Abstract
Microtubule-based transport is required for plasmid translocation to the nucleus during transfections, and having stable structures could enhance this movement. In previous studies in which the cytoskeleton was disrupted, we found that populations of microtubules remain that are stable and highly acetylated. By increasing the levels of acetylated tubulin through inhibition of the tubulin deacetylase HDAC6, we observe more rapid plasmid nuclear localization of transfected plasmids and greater levels of gene transfer. In this study, we sought to understand plasmid movement in cells with enhanced tubulin acetylation. Using variations of a microtubule spin down assay, we found that plasmids bound to hyper-acetylated microtubules to a greater degree than they did to unmodified microtubules. To determine if microtubule acetylation also affects cytoplasmic trafficking, plasmid movement was evaluated in real time by particle tracking in cells with varying levels of acetylated microtubules. We found that plasmids display greater net rates of movement, spend more time in productive motion and display longer runs of continuous motion in cells with highly acetylated microtubules compared to those with fewer modifications. These results all suggest that plasmid movement is enhanced along highly acetylated microtubules, reducing the time spent in the cytoplasm prior to nuclear import. Taken together, these findings provide a foundation for determining how modulation of microtubule acetylation can be used as a means to increase intracellular trafficking of plasmids and enhance gene therapy.
Keywords: trafficking, acetylated microtubules, plasmid, gene transfer
Introduction
Successful non-viral gene transfer requires plasmid DNA to cross several obstacles on its path to the nucleus, resulting in less than optimal transgene expression1-3. Both the plasma and nuclear membranes represent physical barriers that plasmids must overcome for proper transcription of therapeutic genes, and a variety of techniques have been developed to promote cell entry and to enhance nuclear import of plasmids3. Less is known about what happens to plasmids in the cytoplasm or what pathways are used for their trafficking to the nucleus. What we do know is that the microtubule network is required for plasmid movement to the nucleus4,5, even though the cytoskeletal meshwork is another barrier to successful gene transfer6. Therefore, we hypothesize that by modulating the microtubule network, we can enhance plasmid trafficking through the cytoplasm and ultimately, plasmid transgene expression.
Different populations of microtubules exist within a cell that are either dynamic or stable, and post-translational modifications of tubulin have been shown to occur on the more stable polymerized structures7. It is currently unclear however, whether the post-translational modifications occur on microtubules because they are stable, or if they confer stability on the polymerized structures. It has been speculated that some of these alterations impact microtubule half-life and spatial distribution, along with recruitment of protein complexes and microtubule associated proteins8. Therefore, it is assumed that the differently marked microtubules may allow for enhanced binding of regulatory factors, such as motor proteins and plus-end tracking proteins, that can play a role in transient microtubule pausing or stabilization9. This, in turn, could allow for more efficient cargo movement (e.g., plasmids) on these modified structures, which would be beneficial in non-viral gene delivery.
One such population of microtubules that have been observed to remain intact after cytoskeletal disruption by drugs or following mechanical strain (cyclic stretch) and are post-translationally acetylated10,11. We have found that enhanced microtubule acetylation permits increased nuclear localization of plasmids and transfection efficiency10, but the molecular mechanisms behind this improvement have not been elucidated. Others have demonstrated that tubulin acetylation positively influences motor protein binding to microtubules and plays a role in the intracellular trafficking of other types of molecules being moved by these motors such as membrane vesicles12,13. If more motors are recruited to acetylated microtubules, perhaps more plasmids have a better chance of moving towards the nucleus in cells with increased levels of acetylated tubulin. Alternatively, acetylation of microtubules could result in enhanced cargo (e.g., plasmid) movement leading to faster transit times toward the microtubule organizing center and the nuclear envelope. While the tubulin acetyltransferase has not been identified, the main enzyme responsible for removing acetyl groups from tubulin is known. This protein, histone deacetylase 6 (HDAC6), is unique in that it is cytoplasmic and acts on substrates within the cytoplasm (primarily tubulin and HSP90) instead of acting on histones14-16. By knocking down HDAC6, levels of acetylated microtubules can be increased in cells10,15. Therefore, we sought to examine how the cytoplasmic trafficking of plasmids is affected by varying the levels of acetylated microtubules through modification of HDAC6.
In this study, we used a cell-free in vitro microtubule spin-down assay to examine the interactions of plasmids with microtubules and found that more plasmids are bound to acetylated compared to unmodified microtubules. Additionally, real-time particle tracking of fluorescently labeled plasmids microinjected into the cytoplasm showed that plasmids move faster and for longer runs of continuous movement in cells with high levels of acetylated microtubules. Taken together, our findings demonstrate that the acetylation status of tubulin impacts how plasmids move through the cell toward the nucleus, which provides a target for overcoming the barrier of cytoplasmic trafficking in gene transfer.
Results
Plasmid-microtubule interactions are enhanced with acetylated microtubules in vitro
To utilize the microtubule network for intracellular movement, plasmids do not directly interact with microtubules, but form protein complexes in the cytoplasm and must load onto motor proteins as cargo4,5. To directly examine the interactions of plasmid DNA with microtubules, an in vitro spin-down assay was employed17 using taxol-stabilized microtubules and in vitro acetylation was carried out on a subset of these microtubules. Plasmid pCMV-Lux-DTS was incubated with HeLa cell extract (as a source of adapter proteins) and either unmodified or in vitro acetylated microtubules, and then centrifuged to pellet microtubules, associated proteins, and DNA. Quantitative real-time polymerase chain reaction (qPCR) was used to determine how much plasmid was present in the supernatant (unbound) and pellet (microtubule-bound) fractions of each sample. As we have shown previously4, in the absence of microtubules or cell extract, very little, if any, DNA pellets or interacts with the microtubules, but when HeLa cell extract is added along with microtubules, a significant amount of DNA pellets (Figure 1). It should be pointed out that the amount of cell extract was adjusted so that approximately 40% of the input plasmid would associate with unmodified microtubules. When an equal amount of microtubules were acetylated in vitro and used in the assay, there was a twofold increase in the amount of plasmid that interacts and pellets compared to with unmodified microtubules, demonstrating that acetylated microtubules bind more DNA than unmodified microtubules.
Figure 1. Plasmids show greater binding to highly acetylated microtubules than to unmodified ones.
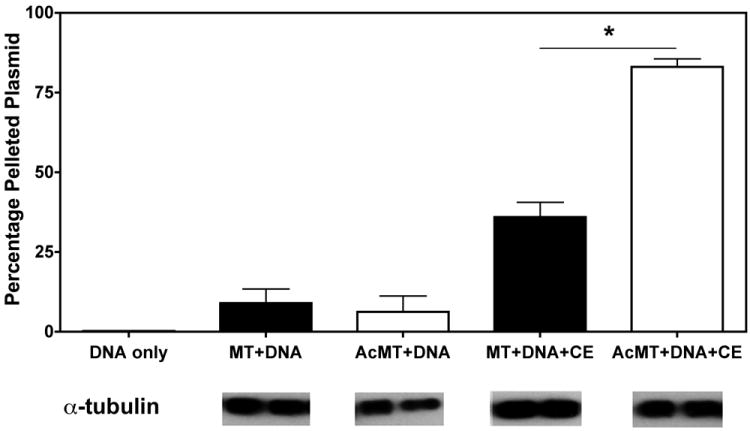
Quantitative analysis of plasmid association with microtubules. Plasmid pCMV-Lux-DTS (DNA) was incubated for 40 minutes either alone, with taxol-stabilized microtubules (MT), or with in vitro acetylated microtubules (AcMT) in the presence or absence of Hela cell extract (CE) as indicated and subsequently separated over a glycerol cushion by centrifugation. The plasmid content of the pellets and supernatants were determined by quantitative PCR, and the percentage of DNA in the pellet was determined by comparing DNA content in pelleted fractions versus total DNA in both supernatant and pellet fractions combined. Mean plasmid DNA concentrations from three independent experiments, preformed in duplicate, are shown + st. dev. *, p<0.001. Western blot images are representative samples from each group probed for total tubulin to ensure that there were not more total microtubules present in the acetylated microtubule samples, which could have accounted for the increased plasmid DNA binding.
To visualize these interactions, a modified version of this assay by Dompierre13 was used in which the microtubule-containing samples were spun onto coverslips. Samples with stable polymerized microtubules or in vitro acetylated microtubules were incubated with and without cell extract or CY3-PNA labeled DNA. In the absence of cell extract, labeled plasmid does not interact with unmodified or acetylated microtubules (Figure 2a), confirming the results from the qPCR experiments. When microtubules are incubated with both cell extract and CY3-DNA before centrifugation, there is some overlap or interaction of CY3-DNA with the microtubules (Figure 2b). However, there is a greatly enhanced interaction of DNA when acetylated microtubules are used. These findings confirm the results seen using qPCR, and demonstrate that acetylated microtubules bind more plasmid DNA than unmodified microtubules (Figure 1).
Figure 2. Immunofluorescence imaging of plasmid interactions with pelleted microtubules.
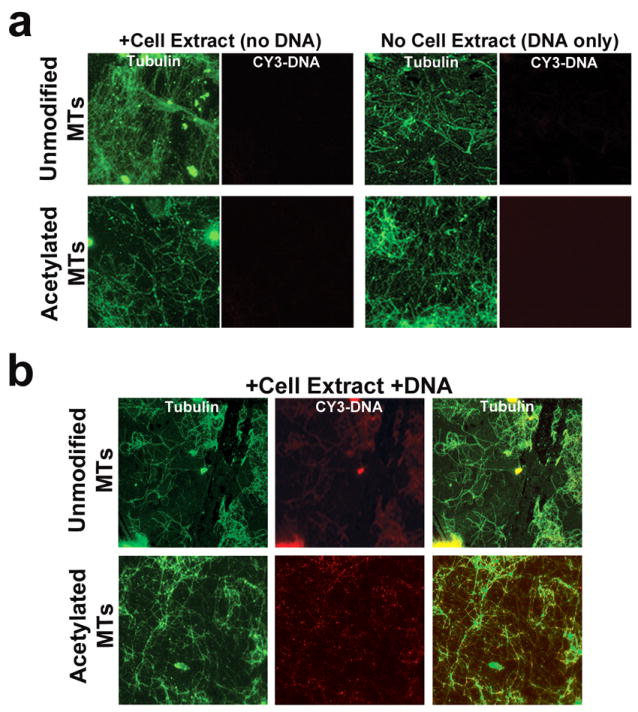
A variation of the microtubule spindown assay in Figure 1 was used to visualize DNA-microtubule interactions. (a) Microtubule or acetylated microtubule controls were incubated with either cell extract or CY3-labeled pCMV-Lux-DTS (CY3-DNA). Following centrifugation onto coverslips, the microtubules were labeled using antibodies against α-tubulin. Images were acquired using fluorescence microscopy and a 100x objective. (b) The same as in a, but microtubule or acetylated microtubule samples were incubated with both cell extract and CY3-DNA before centrifugation. MTs, microtubules.
The net movement of plasmids is enhanced along highly acetylated microtubules
We have shown in past experiments that cells with high levels of acetylated microtubules permit more rapid nuclear accumulation of plasmids compared to cells that have lower levels of acetylated tubulin10. However, it is unknown how differences in microtubule acetylation affect the intracellular trafficking rates of individual plasmid particles. Cells that stably express short-hairpin RNA against the tubulin deacetylase, HDAC6 (knockdown cells) have high levels acetylated microtubules, while wild-type cells have reduced levels of acetylated tubulin by comparison (Figure 3a)10,18. These two different cell lines were microinjected with fluorescently-labeled plasmids and movement was tracked over time. As seen previously, when the empty backbone plasmid pBR322, which does not contain any promoter or enhancer sequences and exhibits only diffusive movement in the cell19, was injected and followed for up to 1 hour after microinjection, most particles failed to show any significant bulk movement and remained largely at the site of injection (Figure 3b and Supplemental data movie 1). Plasmids carrying the Cytomegalovirus promoter (CMViep) and the SV40 enhancer (DNA nuclear targeting sequence) are able to traffic to the nucleus and showed significant directed movement20-22 (Supplemental data movie 2). Individual plasmid velocities were analyzed as the distance moved between each frame (instantaneous velocity) and the net (average) velocity for all tracked frames. When the frequency distribution histograms are plotted for the net velocities of all tracked plasmids in both cell types, there is a shift in the histogram of pCMV-GFP-DTS plasmids, seen as more plasmids moving at higher rates in the HDAC6 knockdown versus wild-type cells (Figure 3c). There was no difference in velocity plots for the negative control plasmid pBR322, which is expected, since they have not been shown to interact with microtubules19. When all net velocities are averaged together for each condition, there was almost a two-fold increase in the velocity of plasmids in HDAC6 knockdown (0.183 ± 0.009 μm/second SEM) versus wild-type (0.108 ± 0.006 μm/second SEM) cells (Figure 3d). The pBR322 plasmid had the same average velocity of all tracked plasmids regardless of the acetylation status of cellular microtubules (0.057 ± 0.001 μm/second SEM). This suggests that plasmids capable of cytoplasmic trafficking have enhanced net movement when microtubules are highly acetylated.
Figure 3. Plasmid cytoplasmic movement is enhanced on highly acetylated microtubules.
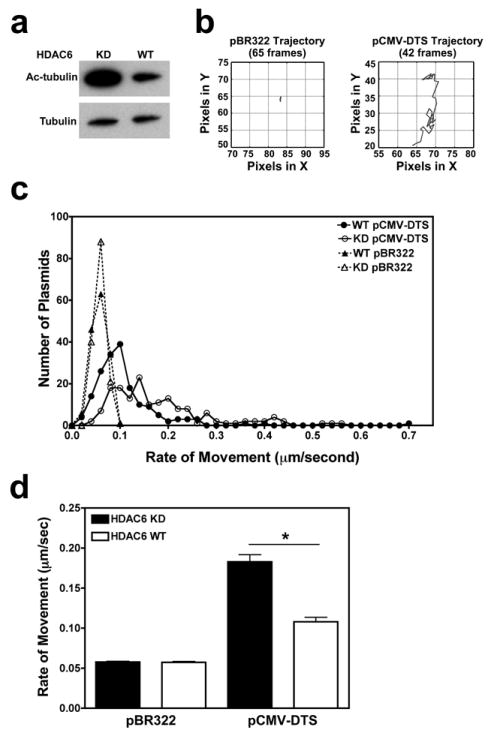
(a) Western blot images show elevated levels of acetylated tubulin in cell lysates from HDAC6 knockdown (KD) compared to HDAC6 wild-type (WT) cells. (b) Representative traces for individual plasmid trajectories. Quantum dot-labeled plasmids were cytoplasmically microinjected into adherent HDAC6 mutant A549 cells and imaged at 0.5-second intervals over 5-10 minutes. The traces of representative negative control (pBR322) and positive control (pCMV-GFP-DTS) plasmids in injected cells are shown. Plasmid trajectories were created using the PolyParticleTracker software downloaded for use in MATLAB, and all plot areas are mapped to 25 × 25 pixel areas. (c) Average cytoplasmic velocity of individual microinjected plasmids. Movement of individual DNA particles, shown in b, were tracked up to 10 minutes using time-lapse imaging (2 frames/second). The net (mean) velocity of each was determined using particle tracking software (PolyParticleTracker, MATLAB), and the frequency distribution histograms are plotted as the number of plasmids moving at certain velocities for each construct. At least 150 particles were tracked per construct in 7 separate experiments. (d) Individual plasmid net velocities were averaged for each of the constructs. Error bars represent means + SEM. *, p<0.001.
Plasmids spend more time in productive movement in cells with highly acetylated microtubules
Upon seeing differences in the net velocities of plasmids moving in HDAC6 knockdown and wild-type cells, we sought to further explore how plasmid movement is altered in the presence of highly acetylated microtubules. Examination of the instantaneous velocities (rate of movement between each captured frame) revealed that plasmids spend less time in diffusive or non-productive movement along highly acetylated microtubules compared to microtubules with fewer modifications (16.9 ± 1.16% versus 32.3 ± 1.76%, p<0.001) (Figure 4a). In other words, plasmids spend significantly more time in active or directed movement in HDAC6 knockdown cells compared to wild-type cells. A frequency distribution histogram demonstrates that in HDAC6 knockdown cells, 50% of tracked plasmids spent 90% or more time in productive motion versus only 19% of tracked plasmids in wild type cells (Figure 4b). Taken together with our findings suggest that plasmids spend a larger percentage of time moving at faster rates in cells with high levels of acetylated microtubules.
Figure 4. Plasmids spend more time in productive motion on highly acetylated microtubules.
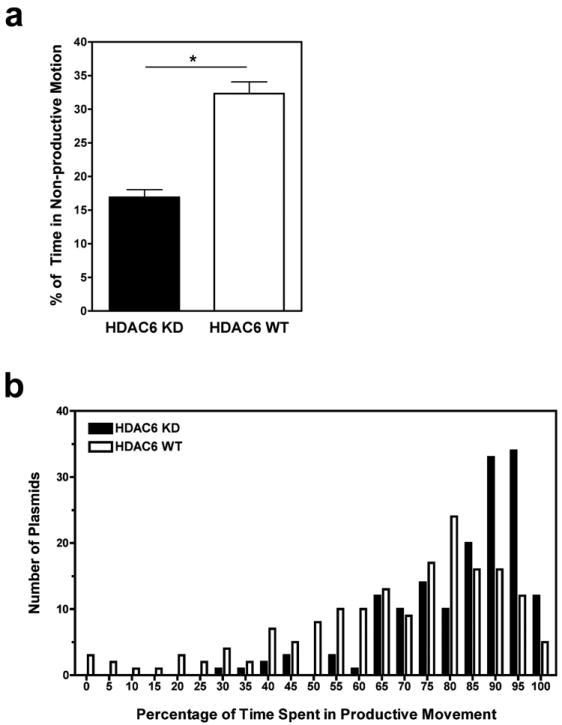
(a) Examination of the instantaneous velocities (rate of movement between each captured frame) of each tracked particle from Figure 3 revealed that pCMV-GFP-DTS plasmids spend less time in non-productive movement along highly acetylated microtubules compared to microtubules with fewer modifications. This was determined by dividing the number of frames showing non-productive motion (velocity <0.057 μm/second) by the total number of frames for individual plasmids in each cell type. Error bars represent means + SEM. *, p<0.001. (b) Frequency distribution histogram plotted as the percentage of time a number of plasmids spent in productive motion in HDAC6 knockdown (HDAC6 KD) and wild-type (HDAC6 WT) cells.
Highly acetylated microtubules permit longer runs of plasmid movement
The retrograde motor protein dynein is involved in microtubule-based movement of many non-viral and viral gene therapy vectors such as plasmid DNA and adenoviral vectors4,23. Besides demonstrating productive, processive movement, dynein has been shown to go through intervals where the motor is paused, inactive, or undergoes diffusive movement24,25. This can affect the processivity and therefore net movement of bound cargo. We hypothesized that the enhanced net velocities of plasmids moving in the HDAC6 knockdown cells (Figure 3d) could be due to improved processivity of the motors moving along acetylated microtubules. To examine how often plasmids were engaged in productive movement, the number of consecutive frames in which the plasmids were in motion was determined for each tracked plasmid in the HDAC6 knockdown and wild-type cells. When the “runs” of continuous motion were averaged for each tracked plasmid, about one-third of the plasmids exhibited a mean productive motion for more than 10 frames in the HDAC6 knock-down cells compared to only one-tenth of plasmids in the wild-type cells (Figure 5a). When the values for all plasmids were averaged for each cell line, the knockdown cells produced intervals of continuous plasmid movement that are almost twice what is seen in wild-type cells (mean of 8 versus 4.8 frames) (Figure 5b), suggesting that perhaps microtubule acetylation can also lead to enhanced motor processivity or fewer pauses.
Figure 5. Highly acetylated microtubules permit longer runs of plasmid movement.
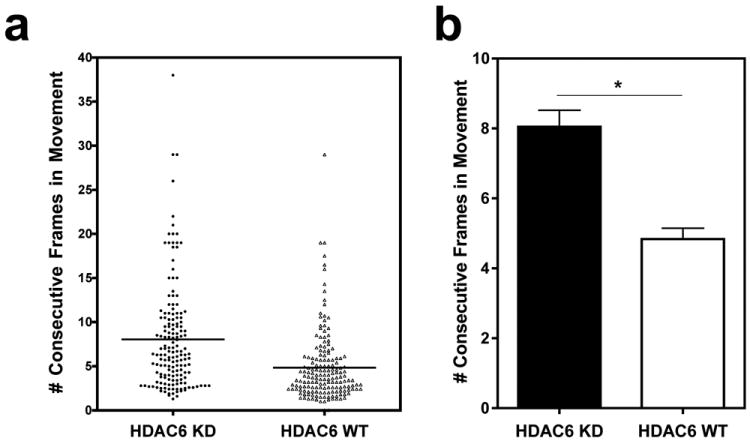
(a) For each particle, the number of consecutive frames in which velocities show active plasmid movement (>0.057 μm/second) were counted for each bout of motion, and these “runs” of continuous movement were averaged for each particle by dividing the number of consecutive frames (per run) by the total number of runs of movement exhibited by the plasmid. Each dot represents the average length of runs for an individual plasmid, and the line indicates the mean for each condition. (b) The data from individual plasmids in a were averaged for all tracked plasmids in HDAC6 knockdown (HDAC6 KD) and wild-type (HDAC6 WT) cells and represented in a bar graph. Error bars represent means + SEM. *, p<0.0001.
Plasmids interact with acetylated microtubules in cells
Based on the particle tracking results, and considering the cell-free microtubule assay, we were interested in looking at plasmid interactions with acetylated microtubules in cells. Wild-type and HDAC6 knock-down cells were microinjected with plasmids, fixed 20 minutes later, and immunofluorescence was carried out using antibodies against tubulin and acetylated-tubulin (Figure 6a). We observed significant overlap or colocalization of Quantum dot-labeled plasmids with both total microtubules (mean Manders overlap coefficient of 93%) and acetylated microtubules (mean Manders overlap coefficient of 73%) in HDAC6 knockdown cells (Figure 6c). Cells that did not receive plasmid were also imaged to ensure that the streptavidin-Quantum dot labeling was specific for biotinylated plasmids (Figure 6b). Also, by determining the mean fluorescence intensity of the two channels for corresponding images, the fraction of tubulin that is positively stained for acetylated tubulin was less in wild-type cells compared to HDAC6 knockdown cells, as expected (Figure 6d). These results provide confirmation that acetylated microtubules can bind plasmids during cellular transfections.
Figure 6. Visualization of plasmid interactions with acetylated microtubules in cells.
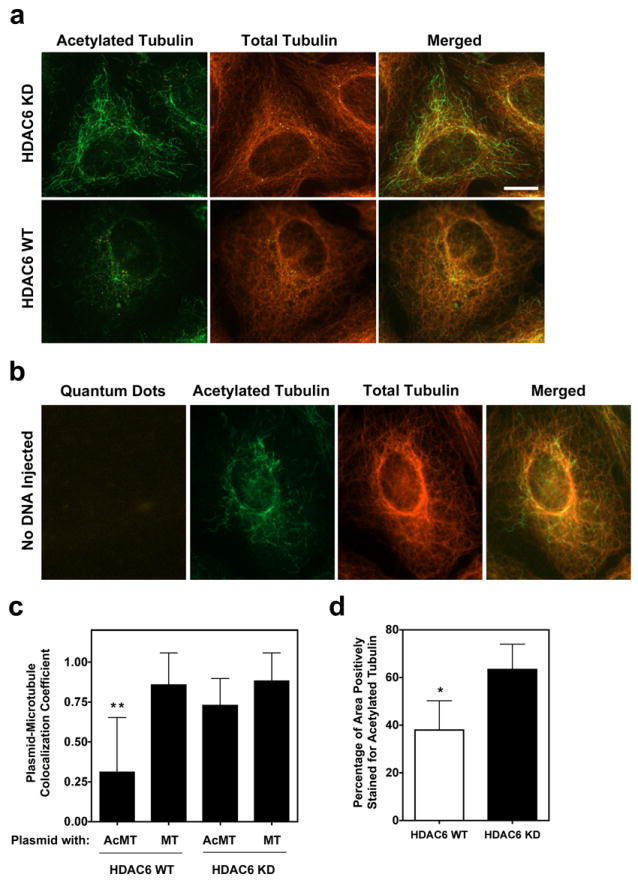
Colocalization of Quantum dot-labeled plasmids with microtubules. (a) HDAC6 knockdown (HDAC6 KD) and wild-type (HDAC6 WT) cells were cytoplasmically microinjected with biotin-PNA-pCMV-GFP-DTS plasmids, fixed 20 minutes post-injection, and labeled with antibodies against β-tubulin and acetylated lysine-40 α-tubulin. Streptavidin-conjugated Quantum dots were used to label the injected biotinylated plasmids. Cells were imaged using a 100x objective by deconvolution microscopy and representative deconvolved Z slices are shown for each cell line. Scale bar, 10 μm. (b) Cells that were not microinjected with plasmid show no cytoplasmic Quantum dots following treatment as above. (c) Manders overlap coefficients were determined for images of at least 20 different HDAC6 knockdown and wild-type cells with at least 100 plasmids. Bars represent the mean overlap coefficients (+ s.d.) for plasmids with either acetylated (AcMT) or total microtubules (MT), with a value of 1 being 100%. *, p<0.0001 for plasmid-AcMT overlap in HDAC6 WT versus HDAC6 KD cells. (d) The average ratio of acetylated to total tubulin staining was determined for 13 different wild-type and HDAC6 knockdown cells by quantifying the grayscale intensity of the positively stained areas for corresponding images. Bars represent the mean + s.d. *, p<0.001.
Discussion
Previous work in our lab has shown that when microtubule acetylation is increased in cells, through the application of cyclic stretch, by chemical inhibitors of HDAC6, or by silencing HDAC6, gene expression from transfected plasmids is improved10,11,26. We have also observed more rapid nuclear localization of microinjected plasmids in cells in which HDAC6 is silenced10. Taken together, we hypothesized that highly acetylated microtubules allow enhanced plasmid movement through the cytoplasm during gene transfer, permitting the higher levels of gene expression that we have observed. In this study, a cell-free microtubule-binding assay, plasmid microinjections, and real-time particle tracking to demonstrate that plasmids show enhanced interaction with and movement on highly acetylated microtubules compared to their unmodified counterparts. We therefore found that plasmids can use modified microtubules to partially overcome the cytoplasmic barrier to gene delivery.
Dompierre et al. recently showed that acetylated microtubules recruit more motors, which results in improved transport efficiency and velocity of moving vesicles in striatal cells13, and previously published work from our lab suggests that plasmids reach the nucleus faster in cells with high levels of acetylated tubulin10. Therefore, it is reasonable to think that highly acetylated microtubules might bind not only more motor proteins, but also a greater number of cytoplasmic plasmids, which are thought to act as cargo utilizing motor proteins to move along microtubules,4,5 leading to increased plasmid transport rates. The analysis of net plasmid velocities indicates that this is the case (Figure 3). We speculate that this enhanced movement likely contributes to the improved gene expression that we see from transfections in which microtubule acetylation is enhanced both in vitro10,11,27 and in vivo26 due to more plasmids eventually reaching the nucleus. This is the first report, to our knowledge, of a tubulin modification being responsible for increased velocities of a gene transfer vector moving on microtubules.
The net rates of movement for the plasmid construct containing promoter and enhancer sequences (0.05 to 0.7 μm/second) are consistent with those measured for various components in the cytoplasm that also move on microtubule-associated motor proteins28-30. For example, mitochondria and other organelles31,32, along with cytoplasmic vesicles33 move at rates within this range. Further, the motor proteins kinesin and dynein are reported to move at similar average rates of around 0.5 to 1.5 μm/second28-30. Studies of the transport rates of other gene delivery vectors have demonstrated similar findings. For example, actively transported DNA complexes within endosomes have been reported to move at rates of about 0.2 μm/second34. Another group reported movement of internalized polyplex non-viral particles at a mean velocity of 0.7 μm/second during bouts of directed particle motion35. These stretches of directed motion were interrupted by diffusive motion and also by back-and-forth movement, which is what we also observed with particles having productive motion. This type of movement, where there are changes in direction and frequent pauses, is typical of cargo moving along microtubules36,37. Higher rates of around 2 μm/second have been observed for adenoviral vectors undergoing smooth, linear translocations over short (10 second) periods of tracking23.
In our microinjection experiments, the plasmids that exhibited movement have the CMViep promoter and the SV40-DTS enhancer sequences, allowing them to move through the cell and be taken up by the nucleus. The Cytomegalovirus immediate early promoter (CMViep) drives gene expression and promotes microtubule-based movement through the binding of several transcription factors, which upon activation in the cytoplasm, translocates to the nucleus. The Simian Virus 40 DTS permits nuclear uptake of plasmids in non-dividing cells22. The only difference between conditions, therefore, is the acetylation status of tubulin, suggesting that a post-translational modification on tubulin is enough to modulate the trafficking of cargoes moving on microtubules. The fact that the negative control plasmid, pBR322, does not exhibit any change in net rates of movement between the two cell lines illustrates that the difference in acetylation status of microtubules only affects the transport of plasmids capable of directed movement (Figure 3d) and not overall diffusion of molecules within the cytoplasm. In fact, recent studies of cytoskeletal micromechanics in our lab have shown that HDAC6 knockdown cells display no difference in cytoplasmic diffusivity or viscosity compared to wild-type cells38.
Others have observed that enrichment of certain microtubule modifications, including acetylation, is correlated with increased movement of kinesin motors and cargo such as vesicles and c-Jun N-terminal kinase-interacting protein (JIP1) in cells, which agrees with our hypothesis13,39,40. The effect of microtubule acetylation on cytoplasmic movement could be due to alterations in the physical structure of microtubules, as it has been hypothesized that acetylation only occurs on polymerized microtubules that are stable41. Further, microtubule dynamics are partially regulated by the hydrolysis of GTP to GDP between tubulin monomers, which causes slight changes in the inter-subunit interactions42,43, and therefore anything that strengthens these interactions can improve the stability and half-life of the structure. It has been proposed that post-translational modifications such as acetylation cause slight changes in tubulin conformation44, which may account for the enhanced stability. Additionally, tubulin acetylation may indirectly impact microtubule stability by enhancing the binding of microtubule associated and plus-end binding proteins, such as Tau, MAP1B, CLIP-170 and EB145,46, which act as stabilizers to control dynamics. Given that motor proteins require polymerized microtubules for long distance transport in the cytoplasm, it makes sense that they would have greater net movement on modified microtubules that are resistant to depolymerization.
Several possibilities could explain why there is enhanced net movement of plasmids in the HDAC6 knockdown cells. One possibility is that motors may move faster along acetylated microtubule tracks, which would allow their cargo (e.g., plasmids) to also have improved kinetics of movement. Examination of all instantaneous and net velocities of pCMV-GFP-DTS plasmids shows no difference in the fastest rates of movement detected in the two cell lines (highest instantaneous velocity of 1.6 μm/second). We simply see longer runs of, and more time spent in, productive transport, giving rise to improved net movement. Further, motor protein kinetics largely depend on ATP hydrolysis47, leading us to believe that microtubule modifications will not change the actual speed of motor “walking” on polymerized structures, although recent evidence has shown that the conformational changes in microtubule structure upon motor binding may play a role in the processivity of motor proteins48,49. Another possibility is that tubulin acetylation may promote stronger interactions of motor proteins, thereby increasing their processivity and leading to more plasmid DNA ultimately reaching the nucleus. Indeed, we found that plasmids spend, in general, less time in diffusive or non-productive motion (Figure 4) in cells with more acetylated microtubules compared to cells with fewer of these structures, suggesting that motor proteins are more actively engaged in productive motion on acetylated microtubules. Further, we found that plasmids moving in HDAC6 knockdown cells, on average, spend more consecutive frames in continual movement than the same plasmids moving in cells with normal levels of acetylated tubulin (Figure 5). This suggests that microtubule acetylation can improve both motor protein recruitment and also processivity.
It is critical in gene transfer that vectors get to the nucleus in a short amount of time to minimize degradation by cytoplasmic nucleases50,51. Thus, it would be more advantageous to the transfection if plasmids spent more time in productive or directed movement and less time spent paused on microtubules. The findings from analyzing instantaneous velocities of individual plasmids correlate with the greater net movement of plasmids seen in HDAC6 knockdown cells compared to wild-type cells (Figure 3). For example, if plasmids have a larger percentage of total movement being productive, and if during active movement they have longer runs of continuous motion, it would generate higher mean net velocities over the whole time they are tracked.
Previous results have demonstrated the necessity of an intact microtubule network for plasmids to reach the nucleus in non-dividing cells10, and our microtubule spin-down assays demonstrate the ability of plasmids to interact with microtubules (Figures 1 and 2). However, no one, to our knowledge has visualized these interactions. The representative immunofluorescence images in Figure 6 confirm this, as there is significant overlap of plasmids with total microtubules in cells, but cells inherently have a very large amount of tubulin, so we expect this number to be high. When we examined plasmid overlap with the acetylated tubulin fraction, we also observed extensive overlap. Given that about two-thirds of the microtubule network is not acetylated in the cells under normal conditions, we aim to enhance this population of modified cytoskeleton in order to increase general plasmid translocation to the nucleus and ultimately, gene transfer.
Post-translational acetylation of tubulin appears to impact microtubule stability and the movement of motor proteins and their cargo in cells, and this offers an advantage for non-viral vectors on their way to the nucleus during gene transfer. In this study we were able to confirm that highly acetylated microtubules can improve the intracellular trafficking of plasmids during gene transfer. These results are important for elucidating the mechanisms behind intracellular movement of not only plasmid DNA, but many different molecules that utilize the microtubule network to be transported through the cell as well. Thus, the recent interest in developing and utilizing HDAC inhibitors may also provide a useful tool for enhancing cellular transfections and gene transfer to animals26.
Materials and Methods
Plasmids, PNA and Quantum dot labeling
The plasmid pBR322 carries no eukaryotic sequences and serves as a backbone for our other vectors20. Plasmids pCMV-Lux-DTS and pCMV-GFP-DTS express luciferase or green fluorescent protein, respectively, from the CMV immediate early promoter/enhancer and contain the SV40 DTS20. All plasmids were purified from Escherichia coli using Qiagen Gigaprep kits as described by the manufacturer (Qiagen, Chatsworth, CA). Plasmids used in the microtubule spin-down assay with immunofluorescence and in microinjection experiments contain the GeneGrip1 PNA binding site (Gene Therapy Systems, San Diego, CA). The PNA (peptide nucleic acid) clamp is a nucleic acid analog that stably hybridizes to a sequence within the plasmid and forms a triplex structure52. This allows fluorophore (CY3) or biotin labeling in a specific region of the plasmid that does not interfere with nuclear translocation or transcription of the plasmid21. Plasmids were labeled with either CY3 fluorophore-conjugated (pCMV-Lux-DTS) or biotinylated PNA (pCMV-GFP-DTS) in Tris EDTA buffer at 37°C for 2 hours, followed by isopropanol precipitation to remove unbound PNA53. Plasmids hybridized to biotin-PNA were further labeled with fluorescent quantum dots (Qdot 605, Invitrogen, Carlsbad, CA) by incubating the streptavidin-conjugated nanocrystals with biotin-labeled plasmids (0.5 mg/ml) at 37°C as previously described19. These pCMV-GFP-DTS plasmids were used in the microinjection experiments.
Microtubule spin-down assays
The microtubule spin-down assay was carried out as previously described19. Half the tubulin sample was in vitro acetylated by adding 1% acetic anhydride to stabilized microtubules and incubated for 10 minutes at 37°C as previously described54, and then incubated with DNA (10 ng of pCMV-Lux-DTS) and/or cell extract (12 μg).
The spin-down assay was modified by centrifuging microtubule samples onto coverslips as described in Dompierre et. al13. The same samples were prepared as above, but the plasmids were labeled with PNA conjugated to a CY3 fluorophore, and 10 μg of tubulin was used. Following a 5 minute fixation with 100% ice-cold methanol, coverslips were incubated with primary antibodies against α-tubulin or acetylated lysine 40-tubulin (1:2000, Sigma Aldrich, St. Louis, MO), followed by Alexa 488 secondary antibody conjugates (Invitrogen, Carlsbad, CA) to label microtubules.
Real-time quantitative PCR
Quantitative, real-time PCR was performed as previously described19.
Cell culture
Stable HDAC6 knockdown A549 cells (human pulmonary adenocarcinoma cells) were a generous gift from T.P. Yao (Duke University)18. These cells, along with wild-type A549 cells (#CCL-185; American Type Culture Collection, Rockville, MD) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1x antimycotic/antibiotic solution (Invitrogen, Carlsbad, CA). One day after microinjection and DNA particle tracking, cells were lysed in Promega lysis buffer (Promega, Madison, WI), and Western blots were performed on lysates to quantify tubulin and acetylated tubulin levels as previously described10.
Microinjections and particle tracking
For microinjections, HDAC6 knockdown and wild-type A549 cells were grown on coverslips in Mattek dishes (Ashland, MA), and Hoechst 33242 nuclear stain (Invitrogen, Carlsbad, CA) was added to the medium prior to injections. Cells were cytoplasmically microinjected with Quantum dot-labeled plasmid constructs (0.5 mg/ml) using an Eppendorf Femtojet system as previously described53. Following injections, fluorescent plasmids were imaged in the cytoplasm using a Leica DMI 6000 B inverted microscope with a 100x objective (N.A. 1.47) and a Hamamatsu EM- CCD camera (Hamamatsu, Japan). Series of images were collected at 2 frames per second for 5 to 10 minutes using Volocity software (Improvision, Waltham, PA). This was repeated for different fields of injected cells for up to 1 hour post-injection. The collected series of images were converted to movie files using ImageJ, and a modified algorithm written in MATLAB was used to track DNA particles and create representative particle trajectories55. Briefly, after choosing the region of interest in the images with fluorescent plasmids, particles were fitted to a fourth order polynomial weighted by a two dimensional Gaussian distribution. The center of the particle was used as the maximum point in the fit. The instantaneous velocities of individual tracked particles were determined for 150 to 168 plasmids per condition, using a minimum of 20 consecutive frames in focus as a cutoff. From the calculated velocities between each frame, the average or net velocity was determined for each tracked plasmid. This was repeated 7 times in at least 30 cells for both cell lines. Non-directed movement was determined by examining instantaneous velocities (frames) less than the average velocity of all tracked negative control pBR322 plasmids, which was calculated to be 0.057 μm/second. The percentage of time engaged in non-directed movement was calculated as the number of frames in which the plasmid was considered to be in diffusive motion divided by the total number of frames. Runs of productive motion were determined as the number of consecutive frames in which instantaneous velocities were greater than 0.057 μm/second, and these “runs” were averaged for each tracked plasmid.
Immunocytochemistry
HDAC6 knockdown and wild-type cells were grown on coverslips and microinjected as above with PNA-biotin labeled plasmids (pCMV-GFP-DTS) and then incubated for 20 minutes at 37°C. Cells were fixed for 10 minutes in 100% ice-cold methanol, coverslips were washed with 1x PBS, and immunofluorescence was carried out using primary antibodies against acetylated-lysine 40-tubulin and β-tubulin (1:1000, Sigma-Aldrich Corp, St. Louis, MO) and secondary Alexa 488 and 647 antibody conjugates (1:200, Invitrogen, Carlsbad, CA) to stain total and acetylated tubulin. Injected biotinylated plasmids were labeled with fluorescent Quantum Dots post-cell-fixation (10 nM Qdot 605, Invitrogen, Carlsbad, CA) by incubating the coverslips with streptavidin-conjugated nanocrystals in 6% bovine serum albumin for 1 hour at room temperature followed by washing with 1x PBS. The coverslips were then mounted onto glass slides using Qmount Qdot mounting medium according to manufacturers instructions (Invitrogen, Carlsbad, CA). Multiple z-series of cells were acquired using a Leica DMI 6000 B inverted microscope with a 100x objective (N.A. 1.47) and a Hamamatsu EM- CCD camera (Hamamatsu, Japan). Volocity software iterative restoration deconvolution (PerkinElmer, Waltham, MA) was used on the resulting images, and colocalization quantified on the 3 dimensional reconstructions by determining the mean Manders overlap coefficient for 20 different cells with over 100 particles for both HDAC6 knockdown and wild-type cells. The mean grayscale intensity was also determined for the corresponding acetylated tubulin and β-tubulin images for each using imageJ. The values were used to determine the ratio of acetylated to total tubulin for each image, which was converted to a percentage.
Supplementary Material
Acknowledgments
We would like to thank Rui Zhou, Erin Vaughan, Mootaz Eldib, and Michael Welte for insightful discussions and technical advice. This work was supported in part by a predoctoral fellowship from the Founders Affiliate of the American Heart Association (MB), and grants EB9903 and ES01247 from the National Institutes of Health.
Footnotes
Supplementary information is available at Gene Therapy’s website.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Lechardeur D, Lukacs GL. Intracellular barriers to non-viral gene transfer. Curr Gene Ther. 2002;2:183–194. doi: 10.2174/1566523024605609. [DOI] [PubMed] [Google Scholar]
- 2.Zhou R, Geiger RC, Dean DA. Intracellular trafficking of nucleic acids. Expert Opin Drug Deliv. 2004;1:127–140. doi: 10.1517/17425247.1.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lechardeur D, Lukacs GL. Nucleocytoplasmic transport of plasmid DNA: a perilous journey from the cytoplasm to the nucleus. Hum Gene Ther. 2006;17:882–889. doi: 10.1089/hum.2006.17.882. [DOI] [PubMed] [Google Scholar]
- 4.Vaughan EE, Dean DA. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol Ther. 2006;13:422–428. doi: 10.1016/j.ymthe.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mesika A, Kiss V, Brumfeld V, Ghosh G, Reich Z. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum Gene Ther. 2005;16:200–208. doi: 10.1089/hum.2005.16.200. [DOI] [PubMed] [Google Scholar]
- 6.Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J Biol Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- 7.Bulinski JC, Gundersen GG. Stabilization of post-translational modification of microtubules during cellular morphogenesis. Bioessays. 1991;13:285–293. doi: 10.1002/bies.950130605. [DOI] [PubMed] [Google Scholar]
- 8.Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R, Gundersen GG. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol. 2008;9:860–873. doi: 10.1038/nrm2522. [DOI] [PubMed] [Google Scholar]
- 10.Vaughan EE, Geiger RC, Miller AM, Loh-Marley PL, Suzuki T, Miyata N, et al. Microtubule acetylation through HDAC6 inhibition results in increased transfection efficiency. Mol Ther. 2008;16:1841–1847. doi: 10.1038/mt.2008.190. [DOI] [PubMed] [Google Scholar]
- 11.Geiger RC, Taylor W, Glucksberg MR, Dean DA. Cyclic stretch-induced reorganization of the cytoskeleton and its role in enhanced gene transfer. Gene Ther. 2006;13:725–731. doi: 10.1038/sj.gt.3302693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, et al. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Dompierre JP, Godin JD, Charrin BC, Cordelieres FP, King SJ, Humbert S, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barlow AL, van Drunen CM, Johnson CA, Tweedie S, Bird A, Turner BM. dSIR2 and dHDAC6: two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp Cell Res. 2001;265:90–103. doi: 10.1006/excr.2001.5162. [DOI] [PubMed] [Google Scholar]
- 15.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 16.Verdel A, Curtet S, Brocard MP, Rousseaux S, Lemercier C, Yoshida M, et al. Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 17.Kelkar SA, Pfister KK, Crystal RG, Leopold PL. Cytoplasmic dynein mediates adenovirus binding to microtubules. J Virol. 2004;78:10122–10132. doi: 10.1128/JVI.78.18.10122-10132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 19.Badding MA, Vaughan EE, Dean DA. Transcription factor plasmid binding modulates microtubule interactions and intracellular trafficking during gene transfer. Gene Ther. 2011 doi: 10.1038/gt.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid nuclear import. Exp Cell Res. 1999;253:713–722. doi: 10.1006/excr.1999.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson GL, Dean BS, Wang G, Dean DA. Nuclear import of plasmid DNA in digitonin-permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J Biol Chem. 1999;274:22025–22032. doi: 10.1074/jbc.274.31.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- 23.Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E, et al. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Sheetz MP. One-dimensional diffusion on microtubules of particles coated with cytoplasmic dynein and immunoglobulins. Cell Struct Funct. 1999;24:373–383. doi: 10.1247/csf.24.373. [DOI] [PubMed] [Google Scholar]
- 25.Mallik R, Petrov D, Lex SA, King SJ, Gross SP. Building complexity: an in vitro study of cytoplasmic dynein with in vivo implications. Curr Biol. 2005;15:2075–2085. doi: 10.1016/j.cub.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman CD, Geiger RC, Dean DA. Electroporation- and mechanical ventilation-mediated gene transfer to the lung. Gene Ther. 2010;17:1098–1104. doi: 10.1038/gt.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor W, Gokay KE, Capaccio C, Davis E, Glucksberg M, Dean DA. The effects of cyclic stretch on gene transfer in alveolar epithelial cells. Mol Ther. 2003;7:542–549. doi: 10.1016/s1525-0016(03)00041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- 29.Paschal BM, Shpetner HS, Vallee RB. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol. 1987;105:1273–1282. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 31.Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li D, Sewer MB. RhoA and DIAPH1 mediate adrenocorticotropin-stimulated cortisol biosynthesis by regulating mitochondrial trafficking. Endocrinology. 2010;151:4313–4323. doi: 10.1210/en.2010-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shubeita GT, Tran SL, Xu J, Vershinin M, Cermelli S, Cotton SL, et al. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci U S A. 2003;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Bruin K, Ruthardt N, von Gersdorff K, Bausinger R, Wagner E, Ogris M, et al. Cellular dynamics of EGF receptor-targeted synthetic viruses. Mol Ther. 2007;15:1297–1305. doi: 10.1038/sj.mt.6300176. [DOI] [PubMed] [Google Scholar]
- 36.Welte MA. Bidirectional transport along microtubules. Curr Biol. 2004;14:R525–537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 37.Posta F, D’Orsogna MR, Chou T. Enhancement of cargo processivity by cooperating molecular motors. Phys Chem Chem Phys. 2009;11:4851–4860. doi: 10.1039/b900760c. [DOI] [PubMed] [Google Scholar]
- 38.Eldib M, Dean DA. Cyclic stretch of alveolar epithelial cells alters cytoskeletal micromechanics. Biotechnol Bioeng. 2011;108:446–453. doi: 10.1002/bit.22941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn S, Morrison EE, Liverpool TB, Molina-Paris C, Cross RA, Alonso MC, et al. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J Cell Sci. 2008;121:1085–1095. doi: 10.1242/jcs.026492. [DOI] [PubMed] [Google Scholar]
- 40.Hammond JW, Huang CF, Kaech S, Jacobson C, Banker G, Verhey KJ. Posttranslational modifications of tubulin and the polarized transport of kinesin-1 in neurons. Mol Biol Cell. 2010;21:572–583. doi: 10.1091/mbc.E09-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 2002;21:6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 43.Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- 44.Regnard C, Audebert S, Boucher D, Larcher JC, Edde B, Denoulet P. Microtubules: functional polymorphisms of tubulin and associated proteins (structural and motor MAP’s) C R Seances Soc Biol Fil. 1996;190:255–268. [PubMed] [Google Scholar]
- 45.Takemura R, Okabe S, Umeyama T, Kanai Y, Cowan NJ, Hirokawa N. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or tau. J Cell Sci. 1992;103(Pt 4):953–964. doi: 10.1242/jcs.103.4.953. [DOI] [PubMed] [Google Scholar]
- 46.Saragoni L, Hernandez P, Maccioni RB. Differential association of tau with subsets of microtubules containing posttranslationally-modified tubulin variants in neuroblastoma cells. Neurochem Res. 2000;25:59–70. doi: 10.1023/a:1007587315630. [DOI] [PubMed] [Google Scholar]
- 47.Xie P. Mechanism of processive movement of monomeric and dimeric kinesin molecules. Int J Biol Sci. 2010;6:665–674. doi: 10.7150/ijbs.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kikkawa M. The role of microtubules in processive kinesin movement. Trends Cell Biol. 2008;18:128–135. doi: 10.1016/j.tcb.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Krebs A, Goldie KN, Hoenger A. Complex formation with kinesin motor domains affects the structure of microtubules. J Mol Biol. 2004;335:139–153. doi: 10.1016/j.jmb.2003.10.039. [DOI] [PubMed] [Google Scholar]
- 50.Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, et al. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 51.Barry ME, Pinto-Gonzalez D, Orson FM, McKenzie GJ, Petry GR, Barry MA. Role of endogenous endonucleases and tissue site in transfection and CpG-mediated immune activation after naked DNA injection. Hum Gene Ther. 1999;10:2461–2480. doi: 10.1089/10430349950016816. [DOI] [PubMed] [Google Scholar]
- 52.Dean DA. Peptide nucleic acids: versatile tools for gene therapy strategies. Advanced Drug Delivery Reviews. 2000;44:81–95. doi: 10.1016/s0169-409x(00)00087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gasiorowski JZ, Dean DA. Postmitotic nuclear retention of episomal plasmids is altered by DNA labeling and detection methods. Mol Ther. 2005;12:460–467. doi: 10.1016/j.ymthe.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogers SS, Waigh TA, Zhao X, Lu JR. Precise particle tracking against a complicated background: polynomial fitting with Gaussian weight. Phys Biol. 2007;4:220–227. doi: 10.1088/1478-3975/4/3/008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


