi J, Mecham RP, Starcher BC, Tellides G, Humphrey JD, Giordano FJ, Niklason LE, Sessa WC. Inhibition of microRNA-29 enhances elastin levels in cells haploinsufficient for elastin and in bioengineered vessels--brief report. Arterioscler Thromb Vasc Biol. 2012;32:756–759. doi: 10.1161/ATVBAHA.111.238113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worth NF, Rolfe BE, Song J, Campbell GR. Vascular smooth muscle cell phenotypic modulation in culture is associated with reorganisation of contractile and cytoskeletal proteins. Cell Motil Cytoskeleton. 2001;49:130–145. doi: 10.1002/cm.1027. [DOI] [PubMed] [Google Scholar]
- 22.Brooke BS, Bayes-Genis A, Li DY. New insights into elastin and vascular disease. Trends Cardiovasc Med. 2003;13:176–181. doi: 10.1016/s1050-1738(03)00065-3. [DOI] [PubMed] [Google Scholar]
- 23.Rolfe BE, Worth NF, World CJ, Campbell JH, Campbell GR. Rho and vascular disease. Atherosclerosis. 2005;183:1–16. doi: 10.1016/j.atherosclerosis.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 25.Parmacek MS. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circ Res. 2007;100:633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 27.Murphy LO, Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem Sci. 2006;31:268–275. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol Cell Biol. 2004;24:144–153. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirano E, Knutsen RH, Sugitani H, Ciliberto CH, Mecham RP. Functional rescue of elastin insufficiency in mice by the human elastin gene: implications for mouse models of human disease. Circ Res. 2007;101:523–531. doi: 10.1161/CIRCRESAHA.107.153510. [DOI] [PubMed] [Google Scholar]
- 30.Li DY, Brooke B, Davis EC, Mecham RP, Sorensen LK, Boak BB, Eichwald E, Keating MT. Elastin is an essential determinant of arterial morphogenesis. Nature. 1998;393:276–280. doi: 10.1038/30522. [DOI] [PubMed] [Google Scholar]
- 31.Goergen CJ, Li HH, Francke U, Taylor CA. Induced chromosome deletion in a Williams-Beuren syndrome mouse model causes cardiovascular abnormalities. J Vasc Res. 2011;48:119–129. doi: 10.1159/000316808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marx SO, Totary-Jain H, Marks AR. Vascular smooth muscle cell proliferation in restenosis. Circ Cardiovasc Interv. 2011;4:104–111. doi: 10.1161/CIRCINTERVENTIONS.110.957332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Descot A, Hoffmann R, Shaposhnikov D, Reschke M, Ullrich A, Posern G. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol Cell. 2009;35:291–304. doi: 10.1016/j.molcel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 36.Kundra V, Escobedo JA, Kazlauskas A, Kim HK, Rhee SG, Williams LT, Zetter BR. Regulation of chemotaxis by the platelet-derived growth factor receptor-beta. Nature. 1994;367:474–476. doi: 10.1038/367474a0. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Hernando C, Jozsef L, Jenkins D, Di Lorenzo A, Sessa WC. Absence of Akt1 reduces vascular smooth muscle cell migration and survival and induces features of plaque vulnerability and cardiac dysfunction during atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29:2033–2040. doi: 10.1161/ATVBAHA.109.196394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.