Abstract
RNA interference (RNAi) is a key regulator of various biological systems including viral infection. Within a virus life cycle gene products can be modulated by the RNA interference (RNAi) pathway which can crucially impact productive virus replication. Herein we explored the RNA interference suppressor protein P19 derived from a plant virus and we found that P19 enhanced adenovirus replication up to 100-fold. Critical factors responsible for this observation were overexpression of adenovirus encoded genes on mRNA and protein levels. To investigate the impact of this phenomenon on recombinant viruses, we exploited its feasibility for therapeutic and genomic applications. We found that P19 significantly increased recombinant adenovirus yields enabling up-scaling for preclinical and clinical studies. Moreover, adenoviruses possessed significantly higher oncolytic activity by expression of P19. Finally, we show that introducing a p19 expression cassette into high-capacity adenovirus provides a strategy to analyze RNAi knockdown in a tissue-specific manner.
The understanding of basic virus host interactions is a key pre-condition to understand basic biology of viruses and to develop and improve viral vector systems for gene therapy. Over the recent years it became clear that the RNA interference (RNAi) system represents a major posttranscriptional regulatory mechanism which is involved in a variety of cellular, physiological and developmental mechanisms. One of the key players of the RNAi system are microRNAs as endogenous non-coding RNAs which were shown to be endogenously expressed within mammalian cells but also from human pathogenic viruses1,2. Within a virus life cycle several gene products can be modulated by host cell factors or mechanisms such as the RNA interference (RNAi) pathway that can crucially influence productive virus replication1. This was shown for numerous viruses including retrovirus primate foamy virus type 1 (PFV-1)3, herpes simplex virus 1 (HSV1)4, Epstein-Barr virus (EBV)5, cytomegalovirus (CMV)6, and simiam virus 40 (SV40)7.
Adenovirus with the ability to infect a wide range of dividing and non-dividing cells has been broadly explored in basic virology and therapeutic approaches and remains to be one of the most potent viruses for efficient DNA transfer, vaccine development and oncolytic applications. However, with respect to the influence of the RNAi pathway on adenovirus infection as well as on the performance of adenovirus vectors virtually no information is available. The only adenovirus products known to suppress the RNAi pathway are represented by adenoviral virus-associated RNAs (VA-RNAs)8. VA-RNAs share the export mechanism with cellular miRNAs, are similarly processed by Dicer into small virus-associated RNAs (sva-RNAs) and are loaded into the RISC complex9. The function of these sva-RNAs is still unknown but very recently the TIA-1 protein could be identified as one target protein9.
P19, which is derived from the tomato bushy stunt virus, binds and inhibits 21 nucleotides long, small-interfering RNAs and was shown to suppress the RNAi pathway. In our previous study we explored the RNAi inhibitor P19 and its influence on transposition activities in mammalian cells10. Herein, we explored the RNAi suppressor protein P19 and its influence on adenovirus infection. To analyze the influence of P19 on adenovirus replication, P19 was either stably expressed in human embryonic kidney cells (B6 cells)10 or directly expressed from the adenoviral vector genome. We found that genome replication and productive virus infection of replication-competent adenovirus was enhanced up to 100-fold and 10-fold, respectively and we observed a massive overproduction of various adenovirus genes on RNA and protein level. As a first step, we translated this finding into increased production of first-generation adenoviral vectors (FgAdV) deleted for the early adenovirus genes E1 and E3 and we observed significantly enhanced production of high-capacity adenoviral vectors (HCA) deleted for all viral coding sequences. The latter vectors combine major advantages compared to FgAdV because they show an improved safety profile as well as long-term transgene expression in small and large animal models9,11,12,13. Furthermore, we found that activity of oncolytic adenoviruses for tumor cell-specific replication and lysis14,15,16 was significantly enhanced in the presence of P19. Finally, we show that the P19 system can also be utilized for RNAi knockdown in mice in a tissue-specific manner.
Results
The RNAi suppressor P19 significantly enhances adenovirus replication
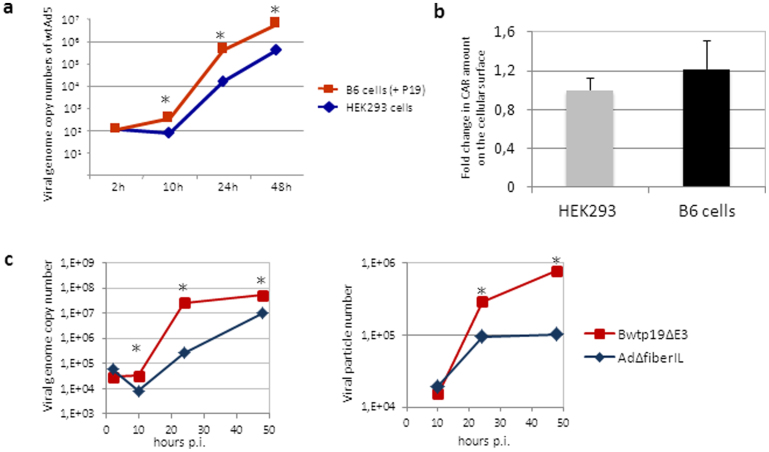
To investigate whether the adenovirus replication cycle is influenced by the RNAi pathway, we explored an RNAi knockdown system based on the RNAi suppressor protein P1917, which is derived from the tomato bushy stunt virus. In our initial experiments we generated stably P19 expressing human embryonic kidney cells (B6 cells) which in contrast to the parental cell line supported up to 10-fold enhanced replication of wildtype adenovirus genome copies (Fig. 1a). Notably, stable expression of the RNAi suppressor protein P19 had no influence on the expression levels of the coxsackie- and adenovirus receptor (CAR) and thus equal infection efficiencies during the early steps of virion uptake can be expected even under RNAi knockdown conditions (Fig. 1b).
Figure 1. The RNAi suppressor P19 enhances adenovirus replication.
(a) Replication of wild type adenovirus serotype 5 (wtAd5) in the RNAi knockdown cell line B6 stably expressing P19 and the parental cell line HEK293. HEK293 and B6 cells were infected at an MOI of 0.05 with wild type adenovirus (wtAd5) and genomic DNA was isolated 2 hrs, 10 hrs, 24 hrs and 48 hrs post infection. Viral genome copy numbers were quantified using hexon specific primers for quantitative real-time PCR and were normalized to expression levels of human B2m. All results are statistically relevant with a p-value < 0.05. (b) Quantification of CAR expression. HEK293 and B6 cells were stained with an anti-CAR antibody labeled with FITC and measured by flow cytometry. Light emission of HEK293 cells after treatment was set to one and the emission of B6 cells was calculated and expressed as fold change. (c) Direct comparison of adenovirus replication and particle production of the P19 expressing adenovirus Bwtp19ΔE3 and the control virus AdΔfiberIL. Viral genome copy numbers (left panel) and infectious viral particle numbers directly correlating with virus production efficiency (right panel) were analyzed.
Since the P19 expression level is critical for its function18 we aimed at achieving high p19 expression upon infection by coupling P19 expression to virus replication. Therefore, we inserted the P19 cDNA into the adenovirus genome under the control of the major late promoter (MLP) by connecting it to the fiber gene via a spacer and an internal ribosomal entry site (IRES) sequence resulting into the recombinant adenovirus Bwtp19ΔE3 (Supplementary Fig. S1a). This adenovirus was reconstituted (Supplementary Fig. S1b) and P19 expression was confirmed by reverse transcription PCR (Supplementary Fig. S1c). P19 expression levels from the P19 expressing virus Bwtp19ΔE3 increased proportionally to the virus replication profile, whereas P19 expression levels from the stably P19 expressing cell line B6 were significantly decreased 48 hours post-infection (Supplementary Fig. S1d), supporting the notion that the amount of P19 molecules within a cell are critical for the repression of the RNAi pathway. Notably, at later time points during infection P19 expression levels derived from the virus Bwtp19ΔE3 were increased up to 100-fold in comparison to the stably p19 expressing cell line B6.
Next, we analyzed replication and productive virus particle yields of the P19 expressing virus Bwtp19ΔE3 in comparison to the respective control virus AdΔfiberIL expressing luciferase instead of P19 (Supplementary Fig. S2). As displayed in Fig. 1c, adenovirus genome replication and virus particle production were enhanced up to 100-fold and 10-fold, respectively, when infecting with the P19 expressing virus. Virus replication was significantly increased 24 hours post-infection and adenovirus particle production showed the highest difference 48 hours post-infection. This result indicated that the RNAi pathway plays a suppressive role during the natural adenovirus life cycle.
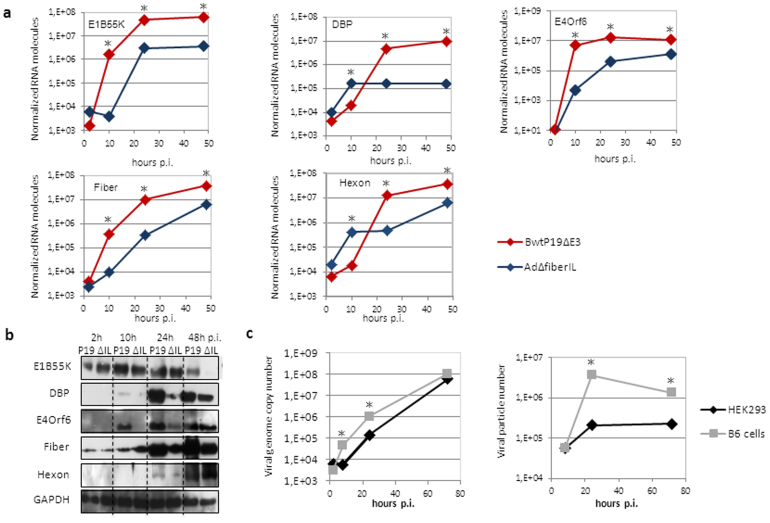
In order to identify critical factors potentially responsible for the up-regulation of virus replication, we investigated expression levels of different structural (Hexon and Fiber) and non-structural viral proteins (E1B55K, DBP, E4Orf6) by quantification of respective mRNA and protein levels. As demonstrated in Fig. 2a we observed a massive increase in mRNA levels of all viral genes tested after infection with Bwtp19ΔE3 compared to the results obtained with the respective control virus AdΔfiberIL not expressing P19 (Supplementary Fig. S2). Significantly increased mRNA expression levels of early adenovirus genes E1B55K and E4Orf6 and the structural protein fiber were observed as early as 10 hours post-infection and highest expression levels of DBP and the structural protein hexon were observed at later time points. In contrast, only protein levels of the early viral transcripts E1B55K, DBP, E4Orf6 and the fiber protein were enhanced whereas the structural protein hexon was equally expressed upon infection with both viruses (Fig. 2b). However, it is known that viral structural proteins are abundantly expressed and therefore one could speculate that their translation may be saturated and subsequently abrogated upon sufficient virus production.
Figure 2. Analysis of adenovirus replication and infection parameters under RNAi knockdown conditions.
(a) Adenoviral early and late RNA expression levels upon infection of HEK293 cells with viruses Bwtp19ΔE3 and AdΔfiberIL. Quantitative real-time PCR was performed using gene specific primers for early adenovirus genes E1B55K, DBP, E4Orf6 and late adenovirus genes fiber and hexon. *: p-value < 0.05. (b) Western Blot analysis for detection of viral early and late proteins after Bwtp19ΔE3 (P19) or AdΔfiberIL (ΔIL) infection. (c) Analysis of genome replication and particle production of an adenovirus deleted for virus-associated RNAs in the RNAi knockdown cell line B6 in comparison to HEK293 cells. *: p-value < 0.05. p.i: post-infection; h: hours.
To further shed light on the mechanism involved in increased virus replication we analyzed expression and fate of the adenoviral virus-associated RNAs (VA-RNAs), which are the only adenovirus products known to suppress the RNAi pathway8. In order to investigate the role of P19 and VA-RNAs during virus replication, we monitored the replication profile of a VA-RNA deleted adenovirus in HEK293 cells and the stably P19 expressing RNAi knockdown cell line B6. As shown in Fig. 2c, virus replication was still enhanced even without the VA-RNAs, suggesting that P19 can substitute the natural function of VA-RNAs in the RNA interference pathway and that during wild type adenovirus (wtAd5) infection P19 complements the function of VA-RNAs. Moreover, after wtAd5 infection and isolation of p19-bound small RNAs, we found that P19 sequestered svaRNAs either from VAI-RNA or VAII-RNA origin and subsequently degraded them (Supplementary Fig. S3a–S3c).This underlines the function of P19 to bind small-interfering RNAs and can be explained by the high copy numbers (up to 108 copies per cell)19 of adenoviral VA-RNAs and svaRNAs during adenovirus infection.
Substantial improved production of recombinant adenoviral vectors in the presence of P19
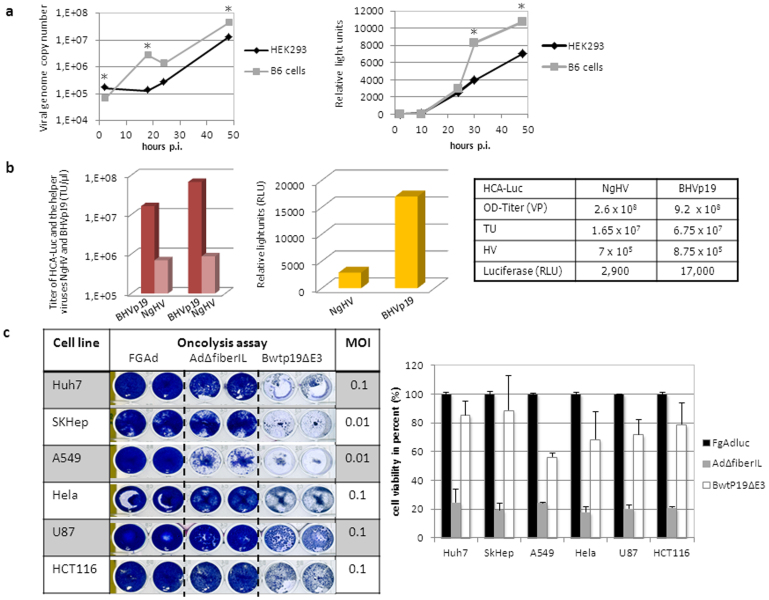
To exploit the full potential of the P19-based RNAi knockdown system and its influence on adenovirus replication we applied it for therapeutic as well as genomic applications. First-generation adenoviral vectors (FgAdV) deleted for the early adenovirus genes E1 and E3 are commonly used as standard vectors for robust gene transfer and were used in various preclinical and clinical studies. For such applications virus production at high titers is a prerequisite and therefore, we addressed the question whether a stably expressing P19 cell line based on the human embryonic kidney cell line HEK293 can be used for increased FgAdV production. In concordance with studies performed with wild type adenovirus we found that a FgAdV expressing luciferase replicates faster (Fig. 3a, left panel) and grows to higher titers (Fig. 3a, right panel) in the presence of the P19 protein. Therefore, our RNAi knockdown cell line B6 can be used as an FgAdV producer cell line enabling superior virus production.
Figure 3. The RNA interference inhibitor P19 improves adenovirus applications and therapeutics.
(a) Increased replication and particle production of a first-generation adenovirus expressing luciferase (FgAdluc) in the RNAi knockdown cell line B6 stably expressing P19. Viral genome copy numbers and infectious virus particle production expressed in luciferase activity (relative light units) were determined. *: p-value < 0.05. (b) Improved high-capacity adenoviral vector large-scale production using the P19 expressing helper virus BHVp19 in comparison to the conventionally used helper virus AdNG163R-2 (NgHV). In final vector preparations infectious units expressed as transducing units (TU, left panel) of HCA-Luc and both helper viruses were determined. In addition, luciferase levels (relative light units, middle panel) were measured for HCA-Luc directly correlating with infectious viral particle production. The table summarizes total viral particle numbers (VP, OD-titer) and infectious units (TU) of HCA-Luc and helper viruses (HV) in the final vector preparation. (c) Bwtp19ΔE3 displays increased oncolytic potential in comparison to the control virus AdΔfiberIL. Different cancer cell lines (liver-derived Huh7 and SKHep cells, lung-derived A549 cells, cervix-derived HeLa cells, glioblastoma-derived U87 cells and colon-derived HCT116 cells) were infected and stained (left panel) and the oncolytic activity was quantified (right panel).
At the time the most advanced version of recombinant adenoviral vectors used in gene-therapy are represented by high-capacity adenoviral vectors (HCA) deleted for all viral coding sequences. However, the production procedure remains a major hurdle for clinical use of this vector system. For generation of HCA a helper virus with a floxed packaging signal is required which provides all adenoviral coding sequences in trans20,21. A schematic outline about HCA production and large-scale amplification in suspension culture is displayed in Supplementary Fig. S4. In the present study we speculated that a helper virus expressing P19 results in production of HCA at higher final titers due to increased expression of viral proteins. Therefore, we generated a p19 expressing helper virus utilizing a cloning strategy based on bacterial artificial chromosomes (BHVp19, Supplementary Fig. S5a). We expected to obtain higher titers of a HCA expressing luciferase (HCA-Luc, Supplementary Fig. S5d) when using the novel helper virus BHVp19 instead of the conventionally used helper virus AdNG163R-221 (NgHV, Supplementary Fig. S5d). Both small (Supplementary Fig. S5e) and large scale amplification (Fig. 3b) of HCA-Luc using the helper virus BHVp19 revealed up to 6-fold enhanced viral titers. Importantly, large scale amplification was associated with low levels of unwanted helper virus contamination in the final vector preparation (Fig. 3b), demonstrating that the purity of the final vector preparation with respect to helper virus contamination levels is comparable to the conventionally used helper virus system for HCA production. Thus, the RNAi suppressor protein P19 is sufficient to enhance the infectious titers of HCA and subsequently improve the production protocol of HCA vectors for clinical use.
Expression of P19 from an oncolytic adenovirus enhances killing of tumor cells
Recombinant adenoviruses have been widely investigated for various clinical applications including tumor treatment and vaccination. Most clinical studies in which adenovirus played a major role aimed at finding novel strategies for cancer treatment. Recombinant adenoviruses used in anti-cancer strategies have been mainly based on oncolytic adenoviruses which allow tumor cell-specific replication and lysis14,15,16. However, one major limitation to overcome is their slow replication rate in cancer tissues, which in many cases is not sufficient to kill all cancer cells. Here, we speculated that an oncolytic adenovirus expressing the RNAi suppressor P19 results in increased virus replication and enhanced viral oncolysis. Thus, we evaluated the P19 expressing adenovirus and directly compared its oncolytic potential with the oncolytic control adenovirus AdΔfiberIL, expressing luciferase instead of P19 (Supplementary Fig. S2a). We analyzed the P19 expressing virus and the control virus in different tumor cell lines derived from liver (Huh7, SKHep cells), lung (A549 cells), cervix (HeLa cells), glioblastoma (U87 cells) and colon (HCT116 cells) (Fig. 3c, left panel) and obtained an up to 6-fold higher oncolytic potential for Bwtp19ΔE3 (Fig. 3c, right panel). We concluded that introducing the P19 gene into an oncolytic background, for example an adenovirus with the Δ24 E1A deletion or targeted E1A expression, represents a promising strategy to improve the therapeutic outcome of oncolytic adenoviruses.
Cell-type and tissue specific knock-down of micro RNAs in vitro and vivo
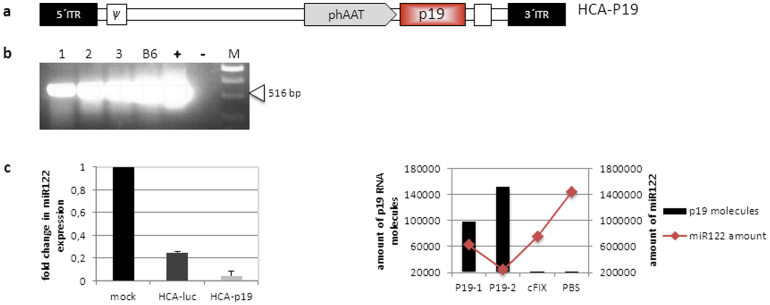
In the latter approaches we aimed at accelerating virus replication and increasing adenovirus production by stably expressing P19 in a cell line or by providing P19 in cis from a replicating adenovirus genome. However, another strategy for using P19 in concert with adenoviral vectors would be adenoviral delivery of P19 for tissue-specific reduction of miRNA expression in vitro and in vivo. In this study we took advantage of P19 which can be delivered and expressed in a tissue-specific manner using recombinant viruses eventually leading to tissue-specific knock-down of the RNAi pathway. Therefore, we generated a high-capacity adenovirus expressing p19 under the liver-specific human alpha-1-antitrypsin promoter22 (HCA-p19, Fig. 4a and 4b). The human hepatoma cell line Huh7 was transduced with this virus and molecules of the liver-specific miR12223 were quantified. Respective control groups were either mock infected of received a control virus HCA-luc expressing luciferase as a marker gene. Quantification of miR122 revealed a 5-fold downregulation of this miRNA in comparison to the control virus (Fig. 4c, left panel). To analyze miRNA suppression in vivo, we infected C57Bl/6 mice with the p19 expressing virus HCA-p19 and the control virus lacking a P19 expression cassette. Two month post-injection mice were sacrificed and the most abundant liver-specific miR122 quantified. As displayed in Fig. 4c (right panel) high P19 expression levels strongly correlated with the miRNA downregulation effect and therefore our approach was sufficient to reduce miR122 levels in murine liver. Since any cell-type specific promoter can be delivered utilizing an adenoviral vector, this system significantly simplifies in vivo knockdown of the RNAi pathway.
Figure 4. Cell-type and tissue specific knock-down of micro RNAs in vitro and vivo using a P19 encoding high-capacity adenoviral vector.
(a) Vector construct used to generate the p19 expressing high-capacity adenoviral vector HCA-P19 expressing P19 under the control of a liver-specific promoter. phAAT: human alpha-1-antitrypsin promoter; p19: p19 cDNA; polyA: polyadenylation signal of the SV40 virus; ITR: inverted terminal repeats; Ψ: packaging signal. (b) Expression of p19 after infection of HEK293 cells with HCA-p19 at an MOI of 3 (lanes 1, 2 and 3). As a positive control for P19 expression, RNA from stably expressing P19 cells (B6 cells) or cells transfected with the plasmid pHCA-p19 (+) was isolated. RNA was extracted and analyzed using P19 specific primers. A band of 519 bp in length indicates functional P19 expression. (c) The P19 expressing high-capacity adenoviral vector HCA-P19 reduces miR122 expression in vitro (left panel) and in murine liver (right panel). For in vitro analysis Huh7 cells were either infected with HCA-P19, the control virus HCA-luc or mock infected. RNA was isolated, polyA-tailed, reverse transcribed and quantitative real-time PCR (qRT-PCR) reactions using miR122 and P19 specific primers were performed. All qRT-PCR data were normalized to 104 human B2m levels. C57BL/6 mice (P19-1, P19-2) were infected with 5*109 TU of the high-capacity adenoviral vector HCA-P19, the negative control virus (cFIX) or PBS as control. Two months post-injection mice were sacrificed, small RNAs were isolated from liver and analyzed by qRT-PCR.
Discussion
In our pivotal study we found that adenovirus replication was enhanced 100-fold (Fig. 1c) in the presence of P19 which was accompanied by overexpression of adenovirus encoded genes on mRNA and protein levels (Fig. 2a and 2b). Although this is a very unique finding, further studies are required to identify the molecular basis responsible for upregulation of virus replication and particle production. For instance mRNA arrays of cellular and viral proteins will help to characterize single genes or a family of genes, which are altered and contribute to the increase in viral replication.
Next, we could show that enhanced adenovirus replication and simultaneous expression of P19 can be translated into enhanced production of recombinant adenoviruses and oncolysis. We obtained higher viral titers of first-generation and high-capacity adenoviral vectors (Fig. 3a and b) and we observed significantly enhanced oncolysis in various tumor cells (Fig. 3c). However, we expressed P19 under the control of the major late viral promoter and it would be of great interest to analyze the replication profile of an adenovirus expressing P19 under the control of an early viral promoter to potentially further accelerate virus replication. Furthermore, the P19-based approach could result in an enhanced spread of the oncolytic virus in tumors of affected patients and it could be used to improve oncolytic activity of other RNA and DNA based oncolytic viruses.
We used our high-capacity adenovirus technology for delivery of the P19 protein and to establish a novel strategy for cell-type and tissue-specific knock-down of the RNA interference pathway (Fig. 4). At the time tissue-specific in vivo knockdown systems of the RNAi pathway are mainly based on disruption of dicer as the key regulator of miRNA processing utilizing the Cre-loxP system24. However, this strategy requires the generation and maintenance of a transgenic mouse with floxed dicer alleles and tissue-specific delivery of Cre recombinase or a second transgenic mouse overexpressing Cre under the control of a cell-type specific promoter. Therefore, an alternative strategy for cell type-specific knockdown of the RNA interference pathway based on P19 provides a valuable alternative. This strategy is less labor intensive and can be used in any tissue because adenovirus targeting strategies25 and cell-type specific promoters26 are broadly available.
Methods
Analysis of adenovirus replication and infection parameters
Analysis of adenovirus replication
Wildtype adenovirus serotype 5 (wtAd5) was obtained from ATCC (Wesel, Germany). Wildtype adenovirus, first-generation adenoviruses and BAC-based viruses were and amplified in HEK293 cells. The VA-RNA deleted virus obtained from Ramon Alemany (Institut Català d'Oncologia/IDIBELL, L'Hospitalet Barcelona, Spain) and the virus AdfiberΔIL were amplified in A549 cells as described previously27,28. For production of the replication-competent adenoviruses Bwtp19ΔE3 please refer to Supplementary Methods 1.
For replication assays cells were seeded into 6 cm dishes to reach 80% confluency for infection. Notably, for infection of B6 cells were cultivated without G418. Infection with wtAd5, FgAdluc or respective BAC derived viruses was performed using a multiplicity of infection (MOI) of 0.05, 0.1 or 3 respectively. After two 10, 24, and 48 hours (if not otherwise stated) DNA and RNA were isolated and cellular and protein lysates were collected, taking 3 independent samples at each time point. To ensure, that only internalized viral particles were analyzed, cells were treated with 5% trypsin for 5 min, centrifuged at 500 g for 3 min and washed once with sterile PBS before preparing for different applications.
For analysis of genome replication and particle production of an adenovirus deleted for virus-associated RNAs, B6 and HEK293 cells were infected at an MOI of 0.5 and virus replication and particle production were monitored.
For analysis of infection parameters such as RNA expression levels of adenoviral genes, vector genome copy numbers, and adenoviral expression on protein level please refer to Supplementary Methods 2.
For analysis of CAR expression on HEK293 cells and stably P19 expressing B6 cells please refer to Supplementary Methods 3.
Improved production of recombinant adenoviruses
Enhanced production of first generation adenoviral vectors
We investigated whether a HEK293 based cell line stably expressing the RNAi inhibitor P19 could lead to enhance replication and production of early generation adenoviral vectors deleted for the adenoviral early gene E1 and E3. Human embryonal kidney cells (HEK293) were obtained from ATCC and used for construction of the p19 expressing cell line B6. This adenovirus producer cell line B6 which substitutes E1 for adenovirus amplification and it was stably transduced with a trangene expression cassette for P1910. As selection pressure for B6 cells which stably express P19, 500 μg/ml G418 was added. For direct comparison of virus production in commonly used HEK293 cells and B6 cells, we analyzed replication and particle production of the previously described first-generation adenovirus FgAdluc expressing luciferase under the control of the SV40 promoter29.
HEK293 cells and the RNAi knockdown cell line B6 were infected with FgAdluc at an MOI of 0.5. Genomic DNA and cellular lysates were harvested 2, 10, 24 and 48 hours post-infection. Total genomic DNA was isolated and for quantification of viral genome copy numbers 50 ng of genomic DNA was subjected to quantitative real-time PCR (qRT-PCR) using hexon specific primers (Supplementary Table S1). For comparison of infectious virus particle production, lysates which were harvested 2, 10, 24 and 48 hours post-infection of HEK293 cells and B6 cells. These lysates were than used to re-infect HEK293 cells and 24 hours post-infection luciferase activity expressed in relative light units was measured by luciferase assay (Promega) which directly correlates with infectious virus particle production.
Enhanced production of high-capacity adenoviral vectors (HCA)
For production of high-capacity adenoviral vectors (HCA) we used a previously described system and protocol allowing small- and large scale vector amplification20,21. This procedure is based on 116 cells which were cultured in MEM medium supplemented with 10% FBS and 1% penicillin-streptomycin (PAA Laboratories, Coelbe Germany). 116 cells are derived from human embryonic kidney cells (HEK293 cells) stably expressing Cre recombinase and can grow as adherent cells and in suspension20,21. The principle of HCA production and a standard protocol applied for small and large scale production of HCA is outlined in Supplementary Figure S4 and was described in detail in a previous study20. In brief, the helper virus which resembles a regular first generation adenovirus vector deleted for the adenoviral early genes E1 and E3 provides all viral gene products required for replication of adenoviral DNA and the capsid in trans. However, the packaging signal of the helper virus is flanked by loxP sites and therefore in 116 cells the packaging signal is excised by Cre recombination prohibiting the helper virus from being packaged. In contrast, the HCA to be produced contains a regular packaging signal allowing the HCA genome to be encapsidated.
For improved production of high-capacity adenoviral vectors (HCA) we generated the novel helper virus BHVp19 (Supplementary Figure S5a). The recombinant DNA of this helper virus was cloned using an improved bacterial artificial chromosome (BAC) based cloning strategy (Supplementary Methods 1). To compare the efficacy of the novel helper virus BHVp19for HCA production with the standard helper virus AdNG163R-221, we analyzed the high-capacity adenovirus HCA-luc (Supplementary Figure S5d, Supplementary Methods 4). Besides the floxed packaging signal this helper virus expresses the RNAi suppressor P19 embedded in a viral transcription unit. The high-capacity adenovirus HCA-luc expressed firefly luciferase under the control of the liver-specific human α-1-antitrypsin promoter (phAAT).
For small scale analysis, cells were seeded in two 15 cm tissue culture dishes and grown to a confluency of 90%. Cells were infected with the HCA-luc (Supplementary Figure S5d) at an MOI of 3 and co-infected either with helper viruses BHVp19 or AdNG163R-2 (NgHV) at an MOI of 1. Two days post infection cells were harvested, viral particles were released by four consecutive freeze and thaw cycles and different amounts of the lysates were used to infect Huh7 cells at 90% confluency seeded in a 24-well tissue culture plate. Twenty-four hours later, these Huh7 cells were harvested and a 1:80 dilution was used for luciferase assay using the dual luciferase reporter assay kit provided by Promega.
For large scale analysis, a stock of high-capacity adenovirus HCA-luc was amplified utilizing a spinner flask system (Supplementary Figure S4). Following the protocol of Jager et al. 200920, 3 liters of suspension of 116 cells were co-infected with HCA-luc and either the helper virus AdNG163R-2 or the new helper virus BHVp19 (Supplementary Figure S5). After an incubation period of 48 hours HCA-luc was purified by CsCl centrifugation. To determine the infectious titer expressed as transducing units per μl (TU/μl), 95% confluent 24-well dishes containing HEK293 cells were infected with 5 μl of the purified virus and cells harvested 2 hours later using trypsin. Subsequently, genomic DNA was isolated and 50 ng were used for qRT-PCR using hexon specific primers (Hexon 5′forw and 3′rev) to quantify helper virus contamination and luciferase specific primers (luc forw and luc rev) to quantify HCA-luc genomes. For normalization human beta globulin was measured. All primer sequences are displayed in Supplementary Table 1. As an alternative for determination of infectious virus particle production of HCA-luc, the purified vector was used to re-infect HEK293 cells. 24 hours post-infection luciferase activity expressed in relative light units was measured by luciferase assay (Promega) which directly correlates with infectious virus particle production. As described previously20, the total physical virus titer (OD-titer, viral particle number) of HCA-Luc was determined by releasing the viral DNA from the purified particles and measuring the absorbance of the supernatant at 260 nm. The following formula was used to calculate the viral particle units (VPU): VPU/ml = (absorbance at 260 nm) × (dilution factor) × (1.1 × 1012) × (36)/(size of HCA in kb).
Oncolysis assay
Various cancer cell lines from different origin were used to perform oncolysis assays which measure the onocolytic potential of virus therapeutics. A549 derived from an alveolar adenocarcinoma, U87 derived from a glioblastoma, and Hela cells from cervical carcinoma were cultured in DMEM medium supplemented with 10% FBS and 1% penicillin streptomycin. Huh7 cells and SKHep cells were cultured in DMEM supplemented with 1% non-essential amino acids, 10% FBS and 1% P/S. McCoy's medium with 10% FBS, 1% P/S and 1% sodium bicarbonate was used to culture HCT116 cells. All cell lines were maintained at 37°C in a humidity atmosphere with 5% CO2. The entire cell culture reagents were obtained from PAA laboratories (Cölbe, Germany).
These cancer cell lines plated in 24-well tissue culture dishes were infected with Bwtp19ΔE3 (Supplementary Figure S2 and Supplementary Methods 1), AdΔfiberIL (Supplementary Figure 3) or an early gene E1 and E3 deleted adenovirus (FGAd) at MOIs of 1, 0.1 and 0.01. At the end of the assay, cells were fixed and stained with crystal violet solution. In order to quantify the oncolytic activity the amount of pixels within a defined area was determined and the amount of pixels for the FgAd control was set to 1. The diagram (right panel) shows the fold-increase of oncolytic activity.
Downregulation of endogenous miRNAs after infection with the high-capacity adenovirus HCA-p19
To analyze endogenous miRNA levels in vitro the human hepatoma cell line Huh7 was either infected with HCA-p19 or the control virus HCA-luc expressing an irrelevant transgene. To evaluate whether the P19 system is sufficient to suppress miRNAs in vivo in a tissue-specific manner, we infected C57Bl/6 mice with the p19 expressing virus HCA-p19 and two month post-injection mice were sacrificed. To determine miR122 levels in vitro and in mouse liver, mice were sacrificed and the liver was harvested. Small RNAs were isolated from the liver sections or Huh7 cell (for in vitro analysis), polyA-tailed and reverse transcribed. 5 μl of this cDNA was then used to quantify the miR122 amount by qRT-PCR using miR122 specific primer miR122 and reverse primer. In addition mRNA was isolated from the liver section using TRIZOL, reverse transcribed and 5 μl of cDNA was used to measure p19 levels from the two HCA-P19 infected mice (p19-1 and p19-2; p19 forw Xho and p19 rev Xba primers). All qRT-PCR measurements in mice were normalized to 104 copies of mouse TATA box binding protein (TBP) levels, in vitro experiments in Huh7 cells were normalized to human beta-2 microglobulin (B2m).
Animal work
Mouse experiments were approved by the Government of Upper Bavaria in Germany. C57Bl/6 mice were kept and animal experiments were performed in accordance with guidelines and regulations of the Government of Upper Bavaria in Germany. Mice were injected via the tail using a total volume of 200 μl. Virus was diluted in Dulbecco's phosphate-buffered saline (DPBS, Invitrogen, Darmstadt, Germany).
Isolation of small RNAs bound to P19 after His-tag purification and Northern blot analysis
For details regarding this method please refer to Supplementary Methods 5.
Statistical analysis
Statistical comparison was made by two-tailed Student's t-test, and a value of p < 0.05 was considered relevant compared with the respective control group.
Author Contributions
C.R. and A.E. conceived and designed the experiments. Experiments were performed by C.R., M.M.-H. and W.Z. and obtained data were analyzed by C.R. and A.E. D.N. and M.H. reviewed and discussed data. A.E. and C.R. wrote the manuscript which was edited and approved by all co-authors. A.E. supervised the project.
Supplementary Material
Supplementary information including figures
Acknowledgments
This work was supported in part by DFG grants SFB 455, EH 192/5-1, EH 192/4-1 (Heisenberg-Programme) and EU Framework Programme 7 (Persistent Transgenesis) to A.E. and a Helmholtz-University Young Investigator Group Grant (VH NG 212) to D.M.N. The authors thank Charles H. Lecellier (Institut de Génétique Humaine, Montpellier, France) for providing the P19 encoding cDNA. We thank Matthew Weitzman (Salk Institute, La Jolla, USA) for providing anti-adenoviral antibodies E1B55K and DBP, Thomas Dobner (Heinrich-Pette-Institut, Hamburg Germany) for the anti-adenoviral antibody E4Orf6, Ramon Alemany (Institut Català d'Oncologia, Barcelona, Spain) for providing the VA-RNA deleted adenovirus.
References
- Grundhoff A. & Sullivan C. S. Virus-encoded microRNAs. Virology 411, 325–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N. & Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews. Genetics 9, 102–114 (2008). [DOI] [PubMed] [Google Scholar]
- Lecellier C. H. et al. A cellular microRNA mediates antiviral defense in human cells. Science 308, 557–560 (2005). [DOI] [PubMed] [Google Scholar]
- Umbach J. L. et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454, 780–783 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. et al. Identification of virus-encoded microRNAs. Science 304, 734–736 (2004). [DOI] [PubMed] [Google Scholar]
- Dolken L. et al. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. Journal of virology 81, 13771–13782 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C. S., Grundhoff A. T., Tevethia S., Pipas J. M. & Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435, 682–686 (2005). [DOI] [PubMed] [Google Scholar]
- Andersson M. G. et al. Suppression of RNA interference by adenovirus virus-associated RNA. Journal of virology 79, 9556–9565 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O., Razquin N., Zaratiegui M., Narvaiza I. & Fortes P. Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production. Journal of virology 80, 1376–1384 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauschhuber C. & Ehrhardt A. RNA interference is responsible for reduction of transgene expression after Sleeping Beauty transposase mediated somatic integration. PloS one 7, e35389 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant S. R. et al. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nature biotechnology 20, 999–1005 (2002). [DOI] [PubMed] [Google Scholar]
- Hu C., Cela R. G., Suzuki M., Lee B. & Lipshutz G. S. Neonatal helper-dependent adenoviral vector gene therapy mediates correction of hemophilia A and tolerance to human factor VIII. Proceedings of the National Academy of Sciences of the United States of America 108, 2082–2087 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiedner G. et al. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nature genetics 18, 180–183 (1998). [DOI] [PubMed] [Google Scholar]
- Yamamoto M. & Curiel D. T. Current issues and future directions of oncolytic adenoviruses. Molecular therapy: the journal of the American Society of Gene Therapy 18, 243–250 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. C., Galanis E. & Kirn D. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nature clinical practice. Oncology 4, 101–117 (2007). [DOI] [PubMed] [Google Scholar]
- Alemany R., Balague C. & Curiel D. T. Replicative adenoviruses for cancer therapy. Nature biotechnology 18, 723–727 (2000). [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Lecellier C. H., Parizotto E. A., Himber C. & Voinnet O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. The Plant cell 16, 1235–1250 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Scholthof H. B. The Tombusvirus-encoded P19: from irrelevance to elegance. Nature reviews. Microbiology 4, 405–411 (2006). [DOI] [PubMed] [Google Scholar]
- Mathews M. B. & Shenk T. Adenovirus virus-associated RNA and translation control. Journal of virology 65, 5657–5662 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager L. et al. A rapid protocol for construction and production of high-capacity adenoviral vectors. Nature protocols 4, 547–564 (2009). [DOI] [PubMed] [Google Scholar]
- Palmer D. & Ng P. Improved system for helper-dependent adenoviral vector production. Molecular therapy: the journal of the American Society of Gene Therapy 8, 846–852 (2003). [DOI] [PubMed] [Google Scholar]
- Miao C. H. et al. Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Molecular therapy: the journal of the American Society of Gene Therapy 1, 522–532 (2000). [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W. & Tuschl T. Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 (2001). [DOI] [PubMed] [Google Scholar]
- Chen J. F. et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proceedings of the National Academy of Sciences of the United States of America 105, 2111–2116 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare R., Chen C. Y., Weaver E. A. & Barry M. A. Advances and future challenges in adenoviral vector pharmacology and targeting. Current gene therapy 11, 241–258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi H. & Hitt M. M. Transcriptionally targeted adenovirus vectors. Current gene therapy 5, 411–427 (2005). [DOI] [PubMed] [Google Scholar]
- Rivera A. A. et al. Mode of transgene expression after fusion to early or late viral genes of a conditionally replicating adenovirus via an optimized internal ribosome entry site in vitro and in vivo. Virology 320, 121–134 (2004). [DOI] [PubMed] [Google Scholar]
- Cascallo M. et al. Deletion of VAI and VAII RNA genes in the design of oncolytic adenoviruses. Human gene therapy 17, 929–940 (2006). [DOI] [PubMed] [Google Scholar]
- Rauschhuber C., Xu H., Salazar F. H., Marion P. L. & Ehrhardt A. Exploring gene-deleted adenoviral vectors for delivery of short hairpin RNAs and reduction of hepatitis B virus infection in mice. The journal of gene medicine 10, 878–889 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information including figures