Abstract
The phytohormone abscisic acid (ABA) regulates many key processes in plants, such as seed germination, seedling growth, and abiotic stress tolerance. In recent years, a minimal set of core components of a major ABA signaling pathway has been discovered. These components include a RCAR/PYR/PYL family of ABA receptors, a group of PP2C phosphatases, and three SnRK2 kinases. However, how the interactions between the receptors and their targets are regulated by other proteins remains largely unknown. In a companion paper published in this issue, we showed that ROP11, a member of the plant-specific Rho-like small GTPase family, negatively regulates multiple ABA responses in Arabidopsis. The current work demonstrated that the constitutively active ROP11 (CA-ROP11) can modulate the RCAR1/PYL9-mediated ABA signaling pathway based on reconstitution assays in Arabidopsis thaliana protoplasts. Furthermore, using luciferase complementation imaging, yeast two-hybrid assays, co-immunoprecipitation assays in Nicotiana benthamiana and bimolecular fluorescence complementation assays, we demonstrated that CA-ROP11 directly interacts with ABI1, a signaling component downstream of RCAR1/PYL9. Finally, we provided biochemical evidence that CA-ROP11 protects ABI1 phosphatase activity from inhibition by RCAR1/PYL9 and thus negatively regulates ABA signaling in plant cells. A model of how ROP11 acts to negatively regulate ABA signaling is presented.
Keywords: ROP11 GTPase, ABA signaling, RCAR1/PYL9A, ABI1, Arabidopsis
Introduction
The phytohormone abscisic acid (ABA) regulates many key processes in plants, including seed germination, seedling growth, abiotic stress tolerance, and especially drought resistance (Guo et al. 2011; Finkelstein et al. 2002). In the past decades, many molecular components involved in ABA signaling were identified (Hubbard et al. 2010). In Arabidopsis, these include the negative regulators ABI1, ABI2, HAB1, HAB2, and PP2CA, which belong to a subfamily of type 2C protein phosphatases (PP2Cs) (Gosti et al. 1999; Merlot et al. 2001; Leonhardt et al. 2004; Saez et al. 2004; Yoshida et al. 2006; Nishimura et al. 2007; Rubio et al. 2009). Downstream of these PP2C proteins are positive regulators that are members of the SNF1-related protein kinase 2 family (SnRK2s), i.e., SnRK2.2, SnRK2.3, and SnRK2.6 (Yoshida et al. 2006; Umezawa et al. 2009; Vlad et al. 2009). The phosphorylated SnRKs can activate transcription factors such as ABF2/AREB1 (Johnson et al. 2002; Furihata et al. 2006), which in turn promote transcription of RD29B and other ABA-responsive genes. In the absence of ABA, the activities of SnRK2s are inhibited by ABI1 and other PP2C phosphatases through dephosphorylation. ABA binds to the RCAR/PYR/PYL family of receptors (Ma et al. 2009; Park et al. 2009; Santiago et al. 2009; Nishimura et al. 2010). The binding of ABA facilitates the formation of complexes between the receptors and PP2Cs, leading to the inhibition of the PP2C phosphatase activities, thus converting the SnRK2 kinases from their inactive form to active form to elicit downstream responses. By using in vitro reconstitution assays in protoplasts, researchers have recently demonstrated that RCAR/PYL/PYR-ABI1-SnRK2.6/OST1 constitute a minimal set of core components of a complete, major ABA signaling pathway (Fujii et al. 2009).
ROPs (Rho GTPase of plants) are a group of plant-specific, Rho-like, small GTPases that work as signaling switches to control cell polar growth and hormone signaling (Nibau et al. 2006). In Arabidopsis, the ROP family consists of 11 members that share high sequence identities (Zheng and Yang 2000). In a companion paper published in this issue, we showed that one member of this ROP family, ROP11, is a negative regulator of multiple ABA responses in Arabidopsis (Li et al. 2012). These responses include ABA-mediated seed germination, seedling growth, stomatal closing, induction of ABA-responsive genes, and tolerance to drought stress. The regulation of multiple ABA responses suggested that ROP11 may act in an early step of ABA signaling.
Since the discovery of the RCAR/PYR/PYL family of ABA receptors, many advances have been made in elucidating the structural basis for interactions between ABA receptor(s) and their downstream PP2C targets (Hubbard et al. 2010). How this interaction is regulated by other protein factors, however, has not been investigated. Here, we report that ROP11 can directly interact with ABI1, a signaling component immediately downstream of the ABA receptor RCAR1/PYL9. This interaction protects ABI1 phosphatase activity from inhibition by the ABA receptor RCAR1/PYL9. A model of how ROP11 acts to negatively regulate ABA signaling is presented.
Results
ROP11 modulates the RCAR1-ABI1-SnRK2-mediated ABA signaling pathway
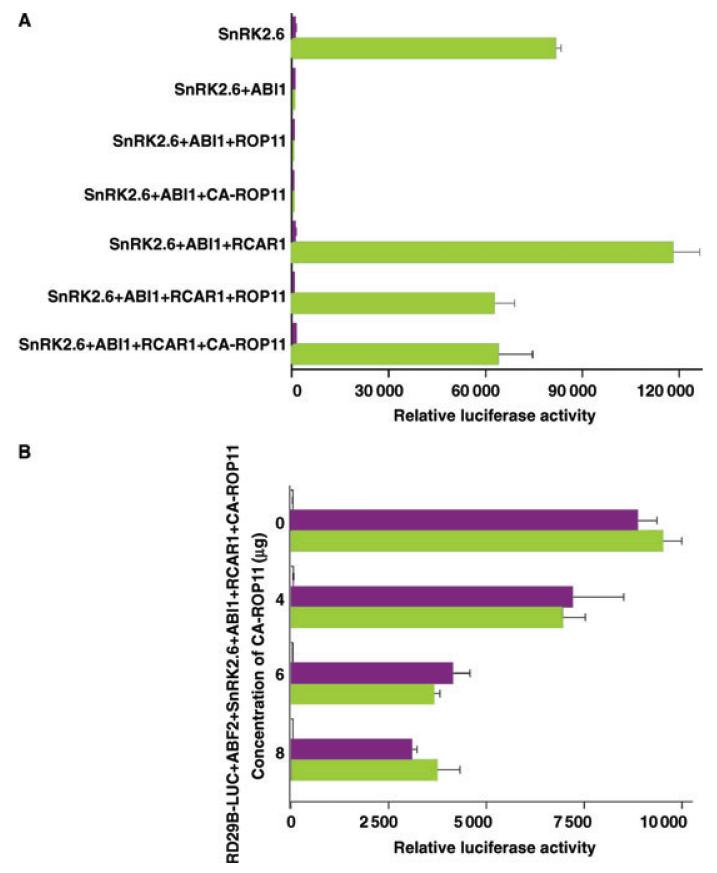
Because ROP11 is located on plasma membranes and negatively regulates multiple ABA-mediated responses (Li et al. 2012), we speculated that ROP11 might act in the early steps of the ABA signaling pathway. Therefore, we used Arabidopsis protoplasts to examine whether ROP11 regulates ABA signaling by modulating the activities of some core components that are involved in the early ABA signaling steps for stress-responsive gene expression. Previously, an in vitro assay system using protoplasts isolated from the snrk2.2/2.3/2.6 triple mutant has been successfully used to reconstruct an ABA signaling pathway (Fujii et al. 2009). In this study, the ABA signaling pathway for stress-responsive gene expression was reconstituted by co-transfection of the RD29B-LUC (Luciferase) reporter gene with ABF2, SnRK2.6, ABI1, and RCAR1 into protoplasts isolated from the snrk2.2/2.3/2.6 triple mutant. As shown in Figure 1A, co-transfection of RD29B-LUC and ABF2 with SnRK2.6 into the protoplast resulted in a high induction of LUC activity by ABA. Addition of ABI1 suppressed the LUC activity. This suppression of LUC activity by ABI1 could be reversed by co-transfection of an ABA receptor gene RCAR1/PYL9 (hereafter referred to as RCAR1). ROP11 or CA-ROP11 did not interfere with the inhibition of SnRK2.6 by ABI1. Both ROP11 and CA-ROP11, however, antagonized the RCAR1-dependent inhibition of ABI1 activity and thus reduced the RCAR1-dependent RD29B-LUC expression in the presence of ABA. The antagonistic effect of CA-ROP11 on RCAR1 activity increased as the concentration of CA-ROP11 plasmid increased from 4 to 8 μg in the protoplast assay (Figure 1B). These results demonstrated that ROP11 could modulate the RCAR1-ABI1-SnRK2-mediated ABA signaling pathway.
Figure 1. The effects of ROP11 on ABA signaling analyzed in protoplast-based assays.
(A) RD29B-LUC, ABF2, SnRK2.6, ABI1, RACR1, ROP11, and CA-ROP11 plasmids were co-transfected into protoplasts isolated from the snrk2.2/2.3/2.6 triple mutant in the indicated combinations. After transfection, protoplasts were incubated for 4.5 h under light in the absence (white bars) or presence (green bars) of 5 μM ABA, and luciferase activity was measured. Error bars indicate SE (n = 3).
(B) Dose-dependent response of CA-ROP11 inhibition of RCAR1 activity. RD29B-LUC, ABF2, SnRK2.6, ABI1, and RCAR1 were co-transfected with 0, 4, 6, or 8 μg of CA-ROP11 plasmid, and the transfected protoplasts were incubated in WI medium supplemented with 0 (white bars), 2.5 (purple bars), or 5 μM ABA (green bars) before luciferase activity was measured. Error bars indicate SE (n = 3).
ROP11 directly interacts with ABI1
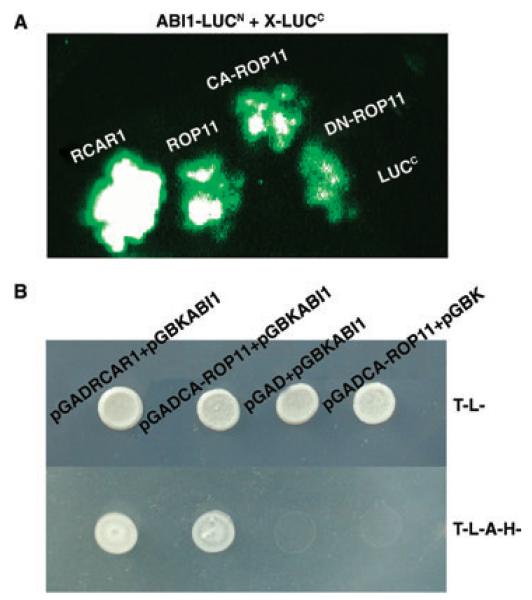
To find the target molecules of ROP11 in the ABA signaling pathway (i.e., to determine whether ROP11 can directly interact with RCAR1 and ABI1), we performed luciferase complementation imaging (LCI) assays (Chen et al. 2008) using leaves of N. benthamiana. The coding sequence of ABI1 was fused to the C-terminal half of the LUC gene (LUCN). Accordingly, the coding sequences of ROP11, CA-ROP11, DN-ROP11, and RCAR1 were fused to the N-terminal half of the LUC gene (LUCC). As a positive control, when LUCN-ABI1 and RCAR-LUCC were transiently co-expressed in the leaves of N. benthamiana, a strong LUC activity was observed due to the reconstruction of the active luciferase proteins through the interaction between ABI1 and RCAR1 (Figure 2A). In contrast, co-expression of LUCN-ABI1 and the empty LUCC vector showed no LUC activity. When LUCN-ABI1 was co-expressed with individual ROP11, CA-ROP11, or DN-ROP11, strong luciferase activity was detected. These results indicated that ABI1 has strong interactions with ROP11 proteins expressed from all three ROP11 constructs. The interaction between CA-ROP11 and ABI1 was then tested using the yeast two-hybrid system. Because the full-length ORF of ABI1 is auto-activating, the coding sequence of ABI1 that only included residues 119-434 was cloned into the bait vector pGBKT7. CA-ROP11 and RCAR1 coding sequences were cloned into the prey vector pGADT7. The resultant constructs were co-transformed into the yeast strain AH109. Transformants selected from SD/-Trp/-Leu medium were transferred onto SD/-Trp/-Leu/-Ade/-His selective medium and allowed to grow for 3–5 days. As shown in Figure 2B, only the yeast cells that were co-transformed with ABI1 and RCAR1 (as a positive control) or with ABI1 and CA-ROP11 could grow on the selection medium. The yeast cells that harbored the ABI1 and empty prey vector pGADT7 or CA-ROP11 and empty bait vector pGBKT7 could not grow on this medium. These results confirmed that CA-ROP11 and ABI1 could interact with each other.
Figure 2. Interactions between ROP11 and ABI1.
(A) LIC assays of the interaction between ROP11 and ABI1. RCAR1, ROP11, CA-ROP11, and DN-ROP11 were cloned into the LUCC vector, and ABI1 and ABI2 were cloned into the LUCN vector. The paired LUCC and LUCN fusion constructs were co-transformed into the leaves of Nicotiana benthamiana, and bioluminescence was captured with a CCD camera.
(B) Yeast two-hybrid analysis of the interaction between CA-ROP11 and ABI1.
In (A) and (B), the interaction between ABI1 and RCAR1 was used as a positive control.
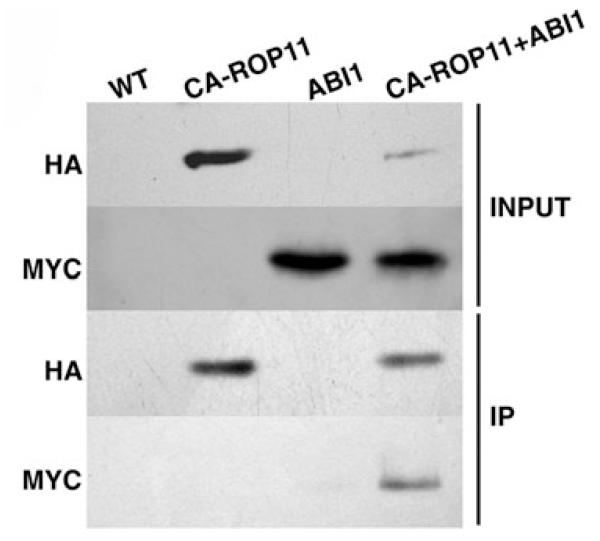
For further determination of whether CA-ROP11 and ABI1 co-exist in the same protein complex, a co-immunoprecipitation (Co-IP) experiment was performed using N. benthamiana leaves that were co-transformed with HA-tagged CA-ROP11 and MYC-tagged ABI1 gene constructs. We first confirmed that the tagged CA-ROP11 and ABI1 proteins could be successfully expressed in N. benthamiana leaves (Figure 3, Input). Using Western blot assays, we detected the expression of CA-ROP11 with anti-HA antibodies in the leaves transformed with CA-ROP11 or co-transformed with a CA-ROP11 and ABI1 construct. Similarly, the expression of ABI1 proteins was demonstrated with MYC antibodies in the leaves of plants transformed with ABI1 or co-transformed with a CA-ROP11 and ABI1 construct. Finally, the proteins extracted from leaves co-transformed with CA-ROP11 and ABI1 gene constructs were subjected to immunoprecipitation with anti-HA antibodies. In this HA-precipitated protein fraction, MYC-ABI1 was detected by anti-MYC antibodies (Figure 3, IP). These results demonstrated that ABI1 and CA-ROP11 co-existed in the same protein complex.
Figure 3. Co-immunoprecipitation analysis of association of MYC-ABI1 with HA-CA-ROP11.
Total proteins were extracted from leaves transiently transformed with the HA-CA-ROP11 construct alone or combined with the MYC-ABI1 construct. The antibodies used in Western blot are indicted on left. Input, total protein extracts; IP, HA-immunoprecipitated proteins.
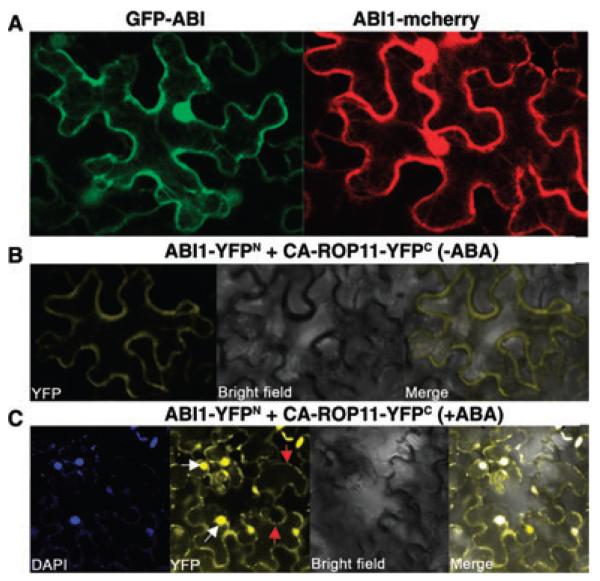
Previously, ABI1 was shown to be located mainly in the cytosol and nucleus (Moes et al. 2008; Umezawa et al. 2009). However, our analysis of expression of GFP- or mCherry-tagged ABI1 in transiently transformed N. benthamiana leaves showed that ABI1 was also strongly associated with plasma membranes (Figure 4A). To investigate where ABI1 and ROP11 or CA-ROP11 interact in plant cells, we conducted a bimolecular fluorescence complementation (BiFC) assay. The coding sequence of ABI1 was cloned into the C-terminal half of the YFP gene (YFPN vector), and the coding sequence of ROP11 or CA-ROP11 was cloned into the N-terminal end of the YFP gene (YFPC vector). The ABI1-YFPN and ROP11-YFPC gene constructs were co-transformed into the leaves of N. benthamiana, and the yellow fluorescence emitted from reconstituted YFP was examined. The results showed that ROP11 can interact with ABI1 both on the plasma membrane and in the nucleus (Figure S1). When ABI1 and CA-ROP11 constructs were co-transformed into the leaves of N. benthamiana, the yellow fluorescence was observed only on the plasma membrane (Figure 4B). After ABA treatment, however, the interaction between CA-ROP11 and ABI1 was also evident in the nucleus (Figure 4C).
Figure 4. Subcellular localization of ABI1 and BiFC analyses of the interactions between ABI1 and CA-ROP11.
(A) The expression of GFP or mCherry-tagged ABI1 in pavement cells of Nicotiana benthamiana leaves.
(B) BiFC analyses of the interactions between ABI1 and CA-ROP11. The fusion YFP constructs used for co-expression in the leaves of N. benthamiana are indicated on top.
(C) The effect of ABA on the interactions between ABI1 and CA-ROP11 as indicated by BiFC assay. The co-transformed Nicotiana benthamiana leaves were treated with 50 μM ABA for 1 h and were then treated with 0.8 M NaCl for 5 min before they were photographed. The cell membranes and nuclei are indicated by white and red arrows, respectively.
CA-ROP11 protects ABI phosphatase activity
Previous studies have shown that when ABA binds to its receptor RCAR1, RCAR1 forms a complex with its downstream component ABI1, resulting in inhibition of ABI1 phosphatase activity. Because ROP11 can interact with ABI1, we wanted to determine the significance of this interaction within the context of ABA signaling. The coding sequences of RCAR1, ABI1, and ROP11 or CA-ROP11 were amplified from the plasmids containing these genes and cloned into the expression vector fused with His-Tag. The resultant constructs were transformed into Escherichia coli cells to produce recombinant proteins. The purity of the recombinant proteins was determined by SDS-PAGE electrophoresis (Figure 5). HPLC analysis showed that purified ROP11 protein was in the GDP-bound form, whereas the CA-ROP11 protein was in the GTP-bound form (data not shown). The detailed procedures for HPLC analysis of the recombinant proteins are described in the Materials and Methods.
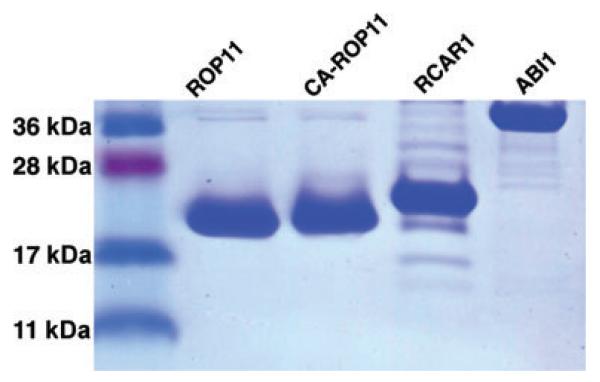
Figure 5. SDS-PAGE gel showing the purified His-tagged ROP11 (residues 1–182), CA-ROP11 (residues 1–182), RCAR1 (residues 1–186), and ABI1 (residues 119–434).
Each lane was loaded with 0.8 μg of protein.
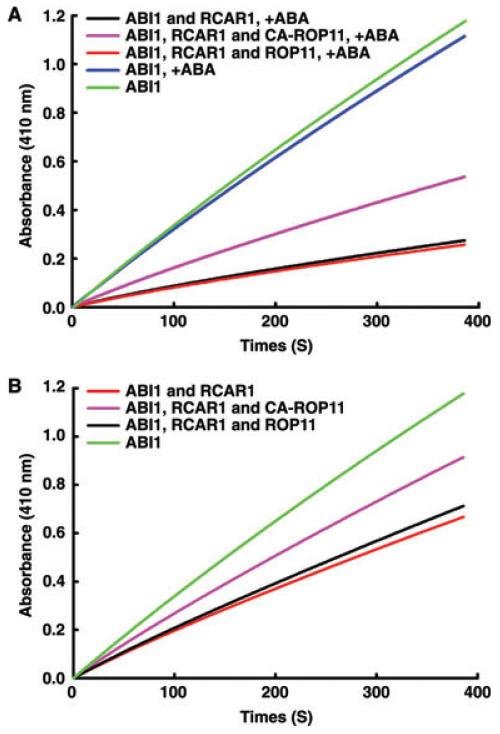
When incubated with phosphatase substrate pNPP, ABI1 exhibited a high phosphatase activity (Figure 6A). The addition of ABA or CA-ROP11 proteins alone did not affect this activity in vitro. When ABI1 proteins were further mixed with RCAR1 in the presence of ABA, however, the ABI1 phosphatase activity was dramatically inhibited as reported previously (Ma et al. 2009; Park et al. 2009; Nishimura et al. 2010). In our experiment, even in the absence of ABA, the RCAR1 could still partially inhibit the phosphatase activity of ABI1 but was less efficient than in the presence of ABA (Figure 6B). When CA-ROP11 proteins were added to the reaction mixtures, the inhibitory effects of RCAR1 on ABI1 phosphatase activity were significantly reduced in the presence of ABA (Figure 6A). This reduction of the RCAR1 inhibitory effect by CA-ROP11 also occurred in the absence of ABA (Figure 6B). However, the purified ROP11 proteins had no observable effect on inhibition of ABI1 by RCAR1 (Figure 6A). These results demonstrated that CA-ROP11 could protect ABI1 phosphatase activity by directly interfering with the inhibitory function of the ABA receptor RCAR1.
Figure 6. In vitro phosphatase activity assay of ABI1 with RCAR1, ROP11, and CA-ROP11 in the presence (A) or absence (B) of ABA.
Y-axis represents the amount of product released from pNPP catalyzed by ABI1, which is expressed as absorbance at 410 nm. The concentration of each component used in the assay was: ABI1, 300 nM; RCAR1, 1 μM; ROP11 or CA-ROP11, 2.5 μM; ABA, 1 mM.
Discussion
The plant hormone ABA is a key regulator of plant growth, development, and response to environmental stress (Cutler et al. 2010). A minimal set of core components for the RCAR/PYR/PYL receptors-mediated ABA signaling pathway was recently defined (Fujii et al. 2009). According to the current model, some ABA responses are mediated in part by the inactivation of several PP2C phosphatases, including ABI1 and ABI2, by ABA-bound receptors. Questions remain, however, about how the interactions between the ABA receptors and PP2C phosphatases are regulated by other protein factors.
In a companion article published in this issue, we showed that ROP 11, a member of the plant-specific RAC/ROP small GT-Pase family, is a negative regulator of multiple ABA responses in Arabidopsis (Li et al. 2012). The ability to regulate multiple ABA responses suggested that ROP11 might act in the early steps of ABA signaling. Our LIC assays in the current study showed that all the forms of ROP11 can directly interact with ABI1 (Figure 2A). That CA-ROP11 and ABI1 interact was further supported by yeast two-hybrid, Co-IP, and BiFC assays (Figure 2B, Figure 3, and Figure 4B,C). Interestingly, the interaction between ABI1 with CA-ROP11 is restricted to the plasma membrane in the absence of ABA but occurs both on the plasma membrane and in the nucleus in the presence of ABA (Figure 4C). This change of interaction pattern between ABI1 and CA-ROP11 is consistent with the subcellular location of CA-ROP11 in the absence or presence of ABA (Li et al. 2012).
Currently, the ABA-bound receptor RCAR1 is thought to inhibit the phosphatase activity of ABI1 and presumably other PP2C phosphatases, thus releasing suppression of downstream ABA responses (Cutler et al. 2010). Using a protoplast-based reconstitution system, we showed that expression of CA-ROP11 or ROP11 can reduce the inhibition of ABI1 activity by the ABA receptor RCAR1. Furthermore, in vitro biochemical assays with purified recombinant proteins demonstrated that the GTP-bound ROP11 (CA-ROP11), but not the GDP-bound ROP11 (proteins expressed from ROP11 construct), can protect ABI1 phosphatase activity from inhibition by the RCAR1/PYL9. The effects of ROP11 in the protoplast-based assay and in the in vitro biochemical assay seem inconsistent. A likely explanation is that the proteins expressed from the ROP11 construct in protoplasts might adopt the GTP-bound form, which is not the case for proteins expressed from the ROP11 construct in E. coli cells. The latter have been shown by HPLC analysis to be in the GDP-bound form, probably due to the hydrolysis of GTP by its intrinsic GTPase activity during purification. Taken together, these data indicate that activation of ROP11 is required for its function in protecting ABI1 phosphatase activity from inhibition by RCAR1. The results thus provide a biochemical explanation of how ROP11 negatively regulates ABA signaling in Arabidopsis.
Based on the work presented here, we propose that, in the absence of ABA, the existing CA-ROP11 may compete with the ABA-free receptors for the interactions with ABI1 and other PP2Cs, blocking the basal level inhibitory effects of the ABA-free receptors and thus protecting the phosphatase activities of ABI1 and other PP2Cs. In fact, we have found that ROP11s also interacted with ABI2 in a LIC assay (Figure S2). The active ABI1 and other PP2Cs together suppress the ABA-mediated physiological responses when ABA concentration is low in plant cells. When the ABA concentration increases, it causes active ROP11 proteins to dissociate from the PP2Cs. The dissociation of active ROP11 exposes ABI1 and other PP2C phosphatases to the ABA-bound receptor RCAR1, which leads to their inactivation and the release of ABA responses. The dissociation of active ROP11 from ABI1 might be directly caused by the increased level of ABA-bound RCAR1, which competes with active ROP11 for binding to the ABI1, or by changes in ROP11 targeting, such as its translocation into the nucleus. In CA-ROP11 transgenic plants, because constitutively active ROP11 is overexpressed from a strong CaMV 35S promoter, ABA-bound RCAR1 may not effectively compete with CA-ROP11 or cause all ROP11 molecules to dissociate from ABI1, thus resulting in decreased ABA sensitivity. In DN-ROP11 plants or rop11 mutants, in contrast, the protection of ABI1 phosphatase activity from inhibition by RCAR1 is reduced, which leads to enhanced plant sensitivity to ABA. Finally, we also want to point out that because there are 14 putative ABA receptors and five PP2C members that are involved in ABA responses in Arabidopsis, the above is still an over-simplified model. Certainly, more biochemical studies are required for further elucidation of the molecular mechanism of ABA signaling.
In summary, we have used multiple approaches to demonstrate that ROP11 negatively regulates ABA signaling by protecting ABI1 phosphatase activity from inhibition by the ABA receptor RCAR1/PYL9. To the best of our knowledge, this work provides the first example of how the interactions between an ABA receptor and its downstream signaling component are regulated by other protein factors. These results increase our understanding of the molecular mechanisms of ABA signal transduction in higher plants.
Materials and Methods
Construction of plant transformation vectors
For LCI assays, the coding sequence of ABI1 was amplified from Arabidopsis thaliana cDNA by PCR and cloned into the LUCN vector. Accordingly, the coding sequences of ROP11, CA-ROP11, DN-ROP11, and RCAR1 were cloned into the LUCC vector. For BiFC assays, the coding sequence of ABI1 was cloned into the YFPN vector, and the coding sequences of ROP11, CA-ROP11, and DN-ROP11 were cloned into the YFPC vector. The sequences of all the primers and enzyme sites used for vector construction are listed in Table S1.
Yeast two-hybrid analysis
Yeast two-hybrid analysis was carried out using the Match-Maker GAL4 Two-Hybrid System 3 (Clontech) according to the manufacturer’s instructions. The sequences of all the primers used for vector construction are indicated in Table S1.
Co-immunoprecipitation assay
For co-immunoprecipitation assays, MYC-ABI1 was amplified from the corresponding pGBKABI1 construct and cloned into the modified pCAMBIA1301 binary vector using BamHI/SpeI double digestion. HA-CA-ROP11 were amplified from pGADCA-ROP11 constructs and cloned into the modified pCAMBIA1301 binary vector through XbaI/SpeI double digestion.
Protein extracts were prepared from Nicotiana benthamiana leaves injected with Agrobacterium suspensions with a given construct. N. benthamiana leaves (2 g) were ground in liquid nitrogen into fine powder. The powders were transferred into 4 mL of ice-cold extraction buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, 1% Triton X-100, 1 mM DTT, 1×cocktail protease inhibitor (Roche), 1 mM PMSF), briefly mixed with a vortex mixer, and incubated at 4 °C for 30 min. The protein extracts were centrifuged at 8 000 g for 10 min to remove the cell debris. Protein concentration in each lysate was adjusted to the same value. Protein extracts (1 mL) were incubated with 1 μg/mL HA monoclonal antibody (Abmart) at 4 °C overnight. Protein G agarose beads (20 μL, Roche) were added to precipitate the antigen/antibody complex. After incubation at 4 °C for 2 h, the beads were collected by centrifugation at 8 000 g for 3 min, and were washed four times with extraction buffer. The beads were resuspended in 50 μL 2×Laemmli buffer and boiled at 100 °C for 3 min to release the proteins. Supernatant was collected by brief centrifugation. Protein samples (15 μL) were fractionated by SDS-PAGE electrophoresis, and immunoblotting was performed using anti-MYC monoclonal antibody (Abmart).
Transient gene expression in tobacco
Agrobacterium-mediated transient expression in N. benthamiana leaves was performed as described by Batoko et al. (2000). An Agrobacterium cell suspension carrying a given construct was infiltrated into young, fully expanded N. benthamiana leaves using a needle-less syringe. For co-infiltration, the two Agrobacterium cultures with the same OD value (OD600 = 0.3) were mixed in a 1:1 ratio. Fifty hours after infiltration, subcellular localization of GFP or mcherry-tagged proteins in leaf pavement cells was determined with a Zeiss LSM 710 confocal microscope. For LCI assays, luciferin was sprayed on the surface of N. benthamiana leaves, and LUC fluorescence chemical signal was captured with a low-light cooled CCD camera (Andor iXon CCD camera, Andor Technology Ltd, South Windsor, CT). The first 5 min of exposure was used to quench background fluorescence, and the second 5 min of exposure was used to collect LUC signal. For BiFC assays, the transiently transformed leaves of N. benthamiana were examined for fluorescence signals using a Zeiss 710 confocal microscope. The excitation wave lengths were 488, 543, and 515 nm and the emission wavelengths were 515, 630, and 530 nm for GFP, mCherry, and YFP, respectively.
Protoplast-based assays for the expression of stress-responsive gene
Protoplast assays for studying the effects of ROP11 and CA-ROP11 on the stress-responsive gene RD29B-LUC expression in Arabidopsis mesophyll protoplasts were conducted according to Fujii et al. (2009).
Recombinant protein production and in vitro ABI1 phosphatase activity assay
The coding sequences of RCAR1 (residues 1–186), ABI1 (residues 119–434), and ROP11 (residues 1–182) were cloned into the pET21b vector fused with His-Tag. The constructs were transformed into Escherichia coli BL21 (DE3) cells. Bacteria were cultured at 37 °C and induced with 0.3 mM IPTG at 20 °C for 16 h. All the proteins were first purified with NiNTA-resin (Qiagen) and further fractionated with an anion exchange column 15Q and gel filtration column of Superdex 200 (Pharmacia). The purity of the recombinant proteins was determined by SDS-PAGE electrophoresis.
ABI1 phosphatase activity was measured by using p-nitrophenyl phosphate (PNPP) as the substrate and by detecting the absorbance at 410 nm. Each reaction was performed in a 1800-μL reaction system with 100 mM MOPS (pH 7.4), 50 mM NaCl, and 300 nM ABI1 in the different combinations of 1 μM RCAR1 or 2.5 μM ROP11 (or CA-ROP11). The reactions were performed in the absence or presence of 1 mM ABA. The data for absorbance at 410 nm were collected and processed using SigmaPlot 9.0. The experiments were repeated three times, and representative results are shown.
Supplementary Material
The interactions between ABI1 and ROP11 shown by BiFC assay.
LIC assays of the interaction between ROP11 and ABI1.
Acknowledgments
We thank Drs. Zhizhong Gong (China Agricultural University) and Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for their critical comments on this manuscript, Dr. Jianming Zhou (National Institute of Biological Science, China) for providing LUCN and LUCC vectors for LIC assays and Dr. Yule Liu (Tsinghua University) for providing YFPN and YFPC vectors for BiFC assays. We are grateful to Lei Huang (Tsinghua University) for assistance in confocal imaging analysis. This work was supported by the 973 National Basic Research Program of the Ministry of Science and Technology of China (2009CB119100) and the National Natural Science Foundation of China (90717121).
Footnotes
The following materials are available in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Batoko H, Zheng HQ, Hawes C, Moore I. A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell. 2000;12:2201–2218. doi: 10.1105/tpc.12.11.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Yang X, Weston DJ, Chen JG. Abscisic acid receptors: Past, present and future. J. Integr. Plant Biol. 2011;53:469–479. doi: 10.1111/j.1744-7909.2011.01044.x. [DOI] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–1910. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Li S, Lord EM, Yang Z. Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell. 2006;18:366–381. doi: 10.1105/tpc.105.036434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kang J, Sui N, Liu D. ROP11 GTPase is a negative regulator of multiple ABA responses in Arabidopsis. J. Integr. Plant Biol. 2012 doi: 10.1111/j.1744-7909.2012.01100.x. doi: 10.1111/j.1744-7909.2011.01100.x. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E. Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J. 2008;54:806–819. doi: 10.1111/j.1365-313X.2008.03454.x. [DOI] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY. RAC/ROP GTPases: ‘Hubs’ for signal integration and diversification in plants. Trends Plant Sci. 2006;11:309–315. doi: 10.1016/j.tplants.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, Yates JR, Schroeder JI. PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 2010;61:290–299. doi: 10.1111/j.1365-313X.2009.04054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007;50:935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA. Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 2005;15:1998–2006. doi: 10.1016/j.cub.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004;37:354–369. doi: 10.1046/j.1365-313x.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009;60:575–588. doi: 10.1111/j.1365-313X.2009.03981.x. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006;281:5310–5318. doi: 10.1074/jbc.M509820200. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng ZL, Yang Z. The Rop GTPase: An emerging signaling switch in plants. Plant Mol. Biol. 2000;44:1–9. doi: 10.1023/a:1006402628948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The interactions between ABI1 and ROP11 shown by BiFC assay.
LIC assays of the interaction between ROP11 and ABI1.