Abstract
Objective(s)
The aim of this study was to investigate the presence of PER-1-type ESBLs in drug resistant Pseudomonas aeruginosa isolates.
Materials and Methods
During one-year period (2008-2009), following isolation and identification of 56 P. aeruginosa, the E-test method was performed for determination of minimal inhibitory concentration of ceftazidim. The isolates that they had MIC≥16 µg/ml against ceftazidim were used for determination of ESBL-producing by combined disk test (CDT) and double disk synergy test (DDST) methods. Bla PER-1 gene was investigated by PCR. P. aeruginosa KOAS was used as positive control.
Results
Twenty-nine (51.78%) out of fifty six isolates had MIC≥16 µg/ml to ceftazidime, twenty two (75.86%) of them were ESBL producers. Some isolates (27.5%) contained bla PER-1 gene.
Conclusion
PER-1-type ESBLs producing P.aeruginosa has not been reported previously in but there has been a rather high prevalence of it.
Key Words: Combined Disk Test, Double Disk Synergy Test, Extended- Spectrum Beta- Lactamase, PER-1gene, Pseudomonas aeruginosa
Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that causes serious infections among immunity-impaired patients. These bacteria are resistant to many antibiotics intrinsically and are able to produce different virulence factors. Intrinsically resistance of P. aeroginosa to different antibiotics is generally due to decreased membrane permeability and efflux pumping (1). Like other Gram- negative bacteria, P. aeruginosa can acquire resistance against beta- lactam antibiotics by induction of beta- lactamases. These enzymes are coded either by chromosomes such as class C (Amp C) cephalosporinases and some of class A extended spectrum beta lactamases (ESBLs) or by plasmids. Amongst different classes of beta lactamases, ESBLs are usually able to hydrolyze extended spectrum cephalosporins and monobactams (2). For the first time class A enzymes were reported in Enterobacteriaceae family but since 1990 presence of these enzymes have been frequently found in P. aeruginosa (3). PER-1- type ESBLs were the first reported ESBLs ones in P. aeruginosa and like most other ESBLs such as TEM and SHV they can hydrolyze different types of beta lactam antibiotics except for carbapenem and cephamycin (4). The PER-1 gene containing isolates were prevalently reported in hospitalized patients from (5). The infection with ESBLs producing P. aeruginosa increases morbidity and mortality among hospitalized patients, bringing out many difficulties in treatment of patients and places, plus a significant economic burden on health services. The aim of this study was to investigate presence of PER-1 type ESBLs gene in P. aeruginosa isolated from different clinical specimens.
Materials and Methods
Bacterial isolates
During a year (from July-2008 to July-2009), cultures were made from all clinical samples taken from microbiology laboratory from ICU wards of Imam Reza Hospital in Tabriz, Iran. Samples were taken from patients who sustained trauma and were under invasive procedures such as intubation and tracheostomy. The isolates were identified using conventional bacteriological tests such as positive oxidase and OF (oxidation and fermentation) tests (, ), growth on Cetrimide agar () at 42 ˚C (6). The isolates were then stored at -20˚C in trypticase soy broth () containing 20% glycerol.
Determination of ceftazidime minimum inhibitory concentration (MIC)
The MIC of all isolates to ceftazidim was determined by E-test method (AB Biodisk, ) and the results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (7). P. aeruginosa ATCC 27853 was used as control for MIC test.
Screening for ESBL-producing isolates
Combined disk test (CDT) (8) and double disk synergy test (DDST) were performed for detection of ESBLs (2). All isolates which showed MIC≥16 µg/ml to ceftazidime were cultured on Muller-Hinton agar (MH) containing cloxacillin (250 µg/ml) (Sigma, USA). DDSTs were performed by placing disks of ceftazidime (CAZ), cefotaxime (CTX), cefepime (CPM), azteronam (ATM) [30 µg each] (MAST Ltd, UK) at a distance of 20 mm (center to center) from a disk containing AMC (amoxicillin/clavulanate 20/10 µg) (MAST Ltd, UK) on MH plates. ESBL production was interpreted when the cephalosporin zone was expanded by the clavulanate. Combined disk tests were also performed by placing disks of ceftazidime, cefotaxime (CTX), cefepime (CPM), CAZ/clavulanic acid, CTX/clavulanic acid and CPM/clavulanic acid [20/10 µg] (MAST Ltd UK) on MH plates. ESBL production was interpreted if the zones produced by the disks with clavulanate were ≥ 5 mm larger than those without inhibitor (8).
PCR amplification of β-Lactamase genes
All ceftazidime resistance P. aeruginosa isolates were investigated for detection of bla PER-1 gene by polymerase chain reaction (PCR) method, DNA extraction was carried out by sodium dodecyl sulphate (SDS) protienase K modified with N,N,N-trimethyl ammonium bromide (CTAB) (9). The DNA amplification program consisted of an initial denaturation step (94 ˚C, 5 min) followed by 35 cycles of denaturation (94 ˚C, 1 min), annealing (52 ˚C, 1 min), and extension (72 ˚C, 1min) steps. A final extension (72 ˚C, 5 min) step was also included in the scheme. PCR was performed in a total volume of 50 µl containing 2.2 mM MgCl2, 0.5 µM each of the forward: (5'-ATGAATGTCATTATAAAAGCT-3') and the reverse:
(5′-TTAATTTGG GCTTAGGG-3′) primers (Bioneer, Germany) (10), 0.2 mM dNTPs, 5 µl PCR 10X buffer and 1µl of DNA template, (all the PCR components were purchased from Fermentas Litvania). Eight micro-liter of PCR products were analyzed in 1.1% agarose (Sigma, USA) and the results were observed under UV light (9). P. aeruginosa KOAS and P. aeroginosa ATCC 27853 (Institute Pasteur, Iran) were used respectively as positive and negative control strains for blaPER-1 gene.
Results
Fifty six isolates of P. aeruginosa were obtained from different clinical specimens such as 23 (41.1%) tracheotomy tube, 17 (30.36%) urine, 7 (12.5%) Bronchoalveolar lavage, 4 (7.14%) blood, 2 (3.57%) sputum, 2 (3.57%) wound and 1 (1.76%) cerebrospinal fluid (CSF). Twenty nine isolates (51.78%) showed MIC≥16µg/ml to ceftazidime. CDT and DDST were carried out on 29 isolates as confirmatory tests for ESBLs production. Twenty two (75.86%) of them showed positive results while 7 (24.14%) isolates were found to not produce ESBLs (Figure-1). It was also found out that eight (27.5%) ceftazidime resistant isolates contained bla PER-1gene by PCR (Figure 2).
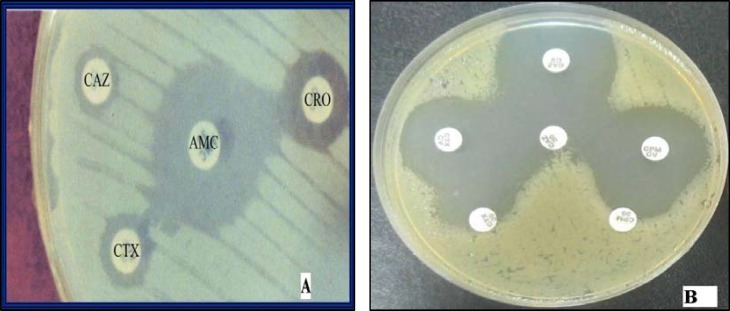
Figure1.
A- DDST using ceftazidime (CAZ), ceftriaxone (CRO(, cefotaxime (CTX) and augmentine disks; ESBLs positive Pseudomonas aeruginosa showing distinct extension of the zone of inhibition towards AMC. B- A representative of P. aeruginosa isolates showing a ≥ 5 mm zone size enhancement in the combined disc (CD) test indicating inhibition of ESBL production. A positive CD using different cephalosporins (CAZ, CTX, cefepime CPM) and the same cephalosporins with clavulanic acid (30 μg/10 μg)
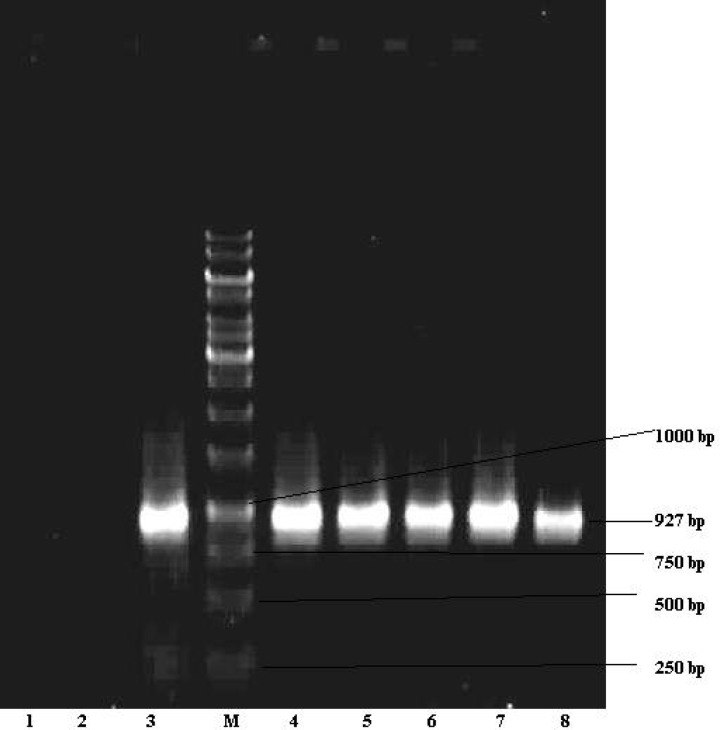
Figure2.
Detection of PER-1 gene in Pseudomonas aeruginosa isolated from clinical specimens (PER-1 specific 927 bp fragment). M; DNA size marker (1 kbp DNA ladder), line 1 negative control (P. aeruginosa ATCC 27853), line 2 PER-1 negative isolate, line 3 Positive control (P. aeruginosa KOAS) and lines 4, 5, 6, 7, 8 PER-1 positive isolates
Discussion
Extended spectrum cephalosporins are one of the most important antibiotics in treatment of infections produced by P. aeruginosa isolates as well as other bacteria in ICU wards (11, 12). Investigations of the ways in which bacteria acquire resistance to these antibiotics are also important and necessary. Phenotypic identification of ESBLs in P. aeroginosa by using clavulanic acid for some reasons such as class Amp C cephalosporinase, production of metalo-beta lactamases and oxacillinases (class B & D beta lactamases) and resistant to inhibitory effect of clavulanic acid in some ESBLs type like GES is more difficult than Enterobacteriaceae and often produce false results (3, 13). In order to stop activity of chromosomal Amp C enzymes in ESBLs detection by phenotypic procedure, cloxacillin could be added to the culture media, and it is possible to decrease the distance of disks from 30 mm to 20 mm to increase positive results too (8). Increase of ESBLs prevalence in last decades among P. aeruginosa isolates has been proven by several reports from in 2001 (28%) and 2003 (20.6%) (12, 14), in 2005 (25.4%) (11), in 2006 (23.4%) (13), and in 2009 (%40) (10). In this study, we obtained 75.8% ESBLs production by P. aeruginosa isolates, which is much higher than the results obtained by others. The reasons for such high results could be the use of improved phenotypic procedure and place of sample collection, as we know in ICU wards antibiotic resistant bacteria are much more prevalent than other wards (12).
Although bla PER-1 gene possessing isolates are very resistant to beta lactam antibiotics, those which lack this gene, due to having other resistance mechanisms such as efflux pumping and decreased permeability, are also resistant to cephalosporins and monobactams. As far as we know, the study is the first report of PER-1-producing P. aeruginosa in Tabriz. For years, PER-1 ß-lactamases were thought to be significant only in Turkey. However, the recent identification of PER-1 producers in several European countries and in the Far East suggests their proceeding dissemination (11, 13, and 14). In Korea, as in Turkey, PER-1 production by Acinetobacter spp. has been reported often, but in Europe, it has been identified mostly in P. aeruginosa.
The result obtained for prevalence of bla PER-I gene in our study is 27.5% (Figure 2), while the results obtained in Turkey have been 11% in 1997 (5) and 55.4% in 2005 (15). Prevalance rate of 13% for bla PER-I gene in Tehran was reported by Mirsalehian et al in 2009 (10). PER-I type ESBLs producing P. aeruginosa isolates are one of the most important challenges in treatment of infections in Turkey (5, 15).
Conclusion
Indiscriminate consumption of antibiotics by patients, the high prevalence of PER-1 gene in Turkey and existence of more communication between Turkey and province of Eastern Azerbaijan (Tabriz) can be the possible reasons for transfer and high prevalence of PER-I gene in this region. Of course demonstration of this hypothesis requires typing of these isolates and comparing them with types which are common in Turkey.
Acknowledgment
This study financially was supported by cooperation of Research Center of Infectious Diseases and Tropical Medicine and vice research chancellor office of Tabriz university of Medical sciences. We would like to thank hospital staff of Imam Reza for their sincerely participation in this study.
References
- 1.Li XZ, Ma D, Livermore DM, Nikaido H. Role of efflux pump (s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother. 1994;38:1742–1752. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad – spectrum β- lactamases conferring transferable resistance to newer β- lactam agents in Enterobacteriaceae hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 3.Weldhagen GF, Poirel L, Nordmann P. Ambler class A extended-spectrum beta-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob Agents Chemother. 2003;47:2385–2392. doi: 10.1128/AAC.47.8.2385-2392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordmann P, Naas T. Sequence analysis of PER-1 extended-spectrum beta-lactamase from Pseudomonas aeruginosa and comparison with class A beta-lactamases. Antimicrob Agents Chemother. 1994;38:104–114. doi: 10.1128/aac.38.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vahaboglu H, Oztürk R, Aygün G, Coşkunkan F, Yaman A, Kaygusuz A, et al. Widespread detection of PER-1-type extended-spectrum beta-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacFaddin JF. Biochemical tests for identification of medical bacteria. 3rded. New York: Liooincott Williams AND Wilkins; 2000. pp. 389–690. [Google Scholar]
- 7.Clinical andLaboratoryStandardsInstituteperformancestandardsforantimicrobialsusceptibilitytesting:DocumentM10–S15.CLSI. PA, USA: Wayne; 2005. [Google Scholar]
- 8.Naas T, Nordmann P, Heidt A. Intercountry transfer of PER-1 extended-spectrum beta-lactamase-producing Acinetobacter baumannii from Romania. Int J Antimicrob Agents. 2007;29:226–228. doi: 10.1016/j.ijantimicag.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 9.Akhi MT, Farzaneh F, Oskouei M. Study of enterococcal susceptibility patterns isolated from clinical specimens in Tabriz, Iran. Pak J Med Sci . 2009;25:211–216. [Google Scholar]
- 10.Mirsalehian A, Feizabadi M, Nakhjavani FA, Jabalameli F, Goli H, Kalantari N. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2009;36:70–74. doi: 10.1016/j.burns.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Park YJ, Kim M, Lee HK, Han K, Kang CS, et al. Prevalence of Ambler class A and D beta-lactamases among clinical isolates of Pseudomonas aeruginosa in Korea. J Antimicrob Chemother . 2005;56:122–127. doi: 10.1093/jac/dki160. [DOI] [PubMed] [Google Scholar]
- 12.Rossolini GM, Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin Microbiol Infect. 2005;11:17–32. doi: 10.1111/j.1469-0691.2005.01161.x. [DOI] [PubMed] [Google Scholar]
- 13.Celenza G, Pellegrini C, Caccamo M, Segatore B, Amicosante G, Perilli M. Spread of bla (CTX-M-type) and bla (PER-2) beta-lactamase genes in clinical isolates from Bolivian Hospitals. J Antimicrob Chemother. 2006;57:975–978. doi: 10.1093/jac/dkl055. [DOI] [PubMed] [Google Scholar]
- 14.Chayakulkeeree M, Junsriwong P, Keerasuntonpong A, Tribuddharat C, Thamlikitkul V. Epidemiology of extended-spectrum beta-lactamase producing gram-negative bacilli at Siriraj Hospital, Thailand, 2003. Southeast Asian J Trop Med Public Health. 2005;36:1503–1509. [PubMed] [Google Scholar]
- 15.Kolayli F, Gacar G, Karadenizli A, Sanic A, Vahaboglu H. Study Group. PER-1 is still widespread in Turkish Hospitals among Pseudomonas aeruginosa and Acinetobacter spp. FEMS Microbiol Lett. 2005;249:241–245. doi: 10.1016/j.femsle.2005.06.012. [DOI] [PubMed] [Google Scholar]