Abstract
Our long term goal is to understand the molecular pathology of otosclerosis and to develop better forms of therapy. Toward this goal, the current study focused on characterizing the molecular factors responsible for the unique biological features of the otic capsule: its minimal rate of remodeling, and lack of healing capacity when fractured. We compared expression levels of 62 genes involved in bone metabolism between the adult murine otic capsule and the tibia and parietal bones; the latter exemplify bones formed by endochondral and intramembranous ossification, respectively. Gene expression levels were measured using real-time quantitative RT-PCR and analyzed using tools of bioinformatics. Expression patterns of key genes were verified with in situ hybridization. The molecular profile of the otic capsule was distinctly different from that of the tibia and parietal bone. Genes found to be most characteristic of the otic capsule were: osteoprotegerin (opg), bone morphogenetic protein receptor 1b (bmpr1b) and bone morphogenetic protein 3 (bmp3). Expression levels were high for opg and bmpr1b, and minimal for bmp3 within the otic capsule. We concluded that opg and bmpr1b likely play important roles in inhibition of remodeling within the otic capsule.
Keywords: Otic capsule, opg, bmpr1b, otosclerosis
Introduction
The bone surrounding the inner ear, known as the otic capsule, is unique for many reasons. First, its rate of remodeling is minimal compared with other bones, as shown in many species including humans, dogs and pigs (Sorensen et al., 1992; Frisch et al., 1998; Soresen et al., 1990). However, under pathologic conditions such as otosclerosis or Paget’s disease, the otic capsule starts to remodel (Schuknecht 1993), which can lead to hearing loss. Second, the otic capsule typically does not heal when fractured, except by fibrous union (Perlman 1939). Third, the otic capsule is the densest bone in the body (Bast and Anson, 1949). Our previous work identified osteoprotegrin, opg, as an important factor in the inhibition of remodeling within the otic capsule (Zehnder et al., 2005). We found expression of opg in the otic capsule to be much higher than in other bones. We also found that Opg protein is produced in extremely high concentrations within the inner ear and that it, most likely, diffuses into the surrounding otic capsule. We demonstrated that opg knockout mice have abnormal otic capsule remodeling that resembles otosclerosis (Zehnder et al., 2006).
We undertook the current study to identify other genes, in addition to opg, that provide the molecular basis for the uniqueness of the otic capsule. We focused on genes that are known to play roles in bone remodeling, including inflammatory cytokines (Goldring 2007), and on genes that encode structural proteins. We compared the expression levels of these genes in the murine otic capsule to those in the parietal bone and tibia, two bones produced by developmentally distinct mechanisms. These mechanisms involve the transformation of preexisting mesenchyme into bone, which occurs directly or indirectly (Gilbert 2000). The direct mechanism, or intramembranous ossification, occurs primarily in flat bones of the skull, including the parietal bone. The indirect mechanism, which involves a cartilage intermediate, is known as endochondral ossification, and it gives rise to the axial and appendicular portions of the skeleton, including the tibia, and the otic capsule.
Material and methods
Collection of tissue, extraction of total RNA and cDNA synthesis
Samples of bone were collected from 7 adult C57BL/6J mice obtained from Jackson Laboratories, Maine. The animals, ranging in age from 8–11 weeks, were sacrificed by intraperiotenal injection of urethane (2.5 mg/kg). Otic capsule bone surrounding the cochlea, parietal bone and diaphysis of tibia were microdissected and placed in RNAlater reagent (Ambion). Care was taken to scrape away mucosa of the middle ear and parts of the membranous labyrinth adherent to the otic capsule. However, some contamination with soft tissues adherent to the otic capsule cannot be definitively excluded.
After removal of RNAlater, total RNA was purified using RNeasy spin-columns (Qiagen) according to the manufacturer’s protocol, with the modification for hypocellular, dense connective tissues (Reno et al., 1997). Specifically, Trizol was used first, followed by chloroform, collection of the aqueous phase, addition of an equal volume of isopropanol, and application of this mixture to RNeasy spin-columns. The RNA quantity and quality was assessed using Agilent 2100 Bioanalyzer (Agilent Technologies) and RNA Pico Kit (Agilent Technologies); samples with RNA integrity number of at least 7, out of possible 10, were used for further analysis. Total RNA that appeared clean and undegraded, based on the Bioanalyzer’s electropherograms, was reverse transcribed with Taqman Reverse Transcription Reagents kit (Applied Biosystems). Typical total RNA yields were slightly less than 1 µg for otic capsule samples, 1.5–2 µg for parietal bone, and 2 µg for tibia. Overall, RNA from 5 otic capsules, 4 parietal bones and 6 tibias met our quality criteria. Two of the tibia samples were pooled together to increase RNA yield; results from the pooled and non-pooled sample were in agreement.
Relative quantitation of mRNA using real-time PCR
Real-time quantitative RT-PCR was used to analyze gene expression levels in a sensitive and high throughput fashion. The 62 genes that were studied (supplemental Table 1) were selected based on their known roles in bone metabolism or inflammation, or suspected roles in bone remodeling based on genetic studies of bone diseases. For all genes, 6-FAM linked fluorescent probes and primers were designed and optimized by Applied Biosystems. The measurements were carried out on an Applied Biosystems 7700 Sequence Detector using 96 well plates. For each well, the 25 µl reaction contained: 1.25 l of the 20× probe/primer mix, 1 µl of cDNA template (approximately 15 ng cDNA/µl), 12.5 µl Universal Master Mix (Applied Biosystems), and 10.25 µl distilled water. All cDNA samples were run in triplicate, and each plate was used to test expression of up to 32 genes from a single sample. Fluorescence data were collected over 45 cycles of PCR, which consisted of an initial denaturation step at 95° C for 10 minutes, followed by 45 cycles of 95°C for 15 seconds and 60°C for 1 minute. Gene expression levels were quantified relative to the 18S rRNA gene and compared between bone types using the Comparative threshold cycle (CT) me thod, i.e. the ΔΔCT method (Livak and Schmittgen, 2001). Statistical significance of the detected changes was determined using the bootstrap method as in Stankovic and Corfas (2003). For a given gene, ΔΔCT was defined the difference of mean ΔCT of all samples from one bone type (e.g. otic capsule) and mean ΔCT of all samples from another bone type (e.g. parietal bone). Data were randomly shuffled 5000 times (using Matlab 5.3, The MathWorks Inc.), and a histogram of “pseudo”ΔΔCT values was built. The actual ΔΔCT was considered to be significant at the 0.05 level if it fell further from the mean than 95% of the histogram values. Statistically significant changes in gene expression were at least two fold.
Bioinformatics analysis
Tools of bioinformatics were applied to simultaneously analyze expression patterns of a relatively large number of genes studied in a relatively small number of samples. Genes were ranked according to signal to noise ratio, SNR, defined as the difference of the mean expression levels of each bone type scaled by the sum of standard deviations. Two unsupervised clustering algorithms (implemented in GenePattern 2.0) were used in the analysis of samples from different bone types and genes within a bone type: self-organizing maps (SOM) (Tamayo et al., 1999) and hierarchical clustering (Eisen et al., 1998). The stability of the identified clusters (i.e. sensitivity of the cluster boundaries to sampling variability) was assessed using consensus clustering (Monti et al., 2003). This method simulates perturbations of the original data using resampling techniques, and then applies the clustering algorithm of choice (SOM or hierarchical) to each of the perturbed datasets to assess the agreement, or consensus, among different runs. The results are typically summarized in a consensus matrix, which is depicted as a square heat map. Consensus matrices were generated assuming 2 to 5 clusters, and the best number of clusters determined using visual inspection of the consensus matrices, and the corresponding summary statistics. The increase in the area under the cumulative distribution function corresponding to the histogram of a consensus matrix peaked at the optimal number of clusters.
The class neighbors analysis (Golub et al., 1999) was used to determine which genes serve as good predictors for a bone type (class), and there was a priori knowledge of which sample came from what bone type. Genes were ranked based on SNR, and the robustness of the bone type predictors was tested using K-nearest neighbors cross validation (Golub et al., 1999). The cross validation technique is a cyclical procedure in which one sample at a time is withheld and a predictor built on the remaining samples; the predictor then predicts the bone type of the withheld sample.
In situ hybridization
Mice were anesthetized and fixed by intracardiac perfusion with 4% paraformaldehyde. Specimens were decalcified in 120 mmol/L ethylenediaminetetraacetic acid (EDTA) for one week. The skulls containing temporal bones were embedded in paraffin and serially sectioned in a horizontal plane at a thickness of 10µm. Selected sections were stained with 0.2% azure B, pH 3 to identify cartilage (Lillie, 1965).
A 535 bp HindIII-XhoI fragment of the mouse opg cDNA was cloned into pBluescript II SK-vector (courtesy of Dr. Abboud Werner, Heinrich et al., 2005). The mouse bmpr1b probe was described in Namiki et al. (1997). To prepare the digoxigenin (DIG)-labeled single stranded antisense and sense RNA probes, the purified fragments were subjected to PCR in the presence of DIG-dUTP (digoxigenin DNA labeling mixture; Roche, Basel, Switzerland) according to the manufacturer’s protocol. The efficiency of probe generation was monitored using a standard DIG labeled probe by membrane hybridization. In situ hybridization was performed as in Sassoon and Rosenthal (1993) with minor modifications. Sections were deparaffinized with xylene, washed in 100% ethanol, placed in 4% formaldehyde for 15 min, washed with PBS, digested with proteinase K (10µg/ml) in PBS for 10 min and washed in PBS. The hybridization mixture containing DIG labeled probe was applied to each section and incubated for 16h at 50°C. After application of 5× saline sodium citrate buffer (SSC), sections were washed in RNase buffer solution at 37°C for 30 min. Consecutive washes at 65°C were performed: twice in 2×SSC, twice in 0.1×SSC. To visualize the hybridized opg probe, the slides were incubated with alkaline phosphatase-conjugated anti-DIG antibody (Roche) for 60 min after applying the blocking buffer (Roche) for 60 min. The colorimetric reaction was developed with nitro blue tetrazolium salt and bromo-4-chloro-3-indolyl phosphate solution (Roche) in the dark for 2h, and stopped with PBS. For bmpr1b probe, fluorescent reaction was used because it gave a better signal than the calorimetric reaction. The slides were exposed to Cy3-conjugated monoclonal anti-DIG-antibody (Jackson Immuno Research, 200-162-156) for 60 min, washed twice with 0.1% Tween, 20mM Tris buffer solution, treated with Cy3-conjugated secondary antibody (AffiniPure Goat Anti-Mouse IgG, Jackson Immuno Research, 1150165-003) for 60 min, washed twice with 0.1% Tween, 20mM Tris buffer solution, and counterstained with nuclear DAPI stain (4’, 6-diamidino-2-phenylinodole, ddihydochloride, Sigma, D9542). Slides were mounted with Aquatex (Merck, Darmstadt, Germany), and evaluated by brightfield (opg) or fluorescent (bmpr1b) microscopy. Serial sections of tissues hybridized with the sense probe served as negative controls.
All animal experiments in this study were approved by the Animal Care and Use Committee of the Massachusetts Eye and Ear Infirmary.
Results
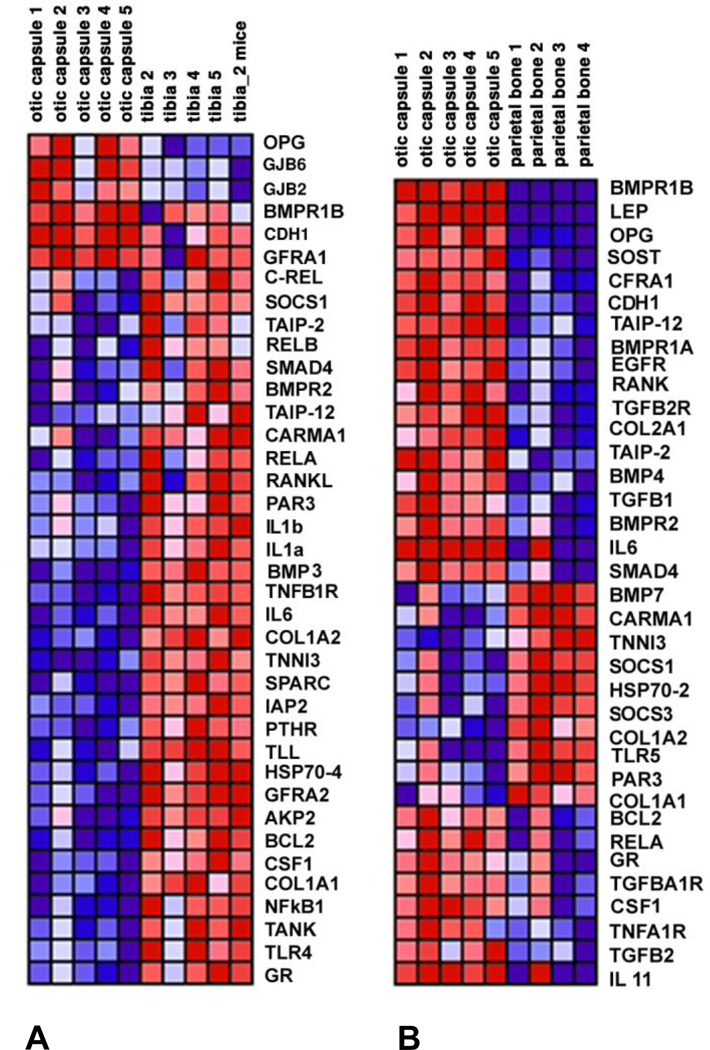
The set of genes we studied was sufficiently rich to allow for robust grouping or clustering of bone types. Clustering was non-overlapping when using hierarchical clustering, and partly overlapping when using SOM clustering; most cross-coupling was present between the otic capsule and parietal bone. Gene expression data are presented in the form of a heat map (Fig. 1) where each column represents a different sample, each row represents a different gene, and color coding reflects row-normalized levels of gene expression with red indicating high, and blue indicating low levels of expression. The otic capsule is compared with tibia (Fig. 1A) and parietal bone (Fig. 1B). The figures illustrate highly reproducible measurements within a bone type as most samples from a given bone type had similar color coding. A majority of the genes was down-regulated (blue in Fig. 1A) in the otic capsule compared to the parietal bone, and up-regulated (red in Fig. 1B) compared to the tibia. Thirty six genes were differentially expressed in the otic capsule compared to parietal bone, and 38 genes were differentially expressed in the otic capsule compared to tibia. Overall, 56 genes were differentially expressed in the otic capsule compared to parietal bone or tibia.
Figure 1.
Heat map of real-time quantitative RT-PCR data comparing the otic capsule with tibia (A) and parietal bone (B). Red indicates high and blue low levels of gene expression.
Genes that differentiate bone types
Genes that are most characteristic of a bone type are characterized by a large difference and a small variance between bone types. Two genes were sufficient at each cross validation step to correctly predict 8 out of 9 samples when comparing otic capsules with parietal bones. These two genes were allowed to vary for each step of cross validation, guided by which sample was withheld to build a predictor. Although there were 18 (i.e. 9×2) distinct possibilities for the two genes, 8 genes were used through all steps of the crossvalidation procedure: bmpr1b was used 7 times, bmp7 was used 3 times, tnni3 was used twice, col1a2 was used twice, and lep, il6, socs1 and carma1 were each used once.
When comparing otic capsules with tibias, two genes were also sufficient at each cross validation step to correctly predict 9 out of 10 samples. Six distinct genes (out of possible 10×2=20) were used through all steps of the cross-validation procedure: opg was used 7 times, bmp3 was used 5 times, tnfb1r was used 3 times, gjb6 was used 3 times, and tnni3 and col1a2 were each used once.
Osteoprotegerin, bmpr1b and bmp3 emerged as key predictors of the otic capsule as they were most frequently used during the cross-validation procedure. Expression of opg in the otic capsule, described as mean +/− standard error of the mean, was 97.8 +/−2.5 times higher than in parietal bone (p=0.003) and 9.6+/−2.5 times higher than in tibia (p=0.007). Expression of bmpr1b was 6.7 +/−2.0 times higher in the otic capsule than the tibia (p=0.015), and was undetectable in parietal bone (p=0.008). Expression of bmp3 in the otic capsule was 5.3+/−1.4 times lower than in the tibia (p=0.008), and not different from expression in parietal bone (p=0.32). The expression data in Fig. 1 are suggestive of this particular choice of predictor genes because the ordering of genes in Fig. 1 is according to the sharpness of the transition between any two bone types. The ordering is performed separately for genes with decreased expression level between the two bone types (“red to blue transition”), and for genes with increased expression level (“blue to red transition”). The most prominent genes in the first ranking are displayed at the top of the map, and the most prominent members of the second ranking are displayed starting in the middle of the map. Given that the two ranking sets are of unequal size in general, the least prominent members of the larger set are used to “fill in” the slots in the other set.
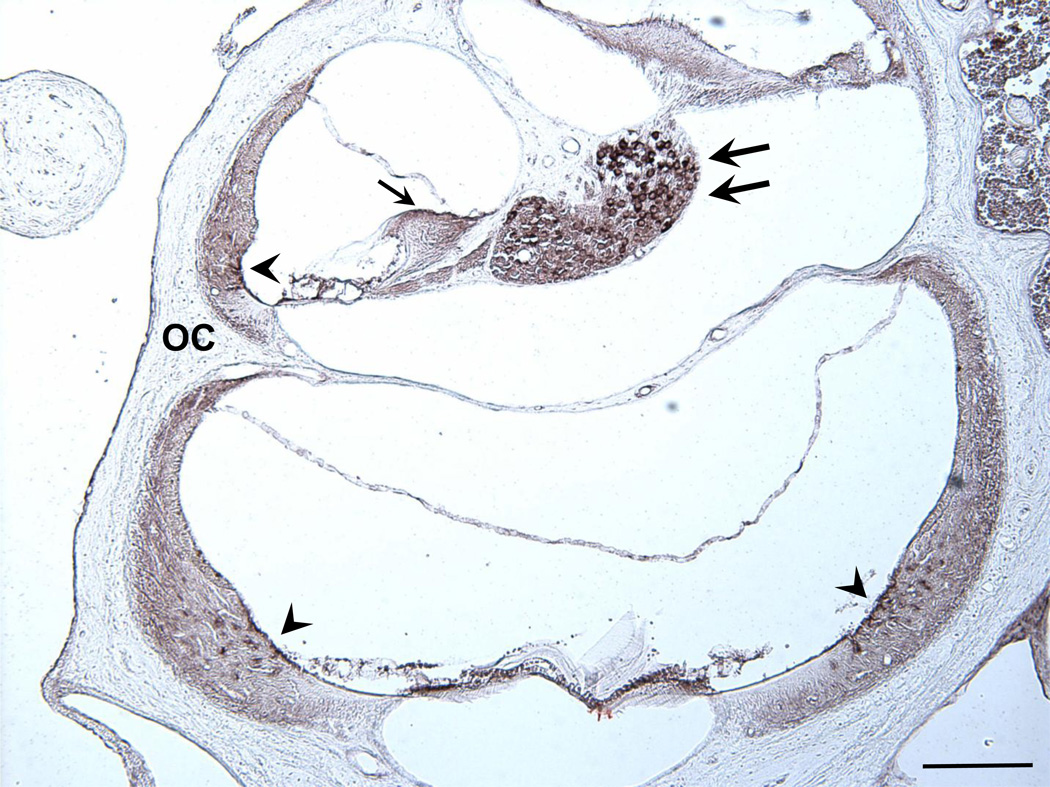
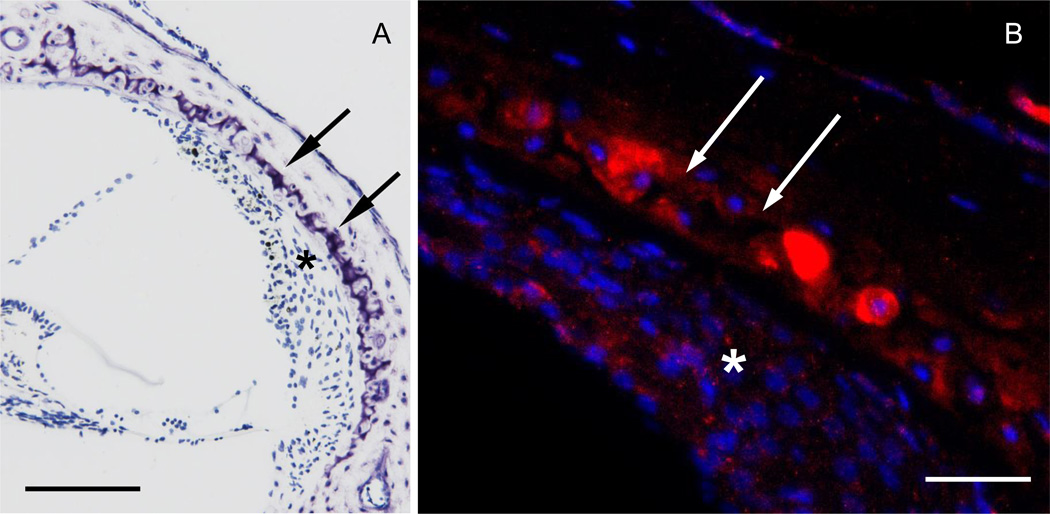
Although in situ hybridization detected opg mRNA in the otic capsule, even higher levels of expression were found in the soft tissues of the cochlea, i.e. spiral ganglion neurons, root cells of the spiral ligament and interdental cells of the spiral limbus (Fig. 2). The colorimetric reaction used to visualize opg mRNA in Fig. 2 produces a specific dark brown signal; diffuse light brown staining, such as in the spiral ligament, is nonspecific, and attributed to electrostatic interaction between the tissue and the probe (Tarenghi, 1998). Bmpr1b was expressed in cartilaginous cell rests of the otic capsule and to a lesser degree in fibrocytes of the spiral ligament (Fig. 3). Hybridization with sense probes produced no signal (not shown).
Figure 2.
A cross section through two cochlear turns hybridized with opg antisense probe. A strong signal (shown in dark brown) is present in root cells of the spiral ligament (arrowhead), interdental cells of the spiral limbus (single arrow) and spiral ganglion neurons (double arrows). A weaker signal is present in the otic capsule (OC). Hybridization with a sense probe produced no signal. Scale bar = 200 µm.
Figure 3.
Bmpr1b localizes to cartilaginous cell rests of the otic capsule. Cochlear cross section stained with azure (A), and with anti sense probe for bmpr1b (B). The red fluorescent signal (B) is most intense in cartilaginous rests (double arrows) and weaker in the spiral ligament (asterisk). Cell nuclei are in blue. Scale bar = 100 µm (A), 50 µm (B).
Clustering of genes within the otic capsule
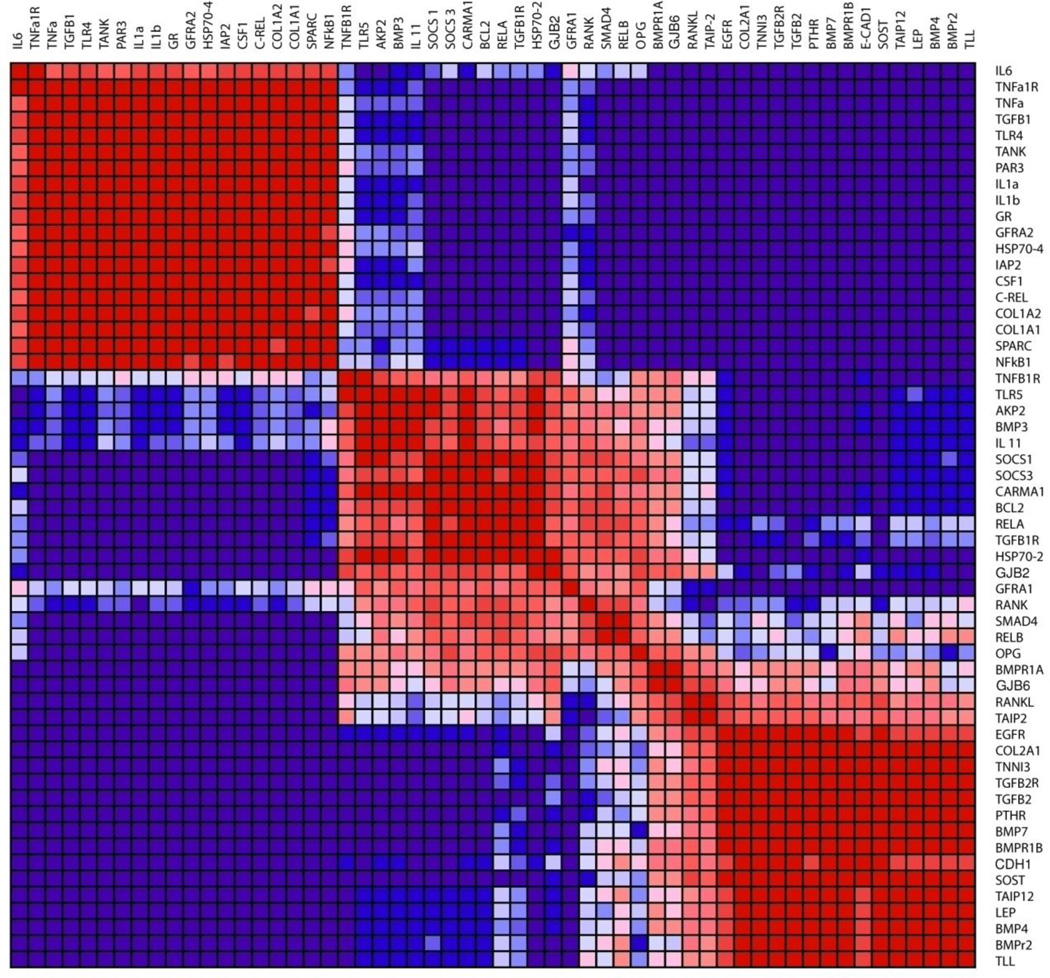
Genes in the otic capsule that share similar patterns of expression are depicted in a square heat map of consensus clustering (Fig. 4). The map illustrates the fraction of instances when any two genes are clustered together across the resampling iterations. When this fraction is 1, genes always cluster, as illustrated by a bright red color in the heat map; when the fraction is 0, genes never cluster, as illustrated by a dark blue color in the heat map. Figure 4 depicts desirable clustering because the entries of diagonal blocks are close to 1 and off-diagonal blocks are close to 0. The figure depicts 3 clusters, which was optimal for our data and resulted in agreement between SOM and hierarchical clustering. The coupling was strong within clusters and minimal between them.
Figure 4.
Heat map of genes that share similar expression patterns within the otic capsule. Consensus SOM clustering was performed assuming three clusters.
The mediators of inflammation belonged to a distinct cluster (Fig. 4), which included pro-inflammatory cytokines tumor necrosis factor alpha (tnfa), interleukin 1 alpha and beta (il1a, il1b), interleukin 6 (il6), as well as tnfa receptor (tnfa1r) and transcription factor nuclear factor of kappa-B subunit 1 (nfκb1). All of these pro-inflammatory mediators were downregulated in the otic capsule compared to tibia (Fig. 1A). In contrast, interleukin 11 (il11), a cytokine with potent anti-inflammatory activity (Trepicchio and Dorner, 1998) was upregulated in the otic capsule compared to parietal bone (Fig. 1B). Within the otic capsule (Fig. 4), il11 contributed to a distinct cluster, which included opg, bmp3 and suppressor of cytokine signaling 1 (socs1) and 3 (socs3). The third cluster included bmpr1b and several genes involved with bone morphogenetic protein (BMP) signaling: BMP receptor 2 (bmpr2), bmp4 and bmp7.
Discussion
The otic capsule has long been recognized as a histologically unique bone (Bast and Anson, 1949) and the current study unravels a part of the molecular basis for such uniqueness. The genes that we identified as being most characteristic of the otic capsule may not be unique because we did not study the whole genome.
Genes most characteristic of the otic capsule
The emergence of opg as the most characteristic gene of the otic capsule is reassuring because our previous work established the importance of that gene in the inhibition of otic capsule remodeling (Zehnder et al., 2005; Zehnder et al., 2006). The current in situ hybridization data are consistent with our reported immunohistochemical data for Opg protein, suggesting that cells most abundantly expressing opg mRNA also produce the highest levels of the secreted Opg protein. We additionally describe expression of opg in spiral ganglion neurons; functional significance of this finding remains to be determined. Higher levels of opg expression in the cochlear membranous labyrinth than the bony otic capsule (Fig. 2) is consistent with our earlier work (Zehnder et al., 2005). Taken together, high levels of opg expression in the otic capsule detected by quantitative RT-PCR may, at least in part, reflect contamination from the surrounding cochlear tissue or blood.
Another characteristic gene, bmpr1b, encodes for a receptor for BMPs that play essential roles in skeletal development and repair (Chen et al., 2004; Wozney and Rosen, 1998). The importance of BMP pathways for the formation of the otic capsule and maintenance of the membranous labyrinth was established in chicken embryos (Chang et al., 1999; Gerlach et al., 2000; Chang et al., 2002). Ectopic expression of the constitutively active form of bmpr1b caused cartilage overgrowth, whereas the dominant negative form of bmpr1b caused loss of cartilage (Chang et al., 2002). Inhibition of BMP signaling by Noggin leads to a loss of not only the otic capsule but also the membranous epithelium (Gerlach et al., 2000). Our results complement earlier work in embryonic avian otic capsule and suggest that bmpr1b continues to play an important role in the adult mammalian otic capsule. Expression of bmpr1b in cartilaginous rests is intriguing because these rests are unique histologic features of the otic endochondral bone, and they have been implicated in the development of otosclerosis (Bast and Anson, 1949). Specificity of bmpr1b expression in the otic capsule is corroborated by lack of bmpr1b expression in the parietal bone – an intramembranous bone whose development does not involve a cartilage intermediate. Transgenic and knockout mice have established that bmpr1b is required for limb chondrogenesis (Yi et al., 2000; Baur et al., 2000) and bone formation (Zhao et al., 2002); otic capsules of these mice have not been studied. The described human mutations in bmpr1b (Lehmann et al., 2003; Demirhan et al., 2005) are typically not associated with hearing loss.
A final gene highly characteristic of the otic capsule was bmp3 by virtue of its low expression in the otic capsule. The product of this gene is a negative regulator of bone density (Daluiski et al., 2001) and the most abundant BMP in the adult trabecular bone. Given that the otic capsule is the densest bone in the body, its low level of expression of a major negative regulator of bone density is not surprising. We analyzed inner ears of bmp3 knockout mice (Daluiski et al., 2001) and bmp3 over-expressor mice (Gamer et al., 2009) and found no gross abnormalities within the otic capsule or cochlear membranous labyrinth. This suggests a minimal role for bmp3 in controlling remodeling of the otic capsule.
Inflammatory mediators are downregulated in the otic capsule
We found pro-inflammatory cytokines to be downregulated and anti-inflammatory cytokines to be upregulated in the otic capsule compared with tibia and parietal bone, respectively. Inhibition of inflammation in the otic capsule may be important for maintenance of normal hearing because elevated levels of pro-inflammatory cytokines in the cochlea are associated with hearing loss (Brodie and Yeung, 2004), and there is anatomic communication between the otic capsule and cochlear perilymph (Zehnder et al., 2005). Pro-inflammatory cytokines promote pathologic formation of new bone within the membranous labyrinth, known as labyrinthitis ossificans, which is associated with profound deafness and loss of vestibular function. Prompt suppression of inflammatory response by steroids can reduce the incidence of labyrinthitis ossificans and the accompanying hearing loss (Hartnick et al., 2001).
Clustering methods suggest mechanistic relatedness among genes
Although expression levels of individual mRNAs are not always predictive of protein expression and function, co-regulation of genes can predict function (Zhang et al., 2004). Our clustering analysis was based on the assumption that all genes are independent, although many genes that we studied share common biologic pathways. The assumption of independence allowed us to not only confirm that genes within the same biologic pathway cluster together, but also to suggest new functional relatedness among genes that were previously not studied in the otic capsule. For example, clustering of bmpr1b with BMPs (Fig. 4) is consistent with prior description of BMPs and their receptors in the otic capsule where they are thought to promote chondrocyte differentiation (Chang et al, 2002). Moreover, variants in human BMP2 and BMP4 genes have been associated with susceptibility to otosclerosis (Schrauwen et al., 2008). Other genes that cluster with bmpr1b include transforming growth factor beta 2 (tgfb2), which has been shown to stimulate chondrogenic differentiation in cultured periotic mesenchyme (Frenz, 2001), and collagen type II, alpha 1 (col2a1), which encodes for type II collagen in cartilaginous cell rests within the endochondral layer (Ishibe and Yoo, 1990). The remainder of the genes that cluster with bmpr1b were not previously described in the otic capsule. However, they are known to play important roles in cartilage and chondrocyte function.
We found that tgfb1 gene, which has been associated with human otosclerosis (Thys et al., 2009), clustered with pro-inflammatory mediators, consistent with known pro-inflammatory effects of tgfb1 in some settings (Blobe et al., 2000). Our data corroborate the view that otosclerosis involves an inflammatory stage, such as due to a host response to an inciting event (Harris and Keithley, 1993).
Implications for otosclerosis
Otosclerosis, a disease of the otic capsule characterized by abnormal bone remodeling, is influenced by genetic and environmental factors (reviewed by Ealy and Smith, 2009). Linkage studies have identified 8 loci that contribute to familial cases of otosclerosis. Specific causative genes within these loci, which contain many genes, remain to be identified. Genetic association studies, however, have implicated specific genes in pathogenesis of otosclerosis: COL1A1 (McKenna et al, 1998), TGFB1 (Thys et al, 2009), BMP2 (Schrauwen et al, 2008), BMP4 (Schrauwen et al, 2008) angiotensin I AGT (Imauchi et al, 2008), angiotensin I converting enzyme (ACE, Imauchi et al, 2008), and reelin (RELN, Schrauwen et al, 2009; Schrauwen et al, 2009b). Only COL1A1 was known at the time we began the current st udy. We found col1a1 to be expressed at lower levels in the otic capsule than other bones (Fig. 1). However, col1a2 – a binding partner of col1a1 to form functional type I collagen – emerged as a more robust predictor of the otic capsule than col1a1 in the current study. Studies have not found association of COL1A2 with otosclerosis (McKenna et al., 1998; Rodriguez et al., 2004).
We found tgfb1 to be expressed at higher levels in the otic capsule than in parietal bone, and not significantly different than in tibia (Fig. 1). Clustering of tgfb1 with pro-inflammatory mediators, as well as col1a1 and col1a2 (Fig. 4), suggests that future studies of pro-inflammatory cytokines in the otic capsule may provide mechanistic insights into pathogenesis of otosclerosis.
It is interesting that BMP2 and BMP4 have recently been associated with otosclerosis, and they bind to BMPR1B, which we have identified to be characteristic of the otic capsule. This suggests that understanding BMP signaling in the human otic capsule may be relevant for understand unique predisposition of this bone to otosclerosis. Mechanisms of AGT, ACE and RELN action in the otic capsule await further characterization.
Summary
We have shown that the molecular profile of the otic capsule is distinctly different from that of the tibia and parietal bone. The genes that we identified as being most characteristic of the otic capsule play a role in inhibition of bone remodeling. Dysregulation of these genes may contribute to the pathogenesis of otosclerosis, which is characterized by pathologic remodeling of the otic capsule.
Supplementary Material
Acknowledgments
We thank Dr. Saumil Merchant for helpful comments on earlier versions of the manuscript. This work was supported by NIDCD Grant 5RO1 DC03401-06 (MJM), American Otological Association (KMS) and Mr. Lakshmi Mittal (MJM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: All authors have no conflicts of interest.
Contributor Information
Osamu Adachi, Email: osamu_adachi@meei.harvard.edu.
Kunikazu Tsuji, Email: kunikazu_tsuji@hsdm.harvard.edu.
Arthur G. Kristiansen, Email: kris@epl.meei.harvard.edu.
Joe C. Adams, Email: jca@meei.harvard.edu.
Vicki Rosen, Email: vicki_rosen@hsdm.harvard.edu.
Michael J. McKenna, Email: michael_mckenna@meei.harvard.edu.
References
- Bast TH, Anson BJ. The Temporal Bone and the Ear. Springfield, IL: Charles C. Thomas; 1949. [Google Scholar]
- Baur ST, Mai JJ, Dymecki SM. Combinatorial signaling through BMP receptor1B and GDF5: shaping of the distal mouse limb and the genetics of distal limb diversity. Development. 2000;127(3):605–619. doi: 10.1242/dev.127.3.605. [DOI] [PubMed] [Google Scholar]
- Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Brodie HA, Yeung AH. Labyrinthitis ossificans. Emedicine. 2004 http://www.emedicine.com/ENT/topic409.htm. [Google Scholar]
- Chang W, Nunes FD, De Jesus-Escobar JM, Harland R, Wu DK. Ectopic noggin blocks sensory and nonsensory organ morphogenesis in the chicken inner ear. Dev. Biol. 1999;216(1):369–381. doi: 10.1006/dbio.1999.9457. [DOI] [PubMed] [Google Scholar]
- Chang W, ten Dijke P, Wu DK. BMP pathways are involved in otic capsule formation and epithelial-mesenchymal signaling in the developing chicken inner ear. Dev. Biol. 2002;251(2):380–394. doi: 10.1006/dbio.2002.0822. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22(4):233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, Stevenson SL, Cox K, Rosen V, Lyons KM. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet. 2001;27(1):84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- Demirhan O, Turkmen S, Schwabe GC, Soyupak S, Akgul E, Tastemir D, Karahan D, Mundlos S, Lehmann K. A homozygous BMPR1B mutation causes a new subtype of acromesomelic chondrodysplasia with genital anomalies. J Med. Genet. 2005;42(4):314–317. doi: 10.1136/jmg.2004.023564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ealy M, Smith RJ. The Genetics of otosclerosis. Hear. Res. 2009 Jul 14; doi: 10.1016/j.heares.2009.07.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenz D. Growth factor control of otic capsule chondrogenesis. Einstein Qaurt. J. Bio. Med. 2001;18:7–14. [Google Scholar]
- Frisch T, Sorensen MS, Overgaard S, Lind M, Bretlau P. Volume-referent bone turnover estimated from the interlabel area fraction after sequential labeling. Bone. 1998;22:677–682. doi: 10.1016/s8756-3282(98)00050-7. [DOI] [PubMed] [Google Scholar]
- Gamer LW, Cox K, Carlo JM, Rosen V. Overexpression of BMP3 in the developing skeleton alters endochondral bone formation resulting in spontaneous rib fractures. Dev. Dyn. 2009;238(9):2374–2381. doi: 10.1002/dvdy.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GenePattern 2.0. http://www.broad.mit.edu/genepattern/. [Google Scholar]
- Gerlach LM, Hutson MR, Germiller JA, Nguyen-Luu D, Victor JC, Barald KF. Addition of the BMP4 antagonist, noggin, disrupts avian inner ear development. Development. 2000;127(1):45–54. doi: 10.1242/dev.127.1.45. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. 6th. part 3. chapter 14. Sinauer Assoicates, Inc.; 2000. [Google Scholar]
- Goldring SR. Inflammatory mediators as essential elements in bone remodeling. Calcif. Tissue Int. 2003;73:97–100. doi: 10.1007/s00223-002-1049-y. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Harris JP, Keithley EM. Inner ear inflammation and round window otosclerosis. Am. J. Otol. 1993;14(2):109–112. [PubMed] [Google Scholar]
- Hartnick CJ, Kim HH, Chute PM, Parisier SC. Preventing labryrinthitis ossificans: the role of steroids. Arch Otolaryngol Head Neck Surg. 2001;127(2):180–183. doi: 10.1001/archotol.127.2.180. [DOI] [PubMed] [Google Scholar]
- Ishibe T, Yoo TJ. Type II collagen distribution in the monkey ear. Am. J. Otol. 1990;11(1):33–38. [PubMed] [Google Scholar]
- Heinrich J, Bsoul S, Barnes J, Woodruff K, Abboud S. CSF-1, RANKL and OPG regulate osteoclastogenesis during murine tooth eruption. Arch. Oral. Biol. 2005;50(10):897–908. doi: 10.1016/j.archoralbio.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Imauchi Y, Jeunemaitre X, Boussion M, Ferrary E, Sterkers O, Grayeli AB. Relation between renin-angiotensin-aldosterone system and otosclerosis: a genetic association and in vitro study. Otol. Neurotol. 2008;29:295–301. doi: 10.1097/mao.0b013e318164d12c. [DOI] [PubMed] [Google Scholar]
- Lehmann K, Seemann P, Stricker S, Sammar M, Meyer B, Suring K, Majewski F, Tinschert S, Grzeschik KH, Muller D, Knaus P, Nurnberg P, Mundlos S. Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2. Proc. Natl. Acad. Sci. U.S.A. 2003;100(21):12277–12282. doi: 10.1073/pnas.2133476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie RD. Histologic Technic and Practical Histochemistry. New York: McGraw-Hill; 1965. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McKenna MJ, Kristiansen AG, Bartley ML, Rogus JJ, Haines JL. Association of COL1A1 and otosclerosis: evidence for a shared genetic etiology with mild osteogenesis imperfecta. Am. J. Otol. 1998;19:604–610. [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: a resampling based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52:91–118. [Google Scholar]
- Namiki M, Akiyama S, Katagiri T, Suzuki A, Ueno N, Yamaji N, Rosen V, Wozney JM, Suda T. A kinase domain-truncated type I receptor blocks bone morphogenetic protein-2-induced signal transduction in C2C12 myoblasts. J. Biol. Chem. 1997;272(35):22046–22052. doi: 10.1074/jbc.272.35.22046. [DOI] [PubMed] [Google Scholar]
- Perlman HB. Process of healing in injuries to the capsule of labyrinth. Arch Otolaryngol. 1939;29:287–305. [Google Scholar]
- Reno C, Marchuk L, Sciore P, Frank CB, Hart DA. Rapid isolation of total RNA from small samples of hypocellular, dense connective tissues. BioTechniques. 1997;22:1082–1086. doi: 10.2144/97226bm16. [DOI] [PubMed] [Google Scholar]
- Rodríguez L, Rodríguez S, Hermida J, Frade C, Sande E, Visedo G, Martín C, Zapata C. Proposed association between the COL1A1 and COL1A2 genes and otosclerosis is not supported by a case-control study in Spain. Am. J. Med. Genet. A. 2004;128A(1):19–22. doi: 10.1002/ajmg.a.30074. [DOI] [PubMed] [Google Scholar]
- Sassoon D, Rosenthal N. Detection of messenger RNA by in situ hybridization. Methods Enzymol. 1993;225:384–404. doi: 10.1016/0076-6879(93)25027-y. [DOI] [PubMed] [Google Scholar]
- Schrauwen I, Thys M, Vanderstraeten K, Fransen E, Dieltjens N, Huyghe JR, Ealy M, Claustres M, Cremers CR, Dhooge I, Declau F, Van de Heyning P, Vincent R, Somers T, Offeciers E, Smith RJ, Van Camp G. Association of bone morphogenetic proteins with otosclerosis. J. Bone Miner. Res. 2008;23(4):507–516. doi: 10.1359/JBMR.071112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Ealy M, Huentelman MJ, Thys M, Homer N, Vanderstraeten K, Fransen E, Corneveaux JJ, Craig DW, Claustres M, Cremers CW, Dhooge I, Van de Heyning P, Vincent R, Offeciers E, Smith RJ, Van Camp G. A genome-wide analysis identifies genetic variants in the RELN gene associated with otosclerosis. Am. J. Hum. Genet. 2009;84(3):328–338. doi: 10.1016/j.ajhg.2009.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Ealy M, Fransen E, Vanderstraeten K, Thys M, Meyer NC, Cosgarea M, Huber A, Mazzoli M, Pfister M, Smith RJ, Van Camp G. Genetic variants in the RELN gene are associated with otosclerosis in multiple European populations. Hum. Genet. 2009b Oct 22; doi: 10.1007/s00439-009-0754-2. [DOI] [PubMed] [Google Scholar]
- Schuknecht HF. Pathology of the ear. Philadelphia, USA: Lea & Febiger; 1993. p. 672. [Google Scholar]
- Sorensen MS, Bretlau P, Jorgensen MB. Human perilabyrinthine bone dynamics. A functional approach to temporal bone histology. Acta Otolaryngol. Suppl. 1992;496:1–27. [PubMed] [Google Scholar]
- Sorensen MS, Bretlau P, Jorgensen MB. Quantum type bone remodeling in the otic capsule of the pig. Acta Otolaryngol. 1990;110:217–223. doi: 10.3109/00016489009122540. [DOI] [PubMed] [Google Scholar]
- Stankovic KM, Corfas G. Real-time quantitative RT-PCR for low-abundance transcripts in the inner ear: analysis of neurotrophic factor expression. Hear. Res. 2003;185(1–2):97–108. doi: 10.1016/s0378-5955(03)00298-3. [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA. 1999;96(6):2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenghi G. Detecting mRNA in tissue sections with digoxigenin-labeled probes. Methods. Mol. Biol. 1998;86:137–142. doi: 10.1385/0-89603-494-1:137. [DOI] [PubMed] [Google Scholar]
- Thys M, Schrauwen I, Vanderstraeten K, Janssens K, Dieltjens N, Van Den Bogaert K, Fransen E, Chen W, Ealy M, Claustres M, Cremers CR, Dhooge I, Declau F, Claes J, Van de Heyning P, Vincent R, Somers T, Offeciers E, Smith RJ, Van Camp G. The coding polymorphism T263I in TGF-beta1 is associated with otosclerosis in two independent populations. Hum Mol Genet. 2007;16(17):2021–2030. doi: 10.1093/hmg/ddm150. [DOI] [PubMed] [Google Scholar]
- Thys M, Schrauwen I, Vanderstraeten K, Dieltjens N, Fransen E, Ealy M, Cremers CW, van de Heyning P, Vincent R, Offeciers E, Smith RH, van Camp G. Detection of rare nonsynonymous variants in TGFB1 in otosclerosis patients. Ann. Hum. Genet. 2009;73(2):171–175. doi: 10.1111/j.1469-1809.2009.00505.x. [DOI] [PubMed] [Google Scholar]
- Trepicchio WL, Dorner AJ. Interleukin-11: a gp130 cytokine. Ann. NY Acad. Sci. 1998;856:12–21. doi: 10.1111/j.1749-6632.1998.tb08308.x. [DOI] [PubMed] [Google Scholar]
- Zehnder AF, Kristiansen AG, Adams JC, Merchant SN, McKenna MJ. Osteoprotegerin in the inner ear may inhibit bone remodeling in the otic capsule. Laryngoscope. 2005;115:172–177. doi: 10.1097/01.mlg.0000150702.28451.35. [DOI] [PubMed] [Google Scholar]
- Zehnder AF, Kristiansen AG, Adams JC, Kujawa SG, Merchant SN, McKenna MJ. Osteoprotegerin knockout mice demonstrate abnormal remodeling of the otic capsule and progressive hearing loss. Laryngoscope. 2006;116(2):201–206. doi: 10.1097/01.mlg.0000191466.09210.9a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, Laurin N, Eftekharpour E, Sat E, Grigull J, Pan Q, Peng WT, Krogan N, Greenblatt J, Fehlings M, van der Kooy D, Aubin J, Bruneau BG, Rossant J, Blencowe BJ, Frey BJ, Hughes TR. The functional landscape of mouse gene expression. J. Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Harris SE, Horn D, Geng Z, Nishimura R, Mundy GR, Chen D. Bone morphogenetic protein receptor signaling is necessary for normal murine postnatal bone formation. J. Cell. Biol. 2002;157(6):1049–1060. doi: 10.1083/jcb.200109012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozney JM, Rosen V. Bone morphogenetic protein and bone morphogenetic protein gene family in bone formation and repair. Clin. Orthop. Relat. Res. 1998;346:26–37. [PubMed] [Google Scholar]
- Yi SE, Daluiski A, Pederson R, Rosen V, Lyons KM. The type I BMP receptor BMPR1B is required for chondrogenesis in the mouse limb. Development. 2000;127(3):621–630. doi: 10.1242/dev.127.3.621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.