Abstract
Objective(s)
Staphylococcus aureus is a common cause of human infection, and emergence of vancomycin-resistance S. aureus is a great concern for treatment of methicillin-resistant S. aureus,(MRSA) in recent years (MRSA). Here, we report the isolation of high-level VRSA.
Materials and Methods
S. aureus was isolated from foot ulcer of a diabetic woman in Tehran, Iran. Antibiotic susceptibility was determined according to CLSI guidelines. VanA gene cluster PCR was carried out and PCR amplicon of vanA was sequenced.
Results
S. aureus had high-level vancomycin-resistant (MIC 512 ≥ µg/ml). Patient's history revealed that VRSA isolate was acquired through community transmission. Only vanA, vanR and vanS genes were amplified in our isolate. Sequencing revealed that the vanA sequence had high similarity to the vanA sequence of Tn1546.
Conclusion
Although VRSA infection continues to be rare, isolation of community–acquired VRSA is a significant issue and it needs the efforts of public health authorities.
Introduction
Staphylococcus aureus is a common cause of hospital and community-acquired infections. Because of the spread of multidrug-resistant Gram-positive bacteria as well as methicilin-resistant S. aureus (MRSA), glycopeptide antibiotics, vancomycin and teicoplanin are used to treat severe staphylococcal infections (1). The first clinical vancomycin-resistance S. aureus (MIC ≥ 32 µg/mL) was reported from Michigan, USA in 2002 (2). It seems that the development of vancomycin-resistant enterococci (VRE) in 1988 led to the emergence of VRSA through acquisition of the VanA gene cluster from Entercoccus spp (3). The first detection of the VRSA in Iran was in 2007 (4) and this report describes clinical isolate of community–acquired vancomycin-resistant S. aureus from a diabetic patient in Iran with the vancomycin MIC 512 ≥ µg/ml.
Materials and Methods
Bacterial isolate
A 51-year-old female with a history of diabetes mellitus was admitted to the Surgery Department of Taleghani Hospital, Tehran. Incision and drainage of the abscess was performed, and discharge was sent to the laboratories for microbiological and molecular investigation. Patient's medical records including antimicrobial drug history and recent bacterial infections were recorded. Isolate was identified based on colony morphology and standard biochemical tests.
S. aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 strains were used as vancomycin-susceptible controls. Vancomycin resistant E. faecium BM4147 was used as positive control.
Antimicrobial agents and MIC determination
Antibiotic susceptibility was determined by disk diffusion on Mueller-Hinton agar (Merck) based on Clinical and Laboratory Standards Institute guidelines (5). The antibiotics (MAST Diagnostics Ltd. Merseyside, England) used for disc diffusion assays included vancomycin, teicoplanin, penicillin, oxacillin, ceftriaxone, erythromycin, clindamycin, amikacin, co-trimoxazole, chloramphenicol, amoxicillin, and imepenem. Minimum inhibitory concentration (MIC) of vancomycin (SERVA FEINBIOCHEMICA GmbH & Co., Germany) was determined by broth microdilution method according to CLSI guidelines (5).
Detection of vanA gene cluster by PCR
VanA gene cluster (vanR, vanS, vanH, vanA, vanX, vanY, vanZ) PCR was carried out with previously published primers (6, 7). The DNA sequence of vanA was determined with an automated sequencer (ABI 377, Applied Biosystems [ABI]) using PCR product to determine sequences of the forward and reverse strands.
Results
The isolate was identified as S. aureus and it was resistant to vancomycin, teicoplanin, penicillin, oxacillin, ceftriaxone, erythromycin, clindamycin, amikacin, co-trimoxazole, chloramphenicol, amoxicillin, and sensitive to imepenem. The isolate showed high-level vancomycin resistance (512 µg/ml).
Based on the resistance to glycopeptide antibiotics, the S. aureus isolate expressed VanA phenotype.
Patient's history revealed that this isolate was a community–acquired VRSA. From VanA gene cluster, only PCR products of vanA, vanR and vanS were amplified from extracted DNA with expected size.
DNA and the inferred amino acid sequences were compared using DNAsis (version 2.5; Hitachi). DNA sequence analysis revealed that this strain's vanA sequence had high similarity to the vanA sequence of Tn1546 (M97297). The partial sequence of vanA gene has been submitted to GenBank (GQ273978).
Table 1.
Primers used in this study
| primer | Sequence (5’ to 3’) | Size of PCR product (bp)* | Ref. |
|---|---|---|---|
|
vanR 1 vanR 2 |
AGCGATAAAATACTTATTGTGGA CGGATTATCAATGGTGTCGTT |
645 | 7 |
|
vanS 1 vanS 2 |
TTGGTTATAAAATTGAAAAATAA TTAGGACCTCCTTTTATC |
1155 | 8 |
|
vanH 1 vanH 2 |
ATCGGCATTACTGTTTATGGAT TCCTTTCAAAATCCAAACAGTTT |
943 | 7 |
|
vanA 1 vanA 2 |
ATGAATAGAATAAAAGTTGCAATAC CCCCTTTAACGCTAATACGAT |
1029 | 7 |
|
vanX 1 vanX 2 |
ATGGAAATAGGATTTACTTT TTATTTAACGGGGAAATC |
609 | 8 |
|
vanY 1 vanY 2 |
ATGAAGAAGTTGTTTTTTTTA TTACCTCCTTGAATTAGTAT |
912 | 8 |
|
vanZ 1 vanZ 2 |
TTATCTAGAGGATTGCTAGC AATGGGTACGGTAAACGAGC |
454 | 9 |
* Based on the sequence of Tn1546 in Enterococcus faecium (GenBank accession no. M97297)
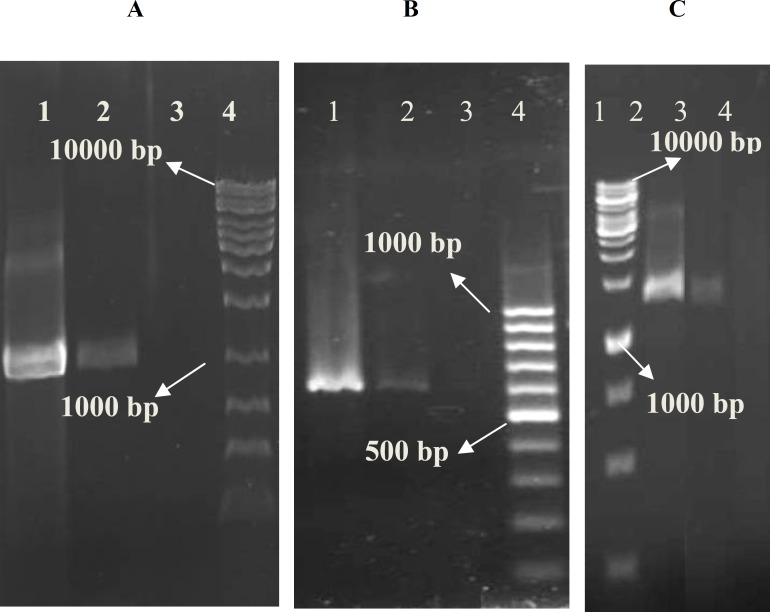
Figure 1.
Specific PCR amplification products of the vanA gene cluster of Staphylococcus aureus. vanA (A: lane 1, vanA-positive control, BM4147; 2, isolated S. aureus; 3, vanA-negative control, ATCC 29213; 4, molecular weight marker, 1kb); vanR (B: lane 1, vanA-positive control, BM4147; 2, isolated S. aureus; 3, vanA-negative control, ATCC 29213; 4, molecular weight marker, 100 bp) and vanS (C: lane 1, molecular weight marker, 1kb; 2, vanA-positive control, BM4147; 3, isolated S. aureus; 4, vanA-negative control, ATCC 29213)
Discussion
There are limited reports about isolation of VRSA from clinical specimens in all over the world and among these isolates a few of them were community–acquired VRSA (8). To the best of our knowledge, it is the first report of community–acquired of vancomycin and methicillin-resistant S. aureus in Tehran, Iran.
According to the in vitro transfer of vanA gene from enterococci to S. aureus (8), we suspect the possibility of transformation of vancomycin resistance gene (vanA) from VRE to Staphylococci spp.
In this study there were significant differences between previously described VanA gene cluster among enterococci and our isolate. Although the vanA, vanR, vanS, vanH and vanX genes are essential for the expression of VanA phenotype, according to the previous study (9), the difficulties with amplification of vanH and vanX in the vanA gene cluster may be due to disruptions of these regions with insertion sequences (ISs).
Prevention of emergence and transmission of VRSA in each community is needed. So, use of proper infection-control practices, appropriate antimicrobial agent management, maintaining a clean environment and increased awareness can control the spread of antimicrobial-resistant microorganisms, including VRSA.
Conclusions
This report describes a community–acquired and multidrug-resistant S. aureus isolated from a diabetic patient in Tehran, Iran. Among antibiotics used in this study, co-resistance to oxacillin and vancomycin is a critical issue because vancomycin is the first-line antimicrobial agent for the treatment of infection with MRSA.
Acknowledgment
This work was financially supported by a grant No.414 from the Research Centre of Gastroenterology and Liver Diseases (RCGLD) in Shahid Beheshti University, Tehran, Iran.
References
- 1.Périchon B, Courvalin P. Heterologous expression of the enterococcal vanA operon in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother . 2004;48:4281–4285. doi: 10.1128/AAC.48.11.4281-4285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Staphylococcus aureus resistant to vancomycin - United States. Morb Mortal Wkly Rep. 2002;51:565–567. [PubMed] [Google Scholar]
- 3.Saha B, Singh AK, Ghosh A, Bal M. Identification and characterization of a vancomycin resistant Staphylococcus aureus isolated from Kolkata (South Asia) J Med Microbiol. 2008;57:72–79. doi: 10.1099/jmm.0.47144-0. [DOI] [PubMed] [Google Scholar]
- 4.Emaneini M, Aligholi M, Hashemi FB, Jabalameli F, Shahsavan S, Dabiri H, et al. Isolation of vancomycin-resistant Staphylococcus aureus in a teaching hospital in Tehran. J Hosp Infect. 2007;66:92–93. doi: 10.1016/j.jhin.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 16th informational supplement, M100-S16. Clinical and Laboratory Standards. USA: Institute Wayne; 2006. [Google Scholar]
- 6.Jung WK, Hong SK, Lim JY, Lim SK, Kwon NH, Kim JM, et al. Phenotypic and genetic characterization of vancomycin-resistant enterococci from hospitalized humans and from poultry in Korea. FEMS Microbiol Lett . 2006;260:193–200. doi: 10.1111/j.1574-6968.2006.00311.x. [DOI] [PubMed] [Google Scholar]
- 7.Miele A, Bandera M, Goldstein BP. Use of primers selective for vancomycin resistance gene to determine van genotype in enterococci and to study gene organization in vanA isolates. Antimicrob. Agents Chemother. 1995;39:1772–1778. doi: 10.1128/aac.39.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitener CJ, Park SY, Browne FA, Parent LJ, Julian K, Bozdogan B, et al. Vancomycin-resistant Staphylococcus aureus in the absence of vancomycin exposure. Clin Infect Dis. 2004;38:1049–1055. doi: 10.1086/382357. [DOI] [PubMed] [Google Scholar]
- 9.Woodford N, Adebiyi AA, Palepou MI, Cookson BD. Diversity of vanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob Agents Chemother. 1998;42:502–508. doi: 10.1128/aac.42.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]