Summary
Schistosomiasis is among the most prevalent human parasitic diseases, affecting more than 200 million people worldwide1. The etiological agents of this disease are trematode flatworms (Schistosoma) that live and lay eggs within the vasculature of the host. These eggs lodge in host tissues, causing inflammatory responses that are the primary cause of morbidity. Because these parasites can live and reproduce within human hosts for decades2, elucidating the mechanisms that promote their longevity is of fundamental importance. Although adult pluripotent stem cells, called neoblasts, drive long-term homeostatic tissue maintenance in long-lived free-living flatworms3,4 (e.g., planarians), and neoblast-like cells have been described in some parasitic tapeworms5, little is known about whether similar cell types exist in any trematode species. Here, we describe a population of neoblast-like cells in the trematode Schistosoma mansoni. These cells resemble planarian neoblasts morphologically and share their ability to proliferate and differentiate into derivatives of multiple germ layers. Capitalizing on available genomic resources6,7 and RNAseq-based gene expression profiling, we find that these schistosome neoblast-like cells express a fibroblast growth factor receptor ortholog. Using RNA interference we demonstrate that this gene is required for the maintenance of these neoblast-like cells. Our observations suggest that adaptation of developmental strategies shared by free-living ancestors to modern-day schistosomes likely contributed to the success of these animals as long-lived obligate parasites. We expect that future studies deciphering the function of these neoblast-like cells will have important implications for understanding the biology of these devastating parasites.
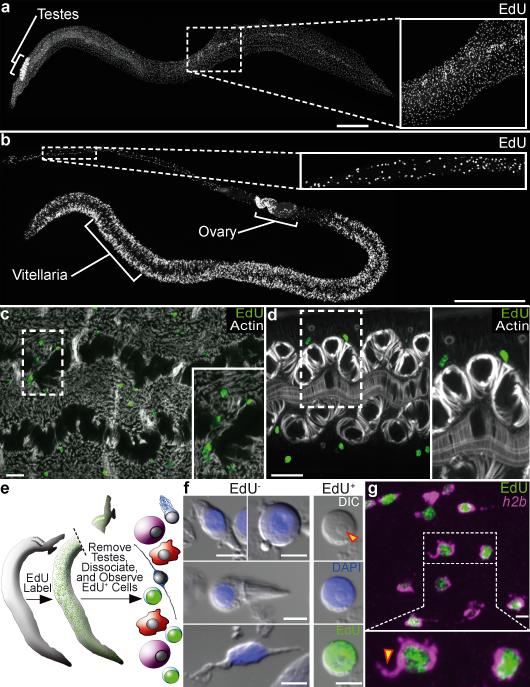
Although classic studies of cell proliferation in Schistosoma focused on reproductive tissues8,9, Den Hollander and Erasmus did note occasional “undifferentiated” somatic cells that incorporated tritiated thymidine in adult parasites. Encouraged that these cells could represent neoblast-like stem cells, we treated adult S. mansoni with the thymidine analogue 5-ethynyl-2'-deoxyuridine (EdU)10 to examine the distribution of S-phase cells in the parasite (Fig. 1a,b). In addition to the expected incorporation in the highly proliferative reproductive organs (testes, ovaries, and vitellaria) (Fig. 1a,b and Supplementary Fig. 1), we observed a population of EdU+ cells throughout the soma of male and female parasites (Fig. 1a–d). Similar distributions of EdU-incorporating cells were observed whether parasites were given EdU during in vitro culture or in vivo by intraperitoneal injection of schistosome-infected mice. Analogous to the neoblasts in free-living flatworms11,12, these proliferating somatic cells (PSCs) were restricted to the mesenchyme of male and female worms (Fig. 1c,d), not associated with reproductive organs, and were often found in clusters near the intestine (Supplementary Fig. 2a). We also observed a conspicuous population of PSCs adjacent to the ventral sucker (Supplementary Fig. 2b). PSCs traversed the cell cycle: they initially expressed the cell cycle-associated transcript histone h2b (Supplementary Fig. 3a–c) and progressed to M-phase within 24 hours following an EdU pulse (Supplementary Fig. 3d).
Figure 1. Proliferation of somatic cells in adult schistosomes.
a–b, EdU labeling in (a) male and (b) female parasites.
c–d, Distribution of mesenchymal PSCs in (c) male and (d) female parasites. Phalloidin staining for actin shows male enteric and dorso-ventral muscles and female enteric and uterine muscles.
e, Strategy to characterize PSC morphology.
f, The morphology of EdU− and EdU+ cells. Arrowhead indicates a nucleolus.
g, FISH for histone h2b with EdU labeling. Arrowhead indicates a cytoplasmic projection.
(a–d, g) are confocal projections; (a–b) are derived from tiled stacks. Scale bars: (a–b) 500 μm, (c–d) 20 μm, (f–g) 5 μm.
Neoblasts are the only proliferating somatic cells in planarians4,11 and they possess a distinct morphology; they are round-to-ovoid mesenchymal cells with a high nuclear-tocytoplasmic ratio, a large nucleolus, and they often extend a cytoplasmic projection3,11,13. To determine if PSCs share similarities with planarian neoblasts, we examined these cells by dissociating male tissues devoid of germ cells (Fig. 1e). In these preparations we observed a number of distinct differentiated cell types that failed to incorporate EdU, including cells with a low nuclear-to-cytoplasmic ratio, neuron-like cells, and ciliated cells (Fig. 1f). By contrast, we found that EdU incorporation was restricted to a neoblast-like population of cells with scant cytoplasm (n=136/137 cells) and often a prominent nucleolus (Fig. 1f). We also inspected PSCs within the mesenchyme using EdU to label nuclei and fluorescent in situ hybridization (FISH) to detect histone h2b mRNA in the cytoplasm of proliferative cells. Consistent with our results from tissue macerates, EdU+ cells possess a narrow rim of cytoplasm surrounding their nucleus, and these cells often display a cytoplasmic projection (Fig. 1g). These observations highlight morphological similarities between proliferating cells in schistosomes and planarian neoblasts.
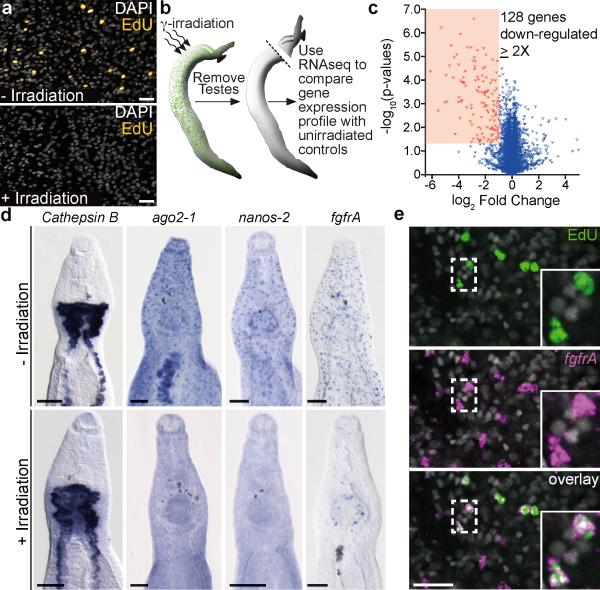
Previous studies have exploited the sensitivity of planarian neoblasts to γ-irradiation as a means to identify neoblast-enriched transcripts14–16. Using this strategy to identify PSC-expressed genes, we exposed parasites to various dosages of γ-irradiation and determined that 100–200 Gy were sufficient to block EDU incorporation (Fig. 2a). Because of their high ratio of somatic tissue to reproductive tissue and their large number of PSCs relative to female worms (compare insets in Fig. 1a and 1b), our remaining studies, unless otherwise noted, focused on male parasites. By comparing the transcriptional profiles of irradiated and non-irradiated parasites by RNAseq (Fig. 2b), we identified 128 genes with significantly down-regulated expression (≥ 2-fold, p < 0.05) 48 hours post-irradiation (Fig. 2c and Supplementary Table 1). Highlighting the efficacy of this approach to identify transcripts specific to proliferating cells, we found that genes expressed in differentiated tissues, such as the intestine (Sm-cathepsin B17), were unaffected by irradiation. By contrast, our list of down-regulated genes was enriched for factors involved in the cell cycle (Supplementary Fig.4 and Supplementary Table 1).
Figure 2. Transcriptional profiling identifies genes expressed in proliferative cells.
a, EdU incorporation is abrogated at D3 following irradiation.
b, Strategy to identify PSC-expressed genes.
c, Volcano plot showing expression differences in control versus irradiated parasites. n = 3 for each group.
d, WISH for various transcripts in unirradiated and D5 post-irradiation parasites. n > 3 parasites.
e, EdU labeling and FISH for Sm-fgfrA. 1988/2000 EdU+ PSCs were Sm-fgfrA+ following a 20–22 hour pulse (n = 20 male parasites). (a, e) are confocal projections. Scale bars: (a, e) 20 μm, (d) 100 μm.
In addition to identifying cell cycle-associated factors, systematic comparison of irradiation-sensitive genes with neoblast-enriched transcripts14,15,18,19 uncovered a number of interesting similarities (Supplementary Table 1). For instance, homologues of genes known to regulate planarian neoblasts such as p53, a sox-family transcription factor, fibroblast growth factor receptors, and argonaute214,20–22, were significantly down-regulated in irradiated schistosomes (Supplementary Fig. 4 and Supplementary Table 1). Another distinctive feature of neoblasts3,14,22, and the somatic stem cells of other invertebrates23, is that they often express post-transcriptional regulators associated with germline development (e.g., vasa, piwi, tudor, and nanos). Although vasa-like genes have been reported in Schistosoma, no true vasa orthologue has been indentified24. Similarly, piwi and tudor genes appear to be absent from schistosomes (data not shown). However, we identified a nanos orthologue (Sm-nanos-2) that was down-regulated in somatic tissue following irradiation (Supplementary Fig. 4, and Supplementary Table 1). Since these genes represented potential regulators of PSC behavior and could serve as useful markers for these cells, we examined their expression by whole-mount in situ hybridization (WISH). We detected Sm-ago2-1, Sm-nanos-2, and Sm-fgfrA transcripts in cells scattered throughout the mesenchyme (Fig. 2d and Supplementary Fig. 5) in a pattern similar to that of cells incorporating EdU (Fig. 1A). This mesenchymal expression was radiation sensitive (Fig. 2d), suggesting these genes are expressed in proliferating cells. Consistent with this idea, we found that following an EdU pulse, >99% of EdU-incorporating somatic cells also expressed Sm-fgfrA (Fig. 2e).
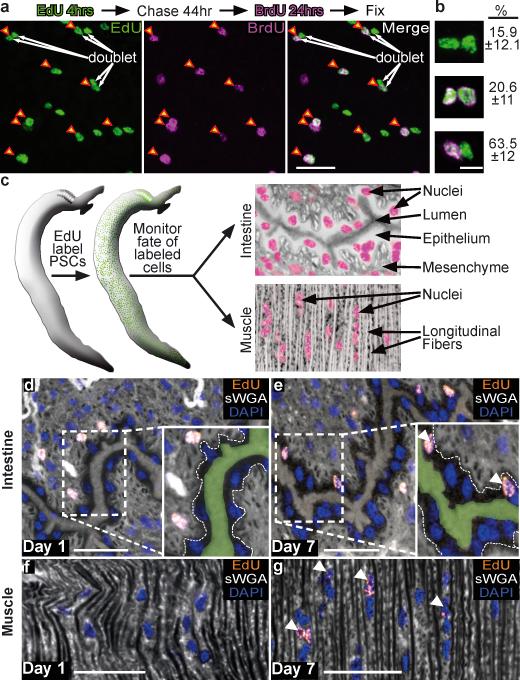
To determine if PSCs are stem cells, we assessed their ability both to self-renew and to produce differentiated cell types. To examine self-renewal, we administered sequential pulses of EdU and 5-Bromo-2'-deoxyuridine (BrdU) to parasites in vitro. Because nearly all PSCs that incorporate EdU are Sm-fgfrA+, the ability of EdU+ cells to incorporate BrdU in subsequent cell cycles would suggest that fgfrA+ PSCs self-renew (i.e., divide and produce more fgfrA+ PSCs). For these experiments we chose a chase period of 44 hours, since this time frame should give many EdU+ PSCs sufficient time to divide (Supplementary Fig. 3d). Consistent with PSCs possessing the capacity for self-renewal, we find that 41% of cells that initially incorporate EdU are BrdU+ 3 days following an initial EdU pulse (Fig. 3a). Furthermore, we observed that many EdU+ cells were distributed in pairs, or “doublets” (Fig. 3a); we suggest a majority of these doublets are the products of cell division (Supplementary Discussion and Supplementary Fig. 6). In these EdU+ doublets, a disproportionately large fraction displayed asymmetric BrdU incorporation (i.e. one nucleus is EdU+BrdU+, while the other is EdU+BrdU−) (Fig. 3b and Supplementary Discussion). This observation suggests that division progeny have an asymmetric capacity to proliferate. Whether this represents stem cell-like asymmetric division or temporal differences in the ability of these cells to reenter the cell cycle requires further experimentation. Nevertheless, these data are consistent with PSCs (or some PSC subpopulation) being capable of self-renewal.
Figure 3. PSCs self-renew and differentiate.
a, EdU-BrdU double labeling. Arrowheads, EdU+BrdU+ nuclei. Arrows, EdU+ “doublets”.
b, Percentage (±s.d.) EdU+ doublets (green) that are BrdU−-BrdU− (top), BrdU+-BrdU+ (middle, BrdU is magenta), or BrdU+-BrdU− (Bottom). n = 21 parasites.
c, Strategy to monitor cellular differentiation.
d–g, EdU and sWGA labeling showing EdU+ cells in (d,e) male intestine or (f,g) dorsal musculature at (d,f) D1 and (e,g) D7 following a pulse. (d,e) Insets, intestinal basal surface (dashed lines) and lumen (green). Arrowheads, EdU+ (e) intestinal cells or (g) muscle cells. Images are confocal projections. Scale bars: (a,d–g) 20 μm (b) 5 μm.
To examine the capacity of PSCs to differentiate, we performed EdU pulse-chase experiments in vivo. For these experiments, schistosome-infected mice were injected with EdU and the distribution of EdU+ cells was monitored at early (D1) and late (D7) time points (Fig. 3c). We successfully used this pulse-chase approach to monitor the differentiation of schistosome germ cells (Supplementary Fig. 7). Visualizing the syncytial epithelium of the schistosome intestine at D1, we did not observe EdU+ intestinal nuclei in male or female parasites (Fig. 3d, 0 EdU+/3151 DAPI+ nuclei, 14 mixed sex parasites, n=5 mice), confirming that cells in the intestine do not proliferate. Following a 7-day chase, however, ~2.5% of the intestinal nuclei were EdU+ (Fig. 3e, 56 EdU+/2189 DAPI+ nuclei, 10 mixed sex parasites, n=3 mice). This observation suggests that cells initially labeling with EdU have the capacity to migrate into the intestine and differentiate into new intestinal cells. Similarly, we were able to monitor the differentiation of new cells in the body wall muscles. At D1 no EdU+ nuclei were observed in the male body wall musculature (Fig. 3f, 0 EdU+/1882 DAPI+ nuclei, 13 male parasites, n=6 mice), whereas at D7 ~10% of the muscle cell nuclei were EdU+ (Fig. 3g, 55 EdU+/584 DAPI+ nuclei, 6 male parasites, n=3 mice). Since virtually all cells that initially incorporate EdU are Sm-fgfrA+, we suggest that these double-positive cells are likely to represent the only source of new intestinal and muscle cells and, thus, represent a collectively multipotent population of neoblast-like stem cells. Whether all Sm-fgfrA+ PSCs are multipotent or whether they exist as lineage-restricted progenitors remains unclear.
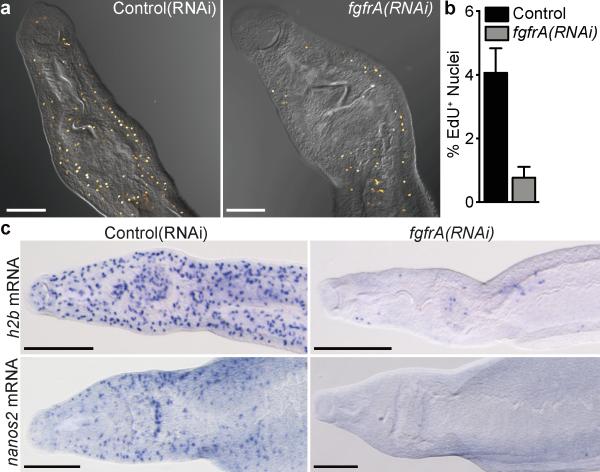
While progress has been made in identifying transcriptional14 and post-transcriptional22 regulators of planarian neoblasts, little is known about the signal transduction networks functioning within these cells. Since the expression of FGF receptor family members in proliferative cells is conserved between planarians14,21 and schistosomes, we speculated that FGF signaling could regulate these cells in S. mansoni. To examine this idea, we disrupted Sm-fgfrA in in vitro-cultured adult parasites using RNA interference (Supplementary Fig. 8). We found that inhibition of Sm-fgfrA resulted in reduced EdU incorporation (Fig. 4a,b and Supplementary Table 2) and down-regulation of cell cycle-associated transcripts (Fig. 4c and Supplementary Fig. 8). To resolve whether this effect is due to reduced cell proliferation or a failure to maintain neoblast-like cells, we monitored the expression of PSC markers Sm-ago2-1 and Sm-nanos-2 in Sm-fgfrA(RNAi) parasites. Sm-fgfrA RNAi treatment resulted in a dramatic reduction in the number of cells expressing Sm-nanos-2 (Fig. 2c) as well as significantly reduced mRNA levels for Sm-ago2-1 and Sm-nanos-2 (Supplementary Fig. 8b). Together, these results suggest that Sm-fgfrA promotes the long-term maintenance of neoblast-like cells in S. mansoni. FGF signaling is known to influence multiple processes, such as cell proliferation, differentiation, and survival; furthermore, it plays key roles in various stem cell populations25. Our results suggest a conserved role for FGF signaling in controlling stem cell behavior in these parasites and demonstrate the feasibility of using RNAi to abrogate adult gene expression and manipulate neoblast-like cells in S. mansoni.
Figure 4. Sm-fgfrA is required for the maintenance of somatic stem cells.
a, EdU labeling and DIC images in control and Sm-fgfrA(RNAi) at RNAi D17.
b, Percentage EdU+ nuclei/total nuclei in dissociated tissues from control(RNAi) (n = 3002 nuclei) and Sm-fgfrA(RNAi) (n = 3642 nuclei) parasites. Error bars, 95% confidence intervals, p < 0.0001 χ2.
c, WISH for histone h2b (top row) and nanos2 (bottom row) transcripts in control (left column) versus Sm-fgfrA(RNAi) (right column) parasites at RNAi D20-21. n > 5 parasites/experiment. Scale bars: (a) 100 μm (c) 200 μm.
Adult schistosomes can modulate growth in response to host immune signals26 and male-female pairing status2,27 and they can regenerate damaged tissues following sub-lethal doses of the anti-schistosomal drug praziquantel28. These observations reveal the developmental plasticity of schistosomes, and suggest that these parasites can utilize distinct developmental programs in response to a range of external stimuli. Future studies characterizing the role of neoblast-like cells in diverse contexts could address long-standing gaps in our knowledge of schistosome biology and may reveal novel therapeutic strategies for treating and eliminating schistosomiasis.
Methods
Parasite Acquisition and Culture
Adult S. mansoni (6–8 weeks post-infection) were obtained from infected mice by hepatic portal vein perfusion31 with 37°C DMEM (Mediatech, Manassas, VA) plus 5% Fetal Calf Serum (FBS, Hyclone/Thermo Scientific Logan, UT). Parasites were rinsed several times in DMEM + 5% FBS and cultured (37°C/5% CO2) in Basch's Medium 16932 and 1× Antibiotic-Antimycotic (Gibco/Life Technologies, Carlsbad, CA 92008). Media was changed every 1–3 days.
EdU labeling
For in vitro labeling, parasites were cultured in Basch's Medium 169 supplemented with 10 μM EdU (Invitrogen, Carlsbad, CA) diluted from a 10 mM stock in DMSO. Unless otherwise noted, animals were pulsed for 18–24 hours. For in vivo labeling, schistosome-infected mice (6–8 weeks post-infection) were given a single intraperitoneal injection (100–200 mg EdU/kg bodyweight) with 5 mg/ml EdU dissolved in PBS and then harvested at various time points after injection.
In situ hybridization
Male and female parasites were separated by incubation (2–3 minutes) in a 0.25% solution of the anaesthetic ethyl 3-aminobenzoate methanesulfonate (Sigma-Aldrich, St. Louis, MO) dissolved in Basch's Medium 169 or Phosphate Buffered Saline (PBS). Relaxed parasites were then killed in a 0.6 M solution of MgCl2 and fixed for 4.5 hrs in 4% Formaldehyde dissolved in PBSTx (PBS + 0.3% Triton X-100). Following fixation, parasites were dehydrated in MeOH and stored at −20°C. Samples were rehydrated by incubation in 1:1 MeOH:PBSTx followed by incubation in PBSTx. Rehydrated samples were bleached for 1–2 hours in formamide bleaching solution (0.5% Formamide, 0.5% SSC, and 1.2% H2O2), rinsed with PBSTx, treated with Proteinase K (2–10 μg/mL, Invitrogen, Carlsbad, CA) for 20–30 minutes at room temperature and post-fixed for 10–15 minutes in 4% formaldehyde in PBSTx. Samples were hybridized at 52–55°C and otherwise processed as previously described29,33. Plasmids used for riboprobe synthesis were generated as described previously29 using oligonucleotide primers listed in Supplementary Table 3.
Immunofluorescence, histological staining, and EdU detection
Parasites were relaxed, killed, fixed, dehydrated and rehydrated as described above and bleached in 6% H2O2 dissolved in PBS for 0.5–2 h. Dehydration and bleaching were omitted for samples labeled with phalloidin. Samples were then treated with Proteinase K and post-fixed as described above. Immunofluorescence, lectin, and phalloidin staining were performed as described previously30. Rabbit anti-Phospho-Histone H3 Ser10 (anti-pH3) (D2C8, Cell Signaling, Danvers, MA), rhodamine-conjugated sWGA (Vector Laboratories Burlingame, CA), and Alexa Fluor 568 phalloidin (Invitrogen, Carlsbad, CA), were used at 1:1000, 1:100, and 1:100, respectively. EdU detection was performed essentially as previously described10,34 with 100 μM Alexa Fluor 488 or Alexa Fluor 594 azide conjugates. All imaging was performed as described previously29,30. To quantify intestinal cell differentiation, the number of EdU+ and DAPI+ intestinal nuclei were determined from 12 consecutive confocal sections imaged from the intestine. To quantify muscle cell differentiation, the number of EdU+ and DAPI+ nuclei were determined from 4 to 9 consecutive confocal sections through the dorsal muscle layer of male parasites.
Tissue dissociation and EdU detection
Following an overnight pulse with 10 μM EdU, the heads and testes of adult male S. mansoni were removed and the remaining tissue added to dissociation solution (Hanks Balanced Salt Solution with 3.5 × Trypsin-EDTA (from 10× stock, Sigma-Aldrich, St. Louis, MO)) and minced with a razor blade. These tissue fragments were incubated in ~4 mL of dissociation solution for 45–60 min at room temperature on a rocker and gentle pipetting was used to break up large tissue fragments. This mixture was passed over two sets of cell strainers (100 and 40 μm, BD, Franklin Lakes, New Jersey) and dissociated cells were collected by centrifugation (250 × g for 5 m). Pelleted cells were fixed in 4% formaldhyde in PBS for 30 min, spotted on Superfrost Plus microscope slides (Fisher Scientific), permeabilized for 30 min with PBSTx, and EdU was detected as described above with 10 μM Alexa Fluor 488 azide.
To quantify the ratio of EdU+ to total DAPI+ nuclei in RNAi knockdowns, 8 male parasites were processed as above and tiled images of EdU and DAPI labeling were captured on a Zeiss LSM 710 (Plan-Apochromat 20×/0.8). Numbers of EdU+ and DAPI+ nuclei were quantified using the Image-based Tool for Counting Nuclei (ITCN) plugin for ImageJ35.
γ-irradiation and transcriptional profiling
Parasites (D43 post-infection) were harvested from mice, suspended in Basch medium 169, and exposed to 200 Gy of γ-irradiation using a Gammacell-220 Excel with a Co60 source (Nordion, Ottawa, ON, Canada). Control parasites were mock irradiated. Parasites were cultured in Basch Medium 169 and 48 hours post-irradiation males were separated from female parasites using ethyl 3-aminobenzoate methanesulfonate. Following separation, the head and testes of males were removed and purified total RNA was prepared from the remaining tissue from pools of 14–18 parasites using Trizol (Invitrogen, Carlsbad, CA) and DNase treatment (DNA-free RNA Kit, Zymo Research, Irvine, CA). Three independent biological replicates were performed for both control and irradiated experimental groups. Individually tagged libraries for RNAseq were prepared (TruSeq RNAseq Sample Prep Kit, Illumina, San Diego, CA), pooled in a single lane, and 100 bp reads were generated using an Illumina HiSeq2000. Library preparation and Illumina sequencing were performed at the W.M. Keck Center for Comparative and Functional Genomics. The resulting reads were mapped to the annotated S. mansoni genome6 (v5.0) and differences in gene expression were determined using CLC Genomics Workbench (CLC bio, Aarhus, Denmark). Statistical enrichment of Gene Ontology terms was determined in CLC Genomics Workbench using a hyper geometric test that is similar to the GOstat test described in previous studies36. To examine similarities between proteins encoded from irradiation-sensitive transcripts in S. mansoni and genes expressed in planarian neoblasts, we compared our schistosome dataset with both neoblast-enriched and “whole” transcriptomes14,15,18,19 using standalone tBLASTn. Schistosome proteins sharing no similarity to translated planarian mRNAs (e-value cut-off > 1e-5) were omitted from analysis. Assignment of whether protein pairs were orthologous, homologous, paralogous, or unrelated was assessed manually on an individual basis. Data and evidence supporting protein similarity is provided in Supplementary Table 1.
EdU/BrdU double labeling
Parasites labeled with 10 μM EdU and BrdU were fixed in Methacarn (6:3:1 methanol:chloroform:glacial acetic acid) or processed for in situ hybridization. Following a 45 min 2N HCl treatment, EdU was detected and parasites were processed for anti-BrdU immunofluorescence (anti-BrdU 1:500, clone MoBU, Invitrogen). We observed no cross-reactivity between this antibody and EdU.
To quantify the level of BrdU/EdU overlap and measure center-to-center distances between nuclei, 3D confocal stacks from EdU/BrdU labeled animals were resampled to give isotropic voxels, and subjected to Gaussian filtering and background-subtraction. Labeled nuclei were segmented with Imaris (Bitplane Inc., South Winsor, CT) using parameters empirically determined to minimize the need for manual corrections; typically, fewer than 5% of the total nuclei required correction. The 3D coordinates of the nuclei were exported and analyzed with MATLAB. Overlapping EdU and BrdU labeled nuclei were defined as nuclei with center-to-center distances < 1 nuclear size (~4 um). Statistical analyses were performed in Origin (OriginLab, Northampton, MA).
RNA interference
Although procedures have been previously described37,38, RNAi experiments with adult parasites were based on methods optimized for schistosomula39. Briefly, in vitro cultured parasites were soaked with 20–30 μg of dsRNA freshly added on days 1–3 and every 5–6 days thereafter. As a negative control, animals were soaked with dsRNA synthesized from the ccdB and camR-containing insert of pJC53.229. dsRNA synthesis was performed as previously described29. Sequences used to generate dsRNAs are provided in Supplementary Fig. 9. To measure mRNA levels, total RNA from control and knockdown parasites (~8 male posterior somatic fragments) was reverse transcribed (iScript cDNA Synthesis Kit, Bio-Rad, Hercules, CA) and quantitative real time PCR was performed on an Applied Biosystems Step One Plus instrument using GoTaq qPCR Master Mix with SYBR green (Promega, Madison, WI). Transcript levels were normalized to the mRNA levels of proteasome subunit beta type-4 (smp_056500). Relative quantities were calculated using the ΔΔCt calculation in the Step One Plus software. Oligonucleotide primer sequences are listed in Supplementary Table 3.
Supplementary Material
Acknowledgments
We thank Rachel Roberts-Galbraith, Melanie Issigonis, and Labib Rouhana for comments on the manuscript; Ryan King for sharing the Cathepsin B plasmid and unpublished protocols; and Alvaro Hernandez for assistance with Illumina sequencing. Schistosome-infected mice were provided by the NIAID Schistosomiasis Resource Center and the Biomedical Research Institute (Rockville, MD) through NIAID Contract N01-A1-30026. This work was supported by: NIH F32 HD062124 (J.J.C.) and NIH R21 AI099642 (P.A.N.). P.A.N. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Full Methods and any associated references are available in the online version of the paper.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions J.J.C., B.W., B.G.L., M.T., H.I., performed experiments. J.J.C. and B.W. analyzed data. J.J.C. and P.A.N. designed the study and wrote the paper.
Author Information Experiments with and care of vertebrate animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Illinois at Urbana-Champaign (protocol approval number 10035). RNAseq analyses have been deposited in the NCBI Gene Expression Omnibus (Accession number: GSE42757). The authors declare no competing financial interests.
References
- 1.Chitsulo L, Engels D, Montresor A, Savioli L. The global status of schistosomiasis and its control. Acta Trop. 2000;77:41–51. doi: 10.1016/s0001-706x(00)00122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basch PF. Schistosomes: Development, Reproduction, and Host Relations. Oxford University Press; 1991. [Google Scholar]
- 3.Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newmark PA, Sánchez Alvarado A. Not your father's planarian: a classic model enters the era of functional genomics. Nat Rev Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- 5.Brehm K. Echinococcus multilocularis as an experimental model in stem cell research and molecular host-parasite interaction. Parasitology. 2010;137:537–555. doi: 10.1017/S0031182009991727. [DOI] [PubMed] [Google Scholar]
- 6.Protasio AV, et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop Dis. 2012;6:e1455. doi: 10.1371/journal.pntd.0001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berriman M, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Den Hollander JE, Erasmus DA. Schistosoma mansoni: DNA synthesis in males and females from mixed and single-sex infections. Parasitology. 1984;88(Pt 3):463–476. doi: 10.1017/s0031182000054731. [DOI] [PubMed] [Google Scholar]
- 9.Nollen PM, Floyd RD, Kolzow RG, Deter DL. The timing of reproductive cell development and movement in Schistosoma mansoni, S. japonicum, and S. haematobium, using techniques of autoradiography and transplantation. J Parasitol. 1976;62:227–231. [PubMed] [Google Scholar]
- 10.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- 12.Forsthoefel DJ, Park AE, Newmark PA. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Dev Biol. 2011;356:445–459. doi: 10.1016/j.ydbio.2011.05.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baguñá J, Romero R. Quantitative analysis of cell types during growth, degrowth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia. 1981;84:181–194. [Google Scholar]
- 14.Wagner DE, Ho JJ, Reddien PW. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 2012;10:299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solana J, et al. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol. 2012;13:R19. doi: 10.1186/gb-2012-13-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caffrey CR, McKerrow JH, Salter JP, Sajid M. Blood 'n' guts: an update on schistosome digestive peptidases. Trends Parasitol. 2004;20:241–248. doi: 10.1016/j.pt.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Onal P, et al. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J. 2012;31:2755–2769. doi: 10.1038/emboj.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labbe RM, et al. A comparative transcriptomic analysis reveals conserved features of stem cell pluripotency in planarians and mammals. Stem Cells. 2012;30:1734–1745. doi: 10.1002/stem.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogawa K, et al. Planarian fibroblast growth factor receptor homologs expressed in stem cells and cephalic ganglions. Dev Growth Differ. 2002;44:191–204. doi: 10.1046/j.1440-169x.2002.00634.x. [DOI] [PubMed] [Google Scholar]
- 22.Rouhana L, Shibata N, Nishimura O, Agata K. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev Biol. 2010;341:429–443. doi: 10.1016/j.ydbio.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 23.Juliano C, Wessel G. Versatile germline genes. Science. 2010;329:640–641. doi: 10.1126/science.1194037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skinner DE, et al. Vasa-Like DEAD-Box RNA Helicases of Schistosoma mansoni. PLoS Negl Trop Dis. 2012;6:e1686. doi: 10.1371/journal.pntd.0001686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- 26.Davies SJ, et al. Modulation of blood fluke development in the liver by hepatic CD4+ lymphocytes. Science. 2001;294:1358–1361. doi: 10.1126/science.1064462. [DOI] [PubMed] [Google Scholar]
- 27.Severinghaus AE. Sex Studies on Schistosoma japonicum. Quarterly Journal of Microscopical Science. 1928;71:653–702. [Google Scholar]
- 28.Shaw MK, Erasmus DA. Schistosoma mansoni: structural damage and tegumental repair after in vivo treatment with praziquantel. Parasitology. 1987;94(Pt 2):243–254. doi: 10.1017/s0031182000053920. [DOI] [PubMed] [Google Scholar]
- 29.Collins JJ, III, et al. Genome-Wide Analyses Reveal a Role for Peptide Hormones in Planarian Germline Development. PLoS Biol. 2010;8:e1000509. doi: 10.1371/journal.pbio.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins JJ, III, King RS, Cogswell A, Williams DL, Newmark PA. An atlas for Schistosoma mansoni organs and life-cycle stages using cell type-specific markers and confocal microscopy. PLoS Negl Trop Dis. 2011;5:e1009. doi: 10.1371/journal.pntd.0001009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis F. Schistosomiasis. Curr Protoc Immunol. 2001;Chapter 19(Unit 19):11. doi: 10.1002/0471142735.im1901s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basch PF. Cultivation of Schistosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. J Parasitol. 1981;67:179–185. [PubMed] [Google Scholar]
- 33.Cogswell AA, Collins JJ, III, Newmark PA, Williams DL. Whole mount in situ hybridization methodology for Schistosoma mansoni. Mol Biochem Parasitol. 2011;178:46–50. doi: 10.1016/j.molbiopara.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neef AB, Luedtke NW. Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides. Proc Natl Acad Sci U S A. 2011;108:20404–20409. doi: 10.1073/pnas.1101126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 36.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 37.Skelly PJ, Da'dara A, Harn DA. Suppression of cathepsin B expression in Schistosoma mansoni by RNA interference. Int J Parasitol. 2003;33:363–369. doi: 10.1016/s0020-7519(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 38.Boyle JP, Wu XJ, Shoemaker CB, Yoshino TP. Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Mol Biochem Parasitol. 2003;128:205–215. doi: 10.1016/s0166-6851(03)00078-1. [DOI] [PubMed] [Google Scholar]
- 39.Stefanic S, et al. RNA Interference in Schistosoma mansoni Schistosomula: Selectivity, Sensitivity and Operation for Larger-Scale Screening. PLoS Negl Trop Dis. 2010;4:e850. doi: 10.1371/journal.pntd.0000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McVeigh P, et al. Discovery of multiple neuropeptide families in the phylum Platyhelminthes. Int J Parasitol. 2009;39:1243–1252. doi: 10.1016/j.ijpara.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faghiri Z, et al. The tegument of the human parasitic worm Schistosoma mansoni as an excretory organ: the surface aquaporin SmAQP is a lactate transporter. PLoS One. 2010;5:e10451. doi: 10.1371/journal.pone.0010451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.