Abstract
Objective(s)
Staphylococcus aureus (S. aureus) is a prevalent pathogen worldwide. Methicillin resistant S. aureus (MRSA), which is usually multi-resistant in hospitals, has been a daunting challenge for clinicians for more than half a century. The aim of this systematic review and meta-analysis is to determine the relative frequency (R.F.) of MRSA in different regions of Iran.
Materials and Methods
Search terms “Staphylococcus aureus”, “Methicillin”, “mecA” and “Iran” were used in PubMed, Scirus and Google Scholar. Two Persian scientific search engines and ten recent national congresses were also explored. Articles/abstracts, which used clinical specimens and had done PCR to detect the mecA gene, were included in this review. Comprehensive Meta-Analysis and Meta-Analyst software were used for statistical analysis.
Results
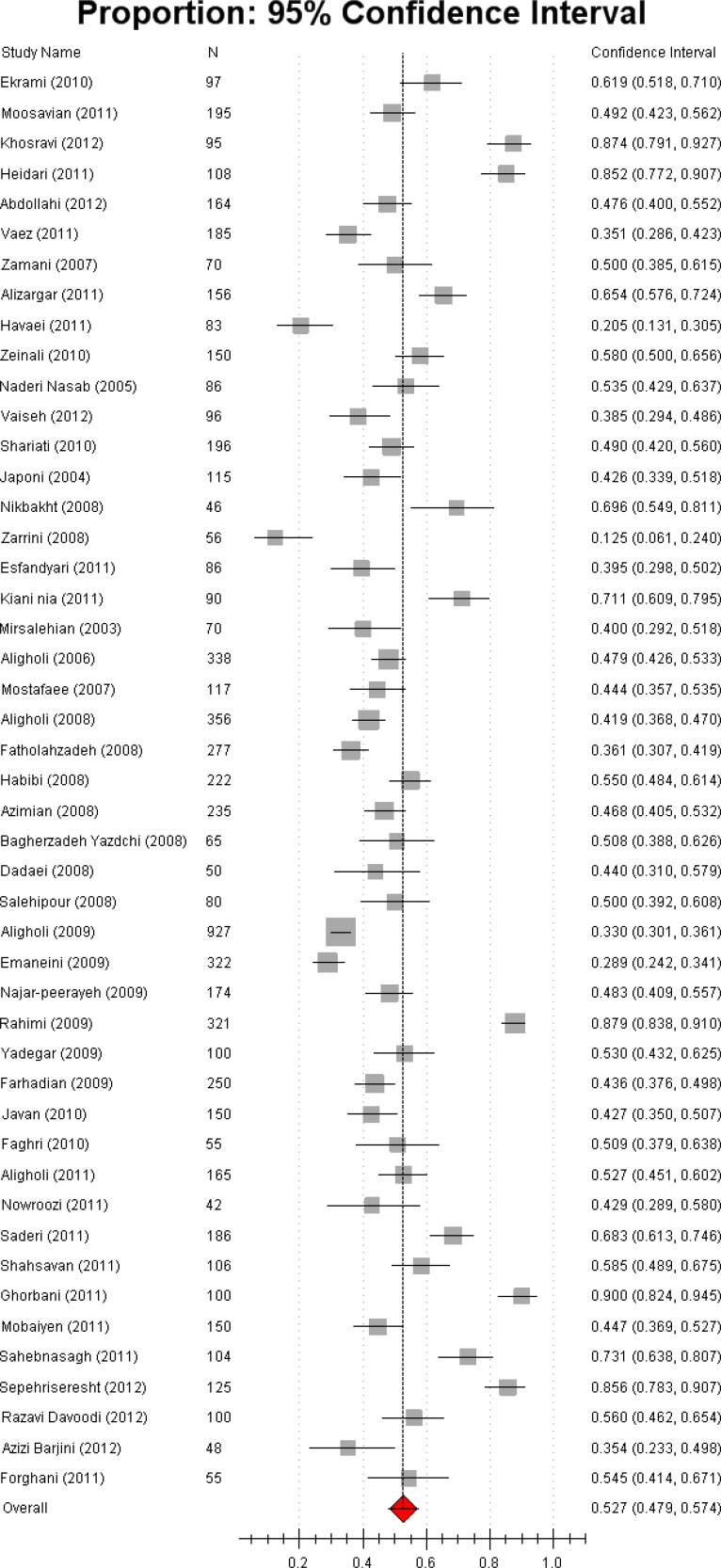
Out of 2690 results found in the mentioned databases, 48 articles were included in the final analysis. These studies were done in Ahvaz, Falavarjan, Fasa, Gorgan, Hamedan, Isfehan, Kashan, Mashhad, Sanandaj, Shahrekord, Shiraz, Tabriz, Tehran and Tonekabon. Pooled estimation of 7464 S. aureus samples showed that 52.7%±4.7 (95% confidence interval [CI]) of strains were mecA positive. MRSA R.F. in different studies varied from 20.48% to 90% in Isfehan and Tehran, respectively. We found a moderate heterogeneity (I2= 48.5%) of MRSA R.F. among studies conducted in Tehran (ranging from 28.88% to 90%, mean 52.7% [95% CI: 46.6%±0.58.8%]).
Conclusion
According to the results of this study, MRSA R.F. in Iran is in the high range. Thus, measures should be taken to keep the emergence and transmission of these strains to a minimum.
Key Words: Iran, mecA gene, MRSA, Staphylococcus aureus, Systematic Review
Introduction
Staphylococcus aureus has been known as a threat to human health for more than a century. This pathogen is responsible for a wide range of maladies from folliculitis and food poisoning to life-threatening conditions such as endocarditis or necrotizing pneumonia (1).
Introduction of penicillin to the market in the 1940s was a cornerstone in treating staphylococcal infections, which was soon followed by the emergence of β-lactamase producing strains. Methicillin, a β-lactamase-resistant antimicrobial agent, was introduced in 1959. The first report of methicillin-resistant Staphylococcus aureus (MRSA) was from London in 1961(2-3).
It has been suggested that the mecA gene is responsible for resistance to methicillin. MecA encodes an altered penicillin-binding protein (i.e. PBP2a) with a low affinity for β-lactam antibiotics (2). The multi-drug resistance phenomenon, seen especially in MRSA strains, is a main cause of treatment failure and increase in treatment costs (4). It is noteworthy that MRSA infections are associated with a higher mortality rate compared to infections with methicillin-susceptible S. aureus (5).
MRSA was previously considered as a nosocomial pathogen, but in the past two decades, reports suggest an increasing trend for community-associated MRSA (CA-MRSA). These clones may replace current health care-associated MRSA (HA-MRSA) clones in the future. This hypothesis is supported not only by mathematical models but also by reports that have shown invasion of CA-MRSA clones to hospitals (6). First described in Minnesota, CA-MRSA has now attracted global attention (1). Since 2004, MRSA related to livestock infections has also been reported. However, this type of MRSA seems to be limited to some countries, especially the ones where pig farms are common (7-8).
Recent studies have revealed an increase in the worldwide prevalence of MRSA. However, some European countries have maintained low rates of MRSA (4, 7). Although there are many reports from different cities of Iran, the average rate of MRSA in Iranian hospitals is still unknown. Our aim in this study is to provide the relative frequency (R.F.) of MRSA in Iran, as detected by the PCR amplification of the mecA gene.
Materials and Methods
Literature Search
“Staphylococcus aureus”, “S. aureus”, “Methicillin”, “MRSA”, “MSSA”, “mecA gene” and Iran (for non-Iranian databases) were searched with special strategies in PubMed, Google Scholar and Scirus search engines. Two Persian scientific search engines “Scientific Information Database" (www. sid.ir), and "IranMedex" (www.iranmedex.com) were searched as well. The keywords were also searched at all Iranian academic domains (i.e. ending with.ac.ir) by “Google advanced search”. Additionally, abstract books of 10 recent congresses (i.e. “1st-5thIranian Congress of Clinical Microbiology”, “4th Congress of Laboratory and Clinic”, “First International and 12th Iranian Congress of Microbiology”, “The First Iranian International Congress of Medical Bacteriology”, “The Congress of Infections and Antibiotic Resistance” and “The Congress of Rational Usage of Antibiotics”) were explored. All common dictation mistakes and possible conditions of mentioned words (in English and Persian) were covered as well. Search strategies were followed until 17th May 2012.
Inclusion criteria
Among English and Persian articles/abstracts found with above strategies, those with the following features were included in the study:
S. aureus Samples were collected from Iranian hospitals.
Clinical specimens were taken from patients. If there were personnel specimens as well, results of the personnel were excluded.
PCR method was done to detect mecA gene. Phenotypic results were not included because: (A) Phenotypic methods had variable sensitivities and specificities in various studies (9). (B) Phenotypic methods were affected by many factors such as pH of media, concentration of NaCl, incubation period of isolates, commercial discs and media used in different studies and also personnel’s/researcher’s skills (10). (C) Generally, avoiding heterogeneity for inclusion of studies is desirable in systematic reviews (11). (D) Breakpoints of phenotypic methods may change over time and make the interpretation of previous results more difficult. For example, Clinical Laboratory Standards Institute revised the breakpoints for cefoxitindisc diffusion and minimum inhibitory concentration in 2007 and 2008, respectively (12, 13).
Exclusion criteria
During observation, studies with at least one of the aspects mentioned below were excluded:
Samples were partially/totally selected from MRSA collections.
Method for detecting MRSA strains could not be discovered from the paper.
Data collection
At this stage, articles/abstracts with the following features were excluded as well:
Any projects published both in English and Persian. (In these cases, the article published later and/or with more detailed results was chosen for analysis.)
Duplicate publications and congress abstracts whose full-text papers were also available.
The origin of samples was not clear, meaning that the reviewer(s) could not find out which region or population (i.e. inpatients, personnel, or out patients) the specimens were gathered from.
Nasal, oral or throat swabs were taken from healthy people or patients/healthcare personnel to detect carriers.
Unclear report of the results, such as studies that mixed results of “Coagulase-negative Staphylococci and S. aureus” or “healthy people and patients”.
Statistical analysis
Statistical analysis was performed by the Meta-Analyst (version 3.13 Beta) and Comprehensive Meta-Analysis (version 2.0) software. Overall relative frequency of MRSA in Iran was pooled by forest plot using the Meta-Analyst software. Statistical heterogeneity of the results was checked using Cochrane Q-test with significance set at P< 0.1. In order to assess possible publication bias, the Begg and Mazumdar’s test was done using the Comprehensive Meta-Analysis software. The Begg and Mazumdar’s rank correlation test reports the rank correlation between the standardized effect size and the variances (or standard errors) of these effects.
Results
Out of 2690 articles/abstracts found by the aforementioned search strategies, 79 results matched inclusion criteria, out of which 48 (29 full-text articles and 19 abstracts) were selected for analysis (Table 1) (14-61). Sample size and 95% confidence interval (CI) of each study was shown in a forest plot (Figure 1). According to heterogeneity test, random model methods were used for meta-analysis tests (P< 0.001). I2 statistics, the proportion of variation due to heterogeneity, was 0.48, indicating moderate heterogeneity.
Table 1.
Sample size and MRSA strains in different studies
| City | Type | Sample size | MRSA1 | Relative frequency of MRSA (%) | Study team (Reference No.) |
Year Published/ Presented |
|---|---|---|---|---|---|---|
| Ahvaz | Article | 97 | 60 | 61 | Ekrami et al (14) | 2010 |
| Abstract | 195 | ≥96 | ≥49.23 | Moosavian et al (15) | 2011 | |
| Article | 95 | 83 | 87.36 | Khosravi et al (16) | 2012 | |
| Falavarjan | Article | 108 | 92 | 85.18 | Heidari et al (17) | 2011 |
| Fasa | Article | 164 | 78 | 47.56 | Abdollahi et al (18) | 2012 |
| Gorgan | Article | 185 | 65 | 35.13 | Vaez et al (19) | 2011 |
| Hamedan | Article | 70 | 35 | 50 | Zamani et al (20) | 2007 |
| Abstract | 156 | 102 | 65 | Alizargar et al (21) | 2011 | |
| Isfehan | Article | 83 | 17 | 20.48 | Havaei et al (22) | 2011 |
| Kashan | Article | 150 | 87 | 58 | Zeinali et al (23) | 2010 |
| Mashhad | Article | 86 | 46 | 53.48 | NaderiNasab et al (24) | 2005 |
| Sanandaj | Abstract | 96 | 37 | 38.5 | Vaiseh et al (25) | 2012 |
| Shahrekord | Article | 196 | 96 | 48.98 | Shariati et al (26) | 2010 |
| Shiraz | Article | 115 | 49 | 42.6 | Japoni et al (27) | 2004 |
| Tabriz | Article | 46 | ≥32 | ≥69.5 | Nikbakht et al (28) | 2008 |
| Abstract | 56 | ≥7 | ≥12.5 | Zarrini et al (29) | 2008 | |
| Abstract | 86 | 34 | 39.5 | Esfandyari et al (30) | 2011 | |
| Abstract | 90 | 64 | 71 | Kianinia et al (31-32) | 2011 | |
| Tehran | Article | 70 | 28 | 40 | Mirsalehian et al (33) | 2003 |
| Article | 338 | 162 | 48 | Aligholi et al (34) | 2006 | |
| Abstract | 117 | 52 | 44.45 | Mostafaee et al (35) | 2007 | |
| Article | 356 | ≥149 | ≥41.85 | Aligholi et al (36) | 2008 | |
| Article | 277 | ≥100 | ≥36 | Fatholahzadeh et al (37) | 2008 | |
| Article | 222 | 122 | 55 | Habibi et al (38) | 2008 | |
| Abstract | 235 | 110 | 46.8 | Azimian et al (39) | 2008 | |
| Abstract | 65 | ≥33 | ≥50.8 | BagherzadehYazdchi et al (40) | 2008 | |
| Abstract | 50 | 22 | 44 | Dadaei et al (41) | 2008 | |
| Abstract | 80 | 40 | 50 | Salehipour et al (42) | 2008 | |
| Article | 927 | ≥306 | ≥33 | Aligholi et al (43) | 2009 | |
| Article | 322 | 93 | 28.88 | Emaneini et al (44) | 2009 | |
| Article | 174 | ≥84 | ≥48.2 | Najar-peerayeh et al (45) | 2009 | |
| Article | 321 | 282 | 88 | Rahimi et al (46) | 2009 | |
| Article | 100 | 53 | 53 | Yadegar et al (47) | 2009 | |
| Abstract | 250 | 109 | ≥43.6 | Farhadian et al (48) | 2009 | |
| Article | 150 | 64 | ≥42.67 | Javan et al (49) | 2010 | |
| Abstract | 55 | 28 | 50.9 | Faghri et al (50) | 2010 | |
| Article | 165 | ≥87 | ≥52.72 | Aligholi et al (51) | 2011 | |
| Article | 42 | 18 | 42.8 | Nowroozi et al (52) | 2011 | |
| Article | 186 | 127 | 68.3 | Saderi et al (53) | 2011 | |
| Article | 106 | 62 | 58.49 | Shahsavan et al (54) | 2011 | |
| Abstract | 100 | 90 | 90 | Ghorbani et al (55) | 2011 | |
| Abstract | 150 | 67 | 44.6 | Mobaiyen et al (56) | 2011 | |
| Abstract | 104 | 76 | 73.1 | Sahebnasagh et al (57) | 2011 | |
| Article | 125 | 107 | 85.6 | Sepehriseresht et al (58) | 2012 | |
| Article | 100 | 56 | 56 | RazaviDavoodi et al (59) | 2012 | |
| Abstract | 48 | 17 | 35.4 | AziziBarjini et al (60) | 2012 | |
| Tonekabon | Abstract | 55 | 30 | ≥54.54 | Forghani et al (61) | 2011 |
1 MRSA strains were detected/confirmed by PCR amplificationofmecAgene
PCR of mecAwas done onlyfor strains resistant to methicillin by phenotypic methods
Results were obtained by comparing references (31) and (32)
Figure 1.
Forrest plot of the current relative frequency of mecA-MRSA among clinical S. aureus isolates in different Iranian studies
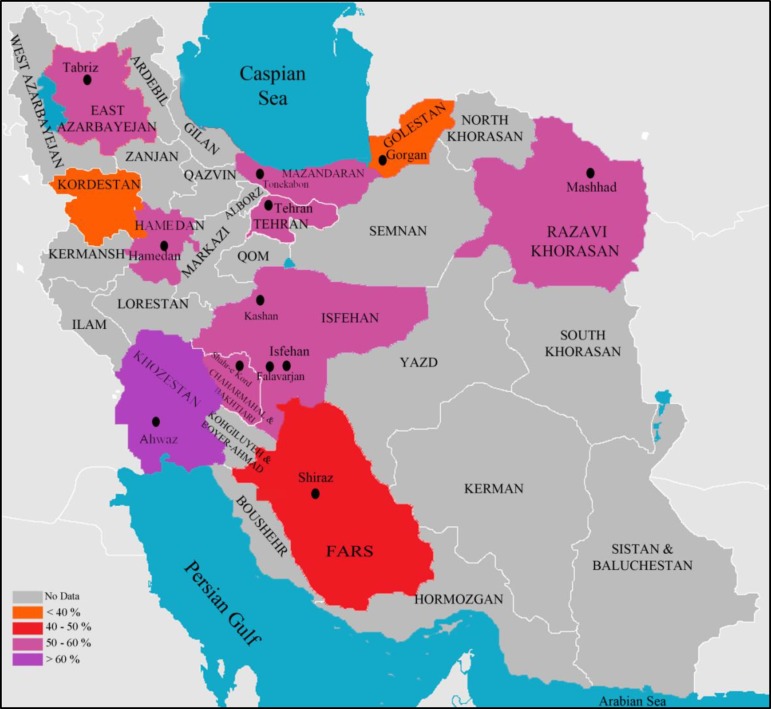
Pooled estimation of 7464 S. aureus samples showed 52.7%±4.7 (95% CI) of strains to be mecA positive. These samples were taken from 14 different Iranian cities (Figure 2). MRSA R.F. varied from 20.48% to 90% in Isfehan and Tehran, respectively (22, 55). Amoderate heterogeneity (I2= 48.5%) of MRSA R.F. In the studies conducted in Tehran, the capital city of Iran (ranging from 28.88% to 90%, mean 52.7% [95% CI: 46.6%-0.58.8%]) (33-60) was found.
A significant correlation suggested that bias exists but does not directly address the implication of bias (Kendall’s tau= 0.21). The results of a Begg and Mazumdar’s rank correlation test supported its possibility (P= 0.039).
Figure 2.
Prevalence of mecA-Methicillin-Resistant Staphylococcus aureus in Iran
Discussion
During the past decade, assays for detection of mecA gene for staphylococci became popular among Iranian researchers. Based on these studies, we reported the cumulative prevalence of MRSA and provided a map to illustrate the epidemiology of MRSA in Iran. In two previous global reports, the prevalence of MRSA in Iran was unknown (2, 7).
According to our study, the mean prevalence of MRSA in Iran was 52.7%±4.7 and was more than fifty percent in many Iranian cities. This finding indicates that physicians may face difficulties in treatment of in more than half of S. aureus infections. Keeping in mind the high prices of newer agents, vancomycin appears to be a suitable agent to fight this pathogen, lthough recent emergence of vancomycin resistance in Iran is really alarming (36, 62).
In a regional perspective, Iran has a higher prevalence of MRSA compared to reports from neighboring countries in the Middle East with the exception of Iraq (2, 7). The ANSORP study which reported HA-MRSA rates for eight Asian countries showed higher percentage of MRSA in those countries compared to Iran. However, judgment cannot be made because most Iranian studies did not clearly divide their S. aureus population to HA- and CA- infections (63).
From an international stand, our data are in the same range as Argentina and Mexico in Latin America (64). Mean Prevalence of MRSA in Iran is moderately higher than Australia and lower than the United States (65, 66). However, recent reports have shown that MRSA rates are declining in United States (67, 68). Prevalence of MRSA in Europe is heterogeneous with average lower than other continents but Portugal seems to have a similar rate of MRSA rates similar to our country (7).
The heterogeneity of MRSA prevalence at national and international level is not completely understood. Possible explanations are different in infection control practices, antimicrobial administration, human population, predominant strain(s), study design and laboratory testing for determining resistance (2, 69).
This study has some limitations. First, it cannot fully represent Iran because there were no data on mecA-MRSA from many parts of the country. However, as described above, this is preferred to mixing the results from different phenotypic methods with genotypic ones. Second, due to limited access to in-press articles and theses, some studies might have been missed, which is also suggested by statistical analysis.
Conclusions
Our study showed that the mean MRSA R.F. among Iranian studies is in the high range. Thus, measures should be taken to keep the emergence and transmission of these strains to a minimum.
Acknowledgment
An abstract of these data has been sent to the International Symposium of Staphylococci and Staphylococcal Infections (ISSSI) 2012, Lyon, France. The authors wish to express their gratitude to Aidin Faroughi, Yeganeh Khazaei, Somayeh Mahdipoor, Zahra Moravvej, Ghazaleh Nouri and Amin Rezaeian for their help in the analyzing step and Najmeh Seifi for her assistance in the search.
References
- 1.Deleo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet . 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet . 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 3.Cookson B. Five decades of MRSA: controversy and uncertainty continues. Lancet. 2011;378:1291–1292. doi: 10.1016/s0140-6736(11)61566-3. [DOI] [PubMed] [Google Scholar]
- 4.Köck R, Becker K, Cookson B, van Gemert-PijnenJE, Harbarth S, Kluytmans J, et al. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro Surveill. 2010;15:1–9. doi: 10.2807/ese.15.41.19688-en. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis . 2003;36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 6.Moellering RC. Why has methicillin-resistant Staphylococcus aureusbecome such a successful pathogen in adults. Infect Dis Clin Pract . 2010;18:286–291. [Google Scholar]
- 7.Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, et al. Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int J Antimicrob Agents . 2012;39:273–282. doi: 10.1016/j.ijantimicag.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Graveland H, Duim B, van DuijkerenE, Heederik D, Wagenaar JA. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Int J Med Microbiol. 2011;301:630–634. doi: 10.1016/j.ijmm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Forbes BA. Issues in the identification and susceptibility testing of Staphylococci. In: Crossley KB, Jefferson KK, Archer GL, Fowler VG, editors. Staphylococci in Human Disease. 2nd ed. Wiley-Blackwell: Wiley-Blackwell; 2009. pp. 235–252. [Google Scholar]
- 10.Karami S, Rahbar M, Vand YousefiJ. Evaluation of five phenotypic methods for detection of methicillin resistant Staphylococcus aureus (MRSA) Iran J Pathol . 2011;6:27–31. [PMC free article] [PubMed] [Google Scholar]
- 11.Radlberger P, Zechmeister I. Innovative framework for evidence-based decision making in healthcare – standardised working in HTA (WP1.2): HTA- Projektbericht. 2011. [2): HTA- Projektbericht]. Available at: http://eprints.hta.lbg.ac.at/932/1/HTA-Projektbericht_Nr.44a.pdf.
- 12.Clinical and Laboratory Standards Institute/NCCLS. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI document M100-S17. Wayne, PA: Clinical and Laboratory Standards Institute; 2007. [Google Scholar]
- 13.Clinical and Laboratory Standards Institute/NCCLS. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI document M100-S18. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 14.Ekrami A, Samarbafzadeh A, Alavi M, Kalantar E, Hamzeloi F. Prevalence of methicillin resistant Staphylococcus species isolated from burn patients in a burn center, Ahvaz, Iran. Jundishapur J Microbiol. 2010;3:84–91. [Google Scholar]
- 15.Moosavian M, Torabipour M. Identification of mecA gene in phenotypic methicillin-resistant Staphylococcus aureus strains isolated from clinical specimens. The First Iranian International Congress of Medical Bacteriology; 4-7 sep; Tabriz, Iran. 2011. 255 pp. [Google Scholar]
- 16.Khosravi AD, Hoveizavi H, Farshadzadeh Z. The prevalence of genes encoding leukocidins in Staphylococcus aureus strains resistant and sensitive to methicillin isolated from burn patients in Taleghani hospital, Ahvaz, Iran. Burns. 2012;38:247–251. doi: 10.1016/j.burns.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Heidari M, Momtaz H, Madani M. Detection of the antibiotic resistance genes in Staphylococcus aureus isolated from human infections and bovine mastitis. Afr J Microbiol Res . 2011;5:5132–5136. [Google Scholar]
- 18.Abdollahi A, Koohpayeh A, Najafipour S, Mansouri Y, Abdollahi S, Jafari S. Evaluation of drug esistance and SCCmec genotype in MRSA strains. Behdad . 2012;1:47–52. [Google Scholar]
- 19.Vaez H, Tabaraei A, Moradi A, Ghaemi EA. Evaluation of methicillin resistance Staphylococcus aureus isolated from patients in Golestan province-north of Iran. Afr J Microbiol Res . 2011;5:432–436. [Google Scholar]
- 20.Zamani A, Sadeghian S, Ghaderkhani J, Alikhani MY, Najafimosleh M, Taghi GoodarziM, et al. Detection of methicillin-resistance (mec-A) gene in Staphylococcus aureus strains by PCR and determination of antibiotic susceptibility. Ann Microbiol. 2007;57:273–276. [Google Scholar]
- 21.Alizargar J, Moravveji A. Prevalence of methicillin resistant Staphylococcus in Kashan. First International and 12th Iranian Congress of Microbiology; 23-26 May; Kermanshah, Iran. 2011. 1328 pp. [Google Scholar]
- 22.Havaei SA, Vidovic S, Tahmineh N, Mohammad K, Mohsen K, Starnino S, et al. Epidemic methicillin-susceptible Staphylococcus aureus lineages are the main cause of infections at an Iranian university hospital. J Clin Microbiol. 2011;49:3990–3993. doi: 10.1128/JCM.05445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeinali E, Moniri R, Safari M, Mousavi GA. Molecular characterization and SCCmec typing in meticillin-resistant Staphylococcus aureus isolated from clinical samples. Feyz, J Kashan Univ Med Sci . 2011;14:439–446. [Google Scholar]
- 24.Naderi NasabM, Tavakolafshar J, Nazem M, Fatehnasab P, Faramarzi P, Khodadoost M. Detection of methicillin-resistant Staphylococcus aureus by phenotypic methods. Med J Mashhad Univ Med Sci. 2005;48:7–16. [Google Scholar]
- 25.Vaiseh P, Ramezanzadeh R, Deilami Z. Identification of class I integron and mecA in Staphylococcus strains typed by REP-PCR method. The Congress of Infections and Antibiotic Resistance; 2-3 May; Gilan, Iran. 2012. 101 pp. [Google Scholar]
- 26.Shariati L, Validi M, Tabatabaiefar MA, Karimi A, Nafisi MR. Comparison of real-time PCR with disk diffusion, agar screen and E-test methods for detection of methicillin-resistant Staphylococcus aureus. Curr Microbiol. 2010;61:520–524. doi: 10.1007/s00284-010-9647-9. [DOI] [PubMed] [Google Scholar]
- 27.Japoni A, Alborzi A, Orafa F, Rasouli M, Farshad S. Distribution patterns of methicillin resistance genes (mecA) in Staphylococcus aureusisolated from clinical specimens. Iran Biomed J. 2004;8:173–178. [Google Scholar]
- 28.Nikbakht M, Nahaei MR, Akhi MT, Asgharzadeh M, Nikvash S. Molecular fingerprinting of meticillin-resistant Staphylococcus aureus strains isolated from patients and staff of two Iranian hospitals. J Hosp Infect . 2008;69:46–55. doi: 10.1016/j.jhin.2008.01.034. [DOI] [PubMed] [Google Scholar]
- 29.Zarrini G, Aein F, Bahari DelgoshaZ. Study of nosocomial methicillin resistant Staphylococcus aureus (MRSA) using disc diffusion, MASTAScreen MRSA + kit and PCR methods. The Second Iranian Congress of Clinical Microbiology; 7-9 October; Shiraz, Iran. 2008. 149 pp. [Google Scholar]
- 30.Esfandyari KalajahiA, Hasani A, Riyazi A, Abbasian S, Pourmohammad A, Hasani A. Detection of virulence genes of Staphylococcus aureus isolated from nasal specimens of patients admitted in high risk wards of University teaching hospital, Tabriz. 4th Congress of Laboratory and Clinic; 21-23 December; Tehran, Iran. 2011. 491 pp. [Google Scholar]
- 31.Kiani niaM. Development of Multiplex PCR for the detection of aph(3)-IIIa and aac(6′)/aph(2′′) genes in methicillin-resistant Staphylococcus aureus (MRSA) isolates from Northwest, Iran. 4th Congress of Laboratory and Clinic; 21-23 December; Tehran, Iran. 2011. 996 pp. [Google Scholar]
- 32.Kiani niaM, Hasani A, Hasani A, Mirza AhmadiS, Sadeghifard M, Deghani L. Nasal colonization of high risk group patients of northwest Iran by MRSA Predictability of resistance and concern about prevention. 4th Congress of Laboratory and Clinic; 21-23 December; Tehran, Iran. 2011. 853 pp. [Google Scholar]
- 33.Mirsalehian A, Jebelameli F, Kazemi B, Alizadeh SA. Comparison of disc diffusion method withpolymerase chain reaction for detectingmethicillin resistancein clinical isolates of Staphylococci. Tehran Univ Med J. 2003;61:420–425. [Google Scholar]
- 34.Aligholi M, Emaneini M, Hashemi FB, Shahsavan S, Jabalameli F, Kazemi B. Determination of antimicrobial resistance pattern of Staphylococcus aureus isolated from clinical specimens. Tehran Univ Med J. 2006;64:26–32. [Google Scholar]
- 35.Mostafaee M, Behzadian-Nejad G, Najar-peerayeh S, Rezaei YazdiH, Tohidpoor A. Prevalence of mecA gene in Staphylococcus aureus strains isolated from Tehran hospitals, 1385. The First Iranian Congress of Clinical Microbiology; 8-10 May; Shiraz, Iran. 2007. 16 pp. [Google Scholar]
- 36.Aligholi M, Emaneini M, Jabalameli F, Shahsavan S, Dabiri H, Sedaght H. Emergence of high-level vancomycin-resistant Staphylococcus aureus in the Imam Khomeini Hospital in Tehran. Med Princ Pract. 2008;17:432–434. doi: 10.1159/000141513. [DOI] [PubMed] [Google Scholar]
- 37.Fatholahzadeh B, Emaneini M, Gilbert G, Udo E, Aligholi M, Modarressi MH, et al. Staphylococcal cassette chromosome mec (SCCmec) analysis and antimicrobial susceptibility patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolates in Tehran, Iran. Microb Drug Resist . 2008;14:217–220. doi: 10.1089/mdr.2008.0822. [DOI] [PubMed] [Google Scholar]
- 38.Habibi M, Saderi H, Owlia P, Asadi KaramM. Detection of methicillin resistance in Staphylococcus aureus by disk diffusion and PCR methods. Iran J Pathol. 2008;3:11–14. [Google Scholar]
- 39.Azimian A, Najar-peerayeh S, Farshchian M, Naderi M, Salehipoor Z, Mostafaee M. Occurrence of the methicillin-resistant Staphylococcus aureus (MRSA) among clinical samples in Tehran and its correlation with the site of infection. The Second Iranian Congress of Clinical Microbiology; 7-9 October; Shiraz, Iran. 2008. 1101 pp. [Google Scholar]
- 40.Bagherzadeh YazdchiS, Pourmand MR, Zaeimi YazdiJ. Anbiotic susceptibility patterns and detection of coa and mecA genes in the Iranian isolates of Staphylococcus aureus. 13th International Congress on Infectious Diseases; 19-22 June; Kuala Lumpur, Malaysia. 2008. e271 pp. [Google Scholar]
- 41.Dadaei T, Eftekhar F. Correlation of biofilm formation in clinical isolates of Staphylococcus aureus by colony morphology and microplate measurement of biofilm formation. The Second Iranian Congress of Clinical Microbiology; 7-9 October; Shiraz, Iran. 2008. 107 pp. [Google Scholar]
- 42.Salehipour Z, Azimian A, Ghazvini K. Phenotypic and genotypic study of drug resistance in Staphylococcus aureus strains isolated from blood culture of septicemia pateints in selected hospitals of Tehran. The Second Iranian Congress of Clinical Microbiology; 7-9 October; Shiraz, Iran. 2008. 172 pp. [Google Scholar]
- 43.Aligholi M, Emaneini M, Jabalameli F, Shahsavan S, Abdolmaleki Z, Sedaghat H, et al. Antibiotic susceptibility pattern of gram-positive cocci cultured from patients in three university hospitals in Tehran, Iran during 2001-2005. Acta Med Iran . 2009;47:329–334. [Google Scholar]
- 44.Emaneini M, Taherikalani M, Eslampour MA, Sedaghat H, Aligholi M, Jabalameli F, et al. Phenotypic and genotypic evaluation of aminoglycoside resistance in clinical isolates of Staphylococci in Tehran, Iran. Microb Drug Resist . 2009;15:129–132. doi: 10.1089/mdr.2009.0869. [DOI] [PubMed] [Google Scholar]
- 45.Najar-Peerayeh S, Azimian A, Mostafaee M, Siadat SD. Identification of methicillin-resistant Staphylococcus aureus by disk diffusion method, determination of MIC, and PCR for mecA Gene. Modares Med Sci J: Pathobiol. 2009;12:61–69. [Google Scholar]
- 46.Rahimi F, Bouzari M, Maleki Z, Rahimi F. Antibiotic susceptibility pattern among Staphylococcus spp. with emphasis on detection of mecA gene in methicillin resistant Staphylococcus aureus isolates. Iran J Clin Infect Dis . 2009;4:143–150. [Google Scholar]
- 47.Yadegar A, Sattari M, Mozafari NA, Goudarzi GR. Prevalence of the genes encoding aminoglycoside-modifying enzymes and methicillin resistance among clinical isolates of Staphylococcus aureus in Tehran, Iran. Microb Drug Resist . 2009;15:109–113. doi: 10.1089/mdr.2009.0897. [DOI] [PubMed] [Google Scholar]
- 48.Farhadian A, Behzadian Nejad, Najar-peerayeh S, Farhadian A, Vaziri Determination of vancomycin and methicillin resistant in isolates of S. aureus in hospitals in the Iran.Third Iranian Congress of Clinical Microbiology; 6-8 October; Shiraz, Iran. 2009. 21 pp. [Google Scholar]
- 49.Javan E, Falahati H, Seifi M, Talebi M, Ebrahimpoor G, Poorshafi M. Detection of mecA gene in oxacillin resistant Staphylococcus aureus strains isolated from Tehran hospitals. Iran J Infect Dis Trop Med . 2010;49:17–22. [Google Scholar]
- 50.Faghri J, Azimian A, Sadighian H, Khosrojerdi M. Occurrence of the methicillin-resistant Staphylococcus aureus (MRSA)among respiratory tract samples in patients of selected Tehran hospitals. 4th Iranian Congress of Clinical Microbiology; 9-11 November; Isfahan, Iran. 2010. 75 pp. [Google Scholar]
- 51.Aligholi M, Mirsalehian A, Halimi S, Imaneini H, Taherikalani M, Jabalameli F, et al. Phenotypic and genotypic evaluation of fluoroquinolone resistance in clinical isolates of Staphylococcus aureus in Tehran. Med Sci Monit . 2011;17:PH71–74. doi: 10.12659/MSM.881920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nowroozi J, Pakzad P, Ebrahimi E, Razavipour R. Detection of biocide resistance genes, qac A/B and smr, among isolated Staphylococcus aureus from clinical and non-clinical sources. Pejouhandeh. 2011;16:83–91. [Google Scholar]
- 53.Saderi H, Emadi B, Owlia P. Phenotypic and genotypic study of macrolide, lincosamide and streptogramin B (MLSB) resistance in clinical isolates of Staphylococcus aureus in Tehran, Iran. Med Sci Monit . 2011;17:BR48–53. doi: 10.12659/MSM.881386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shahsavan S, Emaneini M, Noorazar KhoshgnabB, Khoramian B, Asadollahi P, Aligholi M, et al. A high prevalence of mupirocin and macrolide resistance determinant among Staphylococcus aureus strains isolated from burnt patients. Burns . 2012;38:378–382. doi: 10.1016/j.burns.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Ghorbani S, Imani FooladiAA, Nourani MR. Prevalence of methicillin-resistance (mec-A) gene in Staphylococcus aureus strains from scar by PCR and determination of antibiotic susceptibility. 4th Congress of Laboratory and Clinic; 21-23 December; Tehran, Iran. 2011. 540 pp. [Google Scholar]
- 56.Mobaiyen H, Molaabaszadeh H, Modirrousta S, Reza SoltaniS. Surveying of Staphylococcus aureus methicillin resistant (mec-A) and determine its antibiotic susceptibility with PCR method in Tehran in 2010. International Union of Microbiological Societies 2011 Congress; Sapporo, Japan. 2011. P-BA25-11 pp. [Google Scholar]
- 57.Sahebnasagh R, Saderi H, Owlia P. Detection of methicillin-resistant Staphylococcus aureus strains from clinical samples in Tehran by detection of the mecA and nuc genes. The First Iranian International Congress of Medical Bacteriology; 4-7 September; Tabriz, Iran. 2011. 195 pp. [Google Scholar]
- 58.Sepehriseresht S, Boroumand MA, Pourgholi L, Anvari M, Habibi E, Sattarzadeh M, et al. Emergence of mupirocin-resistant MRSA among Iranian clinical isolates. Comp Clin Pathol doi: 10.1007/s00580-012-1472-z. [Google Scholar]
- 59.Razavi DavoodiN, Vand YousefiA, Harzandi N, Hahrafi A, Rajaei B, Gerayeshnejad S, et al. Molecular detection of methicillin resistantStaphylococcus aureus (MRSA) and methicillinresistant coagulase-negative Staphylococcus (CoNS) inIran. Afr J Microbiol Res. 2012;6:3716–3721. [Google Scholar]
- 60.Azizi BarjiniK, Mousazadeh M, Amani J, Asadi A, Khalili S. Detection of MRSA by Disk diffusion and PCR method and detection of resistance pattern. The Congress of Rational Usage of Antibiotics; 27-28 February; Sari, Iran. 2012. 38 pp. [Google Scholar]
- 61.Forghani F, Alipourfard I, Ghayyomi M, Mahmudi S, Heydari N. Comparative study of methicillin resistant Staphylococcus aureus (MRSA) molecular detection by PCR, with bacteriological methods. First International and 12th Iranian Congress of Microbiology; 23-26 May; Kermanshah, Iran. 2011. 1194 pp. [Google Scholar]
- 62.Dezfulian A, Aslani MM, Oskoui M, Farrokh P, Azimirad M, Dabiri H, et al. Identification and characterization of a high vancomycin-resistant Staphylococcus aureusharboring vanAgene cluster isolated from diabetic foot ulcer. Iran J Basic Med Sci. 2012;15:803–806. [PMC free article] [PubMed] [Google Scholar]
- 63.Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother . 2011;66:1061–1069. doi: 10.1093/jac/dkr024. [DOI] [PubMed] [Google Scholar]
- 64.Guzmán-Blanco M, Hsueh PR, Isturiz R, Alvarez C, Bavestrello L, Gotuzzo E, et al. Epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) in Latin America. Int J Antimicrob Agents. 2009;34:304–308. doi: 10.1016/j.ijantimicag.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 65.Nimmo GR, Pearson JC, Collignon PJ, Christiansen KJ, Coombs GW, Bell JM, et al. Antimicrobial susceptibility of Staphylococcus aureus isolated from hospital inpatients, 2009: Report from the Australian Group on Antimicrobial Resistance. Commun Dis Intell . 2011;35:237–243. [PubMed] [Google Scholar]
- 66.Styers D, Sheehan DJ, Hogan P, Sahm DF. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Ann Clin Microbiol Antimicrob. 2006;5:1–9. doi: 10.1186/1476-0711-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA . 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 68.Ray GT, Morrison MA, Baxter R. Trends and characteristics of culture-confirmed Staphylococcus aureusinfections in a large U.S. integrated health care organization. J Clin Microbiol. 2012;50:1950–1957. doi: 10.1128/JCM.00134-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simor AE, Loeb M. Epidemiology of healthcare-associated Staphylococcus aureus infections. In: Crossley KB, Jefferson KK, Archer GL, Fowler VG, editors. Staphylococci in Human Disease. 2nded. Oxford, UK: Wiley-Blackwell; 2009. pp. 290–309. [Google Scholar]