Abstract
Ferritins, highly symmetrical protein nanocages, are reactors for Fe2+ and dioxygen or hydrogen peroxide that are found in all kingdoms of life and in many different cells of multicellular organisms. They synthesize iron concentrates required for cells to make cofactors of iron proteins (heme, FeS, mono and diiron). The caged ferritin biominerals, Fe2O3•H2O are also antioxidants, acting as sinks for iron and oxidants scavenged from damaged proteins; genetic regulation of ferritin biosynthesis is sensitive to both iron and oxidants. Here, the emphasis here is ferritin oxidoreductase chemistry, ferritin ion channels for Fe 2+ transit into and out of the protein cage and Fe 3+ O mineral nucleation, and uses of ferritin cages in nanocatalysis and nanomaterial synthesis. The Fe2+ and O ferritin protein reactors, likely critical in the transition from anaerobic to aerobic life on earth, play central, contemporary roles that balance iron and oxygen chemistry in biology and have emerging roles in nanotechnology.
Keywords: ferritin, iron biomineral, oxidoreductase, ion channel, nanomaterial, diiron oxygenase
1. Introduction
Ferritins are natural reactors for iron and oxygen chemistry that control the biosynthesis and dissolution of caged hydrated ferric oxides [1, 2]. Caged ferritin minerals can have diameters as large as 8 nm, with thousands of iron and oxygen atoms. The minerals have various amounts of phosphate (P) that reflect local phosphate concentrations; phosphate is low in animal ferritin iron minerals (Fe: P=8:1) and high in plants or microorganisms (Fe: P=1:1) where the ferritin minerals are largely amorphous [1]. Control of ferritin mineral order in animals, where a range of crystallinity occurs [3], depends on the number of protein-based mineral nuclei. The combinatorial structure of hybrid cages assembled from of two types of ferritin gene products, H (catalytically active) and L (catalytically inactive) protein subunits, controls the number of protein-based mineral nuclei [1]. Plant ferritins are localized in plastids; the microbial origin of plastids likely relates to the high phosphate content of plant ferritin minerals. Nevertheless, the plant ferritin protein cage sequence, encoded in the nucleus, is more closely related to animals (55% sequence identity). Ferritins are required to concentrate iron to match requirements for synthesis of proteins with iron cofactors (Fe-porphyrin, Fe-S, and Fe); they also are antioxidants and retrieve iron released from proteins, damaged by oxidation, for future cellular use.
Ferritin protein cages self-assemble into highly symmetrical cages, from polypeptides, after spontaneous folding of each polypeptide into bundles of 4 α helices. Variations are observed in the N-terminal extensions of microbial and eukaryotic ferritins, the location of the active sites, the ferrous oxidants, and the number of ion channels. A central, mineral growth cavity accounts for ~ 60% of the volume of ferritin protein cages (Figure 1). Animal ferritins contrast with all other ferritins (Archaeal, bacterial, and plant) in forming cages from tissue -specific combinations of different H and L subunits; plant ferritin cages are hybrids of different H subunits. Microbes assemble cages that are have identical subunits; the different ferritin gene products are synthesized at different times and assemble into cages containing only one type of subunit (homopolymers) [4].
Figure 1. Eukaryotic ferritin.
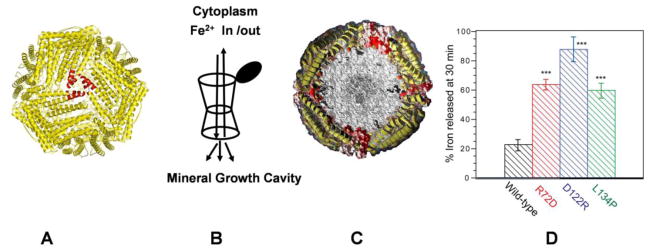
A. The protein cage. B. Outline of one of the eight Fe2+ entry and exit ion channels showing the N-terminal gate (Black) and mid channel constriction [28, 32]. Downward arrows show the direction of Fe2+ transit and distributions to multiple oxidoreductase sites; the upward show Fe2+ leaving the cage after mineral dissolution by external reductant. C. A cross-section of the ferritin protein cage showing a full Fe2O3• H2O mineral; in nature the average filling of the cage is 1000–2000 Fe atoms but the range form a given tissue an be from < 1 iron to 4000 Fe/cage. Helices are shown in yellow; the channels form from helical segments of three polypeptide subunits around the three fold axes of the cage. D. Fe2+ exit increases in ferritin variants with amino acid substitutions at conserved Fe2+ ion channel residues. The figure is constructed from the author’s work in [1] [28], [32]
The multiple, natural, iron-protein interactions occurring within ferritin protein cages throughout the entire length of each subunit: di-Fe2+ sites within each polypeptide where oxidant (O2 or H2O2) produces di-Fe mineral precursors, Fe2+ entry/exit channels formed by three subunits at the 3-fold cage symmetry axes, that feed multiple di-iron catalytic sites, and, specific to animal ferritins, nucleation channels that can enhance mineral order and exit at the 4-fold symmetry axes of the cage [1, 5]; the rates may differ widely as in msec dispersal from single channels to multiple catalytic sites or slow (hours) helix-helix changes as ferric oxo nuclei move through nucleation channels. While localized metal-protein interactions occur throughout individual ferritin polypeptides, cooperative effects on metal ion distribution occur throughout the protein cage [6–8]. Here, we focus in section 2 on oxidoreductase chemistry, in section 3 on Fe 2+ transit into and out of the protein cage, Fe 3+ O mineral nucleation and in section 4 on uses of ferritin cages in nanocatalysis and nanomaterial synthesis. The ancient and widespread occurrence of ferritins Fe2+ and O biological reactors suggests a role in the transition from anaerobic to aerobic life on earth as well as the critical contemporary roles of balancing iron and oxygen chemistry in heath and disease and in emerging roles for nano technology.
2. Oxidoreductase chemistry
The ferritins oxidize Fe2+ and reduce either dioxygen or hydrogen peroxide to synthesize the Fe2O3• H2O, protein-caged biomineral iron concentrates. Active site ligands among the ferritins are diverse and mechanisms vary. The most extensively studied group in the ferritin family is the maxi-ferritin of eukaryotes. In the 24-subunit cages, subunits with active sites use Fe 2+ and dioxygen as substrates; all subunits are catalytically active except in animals where L catalytically inactive subunits, co-assemble with the H active subunits to alter mineral nucleation and crystal order; see section 3.2 and [1]. In microorganisms, there are several types of ferritins; i) bacterial and archaea maxi-ferritins, which are similar to eukaryotic ferritins in terms of the 3D structure [9–11], albeit with low sequence similarity to eukaryotic ferritins. ii) Bacterioferritins (BFRs), which contain heme and are widespread in bacteria; BFR have not been detected in archaea. The 24 subunits of the BFR protein cage contain one b-type heme between dimer pairs [12–14]. iii) Mini-ferritins, called Dps (DNA binding proteins from starved cells) proteins, self-assembled from 12-subunits [15]. Dps proteins like many other ferritins protect cells from oxidant damage by removing iron and oxygen, which in mini-ferritins is predominantly hydrogen peroxide rather than diioxygen, from the cytoplasm and converting into iron biomineral. Ferritin proteins are encoded in genes sensitive to cellular oxidant status. The shared properties of all ferritins are hollow, nanoproteins, of multiple α-helices, containing iron channels and sites for catalytic reactions between two ferrous and oxygen atoms to synthesize caged iron biominerals.
Eukaryotic ferritins catalyze the oxidoreduction reaction for Fe2+ and O2 at the multiple diiron binding sites embedded in the center of a 4-α-helix bundle. Although ferritins utilize Fe2+ as a substrate, the ferritin diiron site shares the structural and functional characteristics with the diiron cofactor site in diiron enzymes. Such diiron cofactor enzymes include methane monooxygenase, ribonucleotide reductase, and Δ9-fatty acid desaturase [16–21]. The first detectable reaction intermediate for the oxidoreduction reaction is a diferric peroxo (DFP) species, which is also detected in the diiron containing enzymes. DFP formation in ferritin has been extensively characterized by kinetics [1, 2] and multiple spectroscopies (UV/vis, resonance Raman, Mössbauer, and EXAFS) [22–24]. DFP decays to Fe3+ oxo or hydroxo dimers or multimers with concomitant production of H2O2; peroxide production is stoichiometric with DFP decay at subsaturating concentrations of Fe2+ [25]. The DFP products move to the central cavity and grow into bulk ferric oxide minerals. The ferritin oxidoreduction reaction at the diiron site is shown in Scheme 1.
| [Scheme 1] |
The mechanism of oxidoreduction reaction in ferritin has been most extensively studied in frog M ferritin, a cage composed of 24 catalytically active (H-type) with favorable kinetics, good yield, and high solubility. The Fe2+ -bound ferritin structure has been elusive because of the millisecond time scale of the reaction. Much structural information on ferritin diiron sites has been obtained by: studying proteins after insertional and deletional site directed mutagenesis [26, 27]. Physical methods used in recent studies of ferritin diiron sites include variable temperature and magnetic field magnetic circular dichroism/circular dichroism (VTVH MCD/CD)[6] and X-ray crystallography of co-crystals with Fe2+ analogues [21, 28]. The structure of the diiron substrate binding site in eukaryotic ferritin is shown in Figure 2, contrasted with sites in diiron oxygenase enzymes where the iron is a cofactor. The O-O bridge is preserved during decay of the diferric peroxo intermediate in ferritin, when there are fewer than one turnover/site on average based on the appearance of H2O2 with 1– 1.5 Fe2+/diiron site [25]. Whether the O-O bond in ferritin diferric peroxo is cleaved when there are multiple turnovers, as it is during catalysis by the di-iron oxygenases, is unknown.
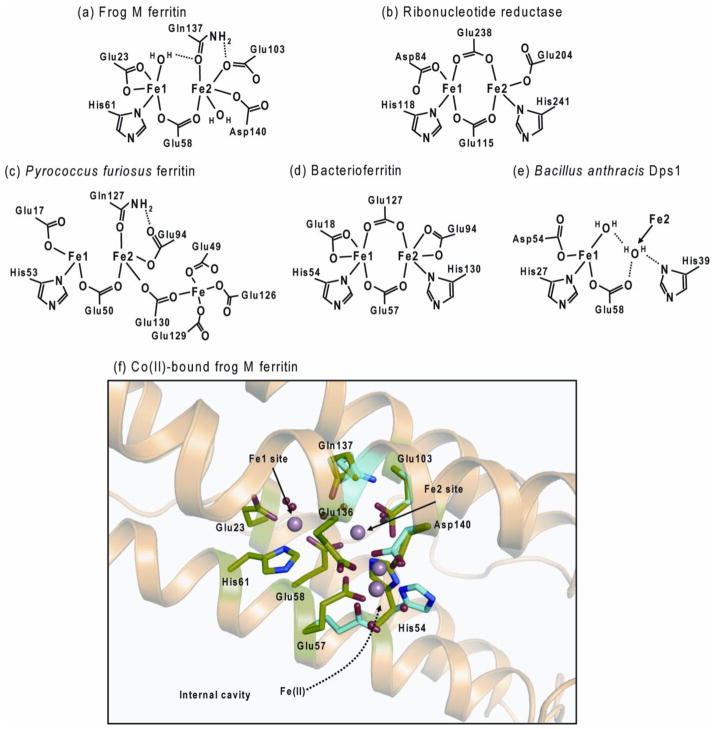
Figure 2. Diiron site structures of different types of ferritins and related diiron protein.
(a) Frog M ferritin based on the spectroscopic analysis and Mg-bound structure (1MFR), (b) Ribonucleotide reductase (1PFR), (c) Microbial ferritin from Pyrococcus furiosus (2JD7), (d) E. coli bacterioferritin (3E1M), (e) Dps1 from Bacillus anthracis (1JI5), and (f) Co2+-bound frog M ferritin (3KA4). Four Co2+ were observed around oxidoreductase site. Several residues near Fe2 site showed two conformations.
One of the iron sites in the ferritin oxidoreductase centers of ferritin, Fe1, has similar coordination structure to the Fe at each dioxygenase site: E, ExxH. However, a distinct feature of the ferritin diiron site, except in BFR [4], is the weaker Fe2+ ligands in Fe2 site, compared with the diiron cofactor sites. The Fe2+ ligands for diiron enzymes are Glu, Glu-x-x-His, but the Fe2 site residues in ferritin are Glu103, Gln137-x-x-Asp140. (The residue at position 140 is not fully conserved in ferritins. For example, frog H and mitochondrial ferritins have Ser, and human H ferritin has Ala at that position). It is noteworthy that anaerobic VTVH MCD/CD revealed cooperative binding of Fe2+ to the ferritin diiron sites, with a Hill coefficient ~3 that was iron independent, and because of the anaerobic conditions, oxygen independent. Rather, the observation reflects cooperative interactions among a subset of 24 subunits, indicating that the ferritin function is regulated by communication among subunits [6].
The importance of the ferritin-specific Fe2 site residues in the oxidoreduction reactions of eukaryotic ferritins, was demonstrated by the substitution of the ferritin Fe2 site residues with the corresponding residues in the diiron enzymes [29]. The substitution of Gln137 with Glu changed the reaction pathway: DFP was absent and a different ferric intermediate formed, yet to be identified. When Fe3+ was measured at a nonspecific absorption band for the mixture of DFP, DFP decay products and mineral, the oxidation rate of the Gln137Glu ferritin was similar to wild-type protein, but only reached an absorbance value 50% of the wild type. The substitution of Asp 140 by His, by contrast, inhibited any Fe2+ oxidation, even though in natural, active ferritins residue 140 can vary among Asp, Ser or Ala; variation among Asp or Ala or Ser only impacts kinetic parameters, Kapp and Vmax [29]. These results indicate that the Fe-2 site, which is ferritin- specific and has weaker iron ligands than in diiron oxygenase proteins such as methane monooxygenase, ribonucleotide reductase, and fatty acid desaturase, control the iron mineralization pathway.
The use of Co2+ in protein cocrystals revealed multiple ligand conformations, especially around the Fe2 site, even when crystals were studied at cryogenic temperatures [28]. Co2+, which is not oxidized in ferritin, inhibits the Fe2+/O2 oxidoreduction reaction by ~70%; the Co2+ sites likely trace transient Fe (II) binding sites and outline a reasonable Fe2+ binding pathway from inner cavity surface to the diiron site. The pivotal role of flexibility at the Fe2 site in ferritin diiron sites for Fe 2+ binding and catalysis is illustrated by the effect of insertion, at the ferritin-specific Fe2 site, of histidine at position 140 and Glu at position 137, creating the diiron oxygenase enzyme sites. In the variant ferritin all Fe2+/O2 oxidoreduction activity was eliminated, and flexibility of the diiron site was lost; only one conformation was present at the diiron site in Co2+-protein crystals of the variant ferritin [28]. These observations indicate the dynamic site properties conferred by weaker Fe2 ligands in ferritin, which complement the changes in Fe-Fe distance from 2.5 to 3.4 Å and the contribution of Fe-protein site flexibility during ferritin catalysis [24] that contrasts with diiron oxygenase enzymes.
The location of oxidoreductase sites in microbial ferritins can differ from animal and plant ferritins. While metal binding sites in microbial ferritins closely resemble those of plant and animal ferritins [9–11] in general, and exception is the Fe2 site ligands at one site. In microbial ferritins there is always Glu at the position equivalent to the variable of Asp/Ala/Ser in eukaryotic ferritins (Figure 2) where the variations impact oxidoreductase kinetics in a biologically significant (tissue-specific) manner. In addition, microbial ferritins have a third iron binding site, called Site C, which is located close to the diiron site (Figure 2). In the microbial ferritin BFR the diiron site is symmetric and has the same ligands as diiron enzyme cofactors (Figure 2): E, ExxH and E, ExxH. The retention of iron at BFR diiron sites during turnover and the absence of the DFP intermediate further emphasize the distinctions between eukaryotic (Scheme 1) and BFR oxidoreductase mechanisms [4, 12, 14]. Dps miniferritins oxidize Fe2+ to form bio-mineral, preferentially using H2O2 rather than O2 as a second substrate [15]. The Fe2+ oxidation sites are usually between two subunits on the inner surface of the cage and the order of binding can be different. In several Dps mini-ferritins, only one Fe2+ binds in the absence of H2O2 oxidant, observed by Trp quenching in Listeria Dps [30] or by VTVH MCD in B. anthracis, whether the oxidant was O2 (Dps 1) or H2O2 (Dps2) [31] (Figure 2). Further mechanistic studies on these microbial ferritins will provide more insights into the iron and oxygen chemistry among members of the ferritin family.
3. Iron transit-Ion channels
Ferritin protein cages move iron around between the multiple Fe-protein interactions sites. Fe2+ entry/exit channels are used for the exchange of iron to and from the caged ferritin iron biomineral and the cytoplasm. In eukaryotic ferritins, at least, the Fe2+entry channels are related to membrane ion channels with cytoplasmic N-terminal gating [32]. Eukaryotic ferritins also contain Fe3+ nucleation channels that connect the oxidoreductase sites in each subunit to the mineral growth cavity.
3.1 Fe2+ entry
The ferritin function of concentrating iron for synthesizing proteins with iron cofactor requires transfer of Fe2+ from the cellular cytoplasm to the ferritin oxidoreductase sites in the protein cage. In addition, after catalysis, Fe3+O catalytic products are transferred through the ferritin protein cage to the mineral growth cavity.
Fe2+ enters the ferritin proteins cages through pores in the external surface of the proteins cage. Fe2+ entry channels are ~ 15Å length. The pores on the external cage surface are gated by extensions of the four-helix bundle subunits. The internal exits of the Fe2+ ion channels are pores on the internal cage surface, which are about 15–20Å away, from oxidoreductase site residues. A set of conserved carboxylate residues in the channels create an ion gradient that directs the Fe2+ ions toward the internal pores [8, 15]; the internal channel exits can accommodate three metal ions [28]. Fe2+ exiting the ion channels on the inner surface of the cage are destined for one of three different catalytic sites, which are either buried in the middle of the helix bundles of each subunit (eukaryotic and some microbial ferritins), near the inner cage surface (BFR) or on the inner cage surface of microbial miniferritins (Dps proteins). The channel carboxylates in 24 subunit ferritins selectively control Fe2+ entry [8], contrasting with channels in the 12 subunit mini-ferritins where the same carboxylate groups control both Fe2+ entry and Fe2+ exit [33].
Carboxylate residues in each subunit near the channel exits direct the exiting Fe2+ to each oxidoreductase site (R.K. Behera and E.C. Theil, to be published). How the distribution of Fe2+ from one channel among the three di-iron sites is determined, remains unknown. However, a protein :protein dependent Hill coefficient of 3 has been observed for Fe2+ binding at the oxidoreductase sites [6] that may relate to cooperative distributions of Fe2+ from the channels to insure filing of all vacant, diiron, oxidoreductase sites.
3.2 Fe3+ nucleation
Iron, during ferritin mineral formation, moves through several reasonably well characterized pathways: i.Fe2+ entry into and through the channels, ii. Transit to the oxidoreductase sites. iii. Reaction at the active sites with dioxygen or hydrogen peroxide (12 subunit ferritins) to synthesize Fe3+O multimers. In the case of eukaryotic ferritins, the first reaction intermediate characterized is a diferric peroxo intermediate followed by diferric oxo product [1]. Variations in microorganisms include retention of diiron as a cofactor and involvement of a third iron site as in E. coli bacterioferritin and other microbial ferritins, and binding of hydrogen peroxide before binding of the second Fe2+ [4, 12, 15, 34].
A long, 20Å channel connects the oxidoreductase sites to the mineral growth cavity. The channel, which was unpredicted from protein crystal structures, was detected using the paramagnetic effects of Fe3+ on 13C NMR spectra of ferritin [7]. As (Fe3+)2O moves through the nucleation channels, the diferric oxo products of multiple catalytic turnovers interact to form tetramers inside the channels [7]. Interestingly, in an older study, diferric peroxo intermediates were observed to decay to diferric oxo and multiferric (N>2) oxo species in ferritin by Mössbauer spectroscopy [25]. The multimers may relate to the tetrameric Fe3+O species observed by magnetic susceptibility measurements after two additions of saturating amounts of Fe (2 Fe/site for the 24 sites) [7].
Ferritin Fe3+O nucleation channels open onto the internal surfaces of ferritin protein cages at the four fold symmetry axes of the ferritin protein cage. Thus, mineral nuclei of 16 Fe3+O can form, oriented by four nucleation channels in close proximity, leading to highly ordered ferritin mineral. Such minerals are observed in human tissues like the heart [3]. In plants ferritin mineral order is disrupted by phosphate. In animals, ferritin mineral disorder can be introduced into ferritin minerals by the coassembly of large numbers of catalytically inactive subunits (L subunits) only found in animals. The catalytically inactive L subunits coassemble with catalytically active H subunits. L subunits constitute a large fraction of the ferritin cage in liver ferritin, which synthesizes a much more disordered mineral than heart ferritin [1, 3]; heart ferritin cages contain a large fraction of the H subunits with catalytic sites and nucleation channels.
3.3 Fe2+ exit
The role of the Fe2+ ion channel in iron exit is detected by measuring rates of formation of Fe2+-bipyridyl from ferritin minerals in wild type and variant proteins cages. The Fe2+-bipyridyl is outside the protein cage when measured. Dissolution of the caged, solid iron-oxy ferritin mineral is, as for all minerals, surface limited. The dissolution of the ferritin mineral under aqueous, physiological conditions, requires reductants such as physiological NADH/FMN or chemical dithionite. Typical chelators used are bipyridyl, or desferioximine, a therapeutically used chelator in humans; since the reactions are usually run in air it is the Fe3+desferioxamiine complex that is measured.
When ferritin mineral dissolution is studied under physiological conditions, many properties of the complex reaction remain unknown. Such unknowns include the location of electron transfer that reduces the ferric mineral (direct or through the protein), how rehydrating water reaches Fe2+ on the surface of the caged mineral, and where the chelators bind the Fe2+ (outside the protein or in the channels/cage). What is clear, however is that the ferritin protein pores are mostly closed to minimize uncontrolled reactions between the ferric ferritin mineral and external, cytoplasmic reductants. Manipulation of a specific, group of conserved channel amino acids increases unfolding of the channel or gate structures and selectively increases mineral dissolution and Fe2+ exit with no or small effects on Fe2+entry [32, 35] (Figure 1D).
The ferritin pore region is differentially sensitive to environmental changes. Pore/channel opening occurs at low temperatures far below cage unfolding, at physiological (mM) concentrations of urea [26] and to stoichiometric concentrations of highly selected peptides [26, 36]. The channel amino acid that control folding of ferritin cage pores and channels are both hydrophobic and hydrophilic residues, based on the higher Fe2+ exit rates/mineral dissolution, both in solution (Figure 1D) and in cultured human cells [37] when the amino acids are changed. Unfolding is also observed by protein crystallography of the variant proteins [32, 35]. Such observations suggest that Fe2+ exit, and possibly Fe2+ delivery and entry, are regulated by external cellular molecules that change the rate of pore folding in ferritin protein cages.
4. Ferritin in Nanochemistry and Technology
The hollow, ferritin nanocages, which have 12/8 and 8/5 nm external/internal diameters respectively[1, 4, 38, 39] are used as templating and restricting devices in nanocatalysis of polymers and nanomaterials [40]. Ferritin nanocages are water soluble and at neutral pH, the external surface has a net negative charge with inner and outer surfaces that may be modified for different applications. The symmetrically placed, hydrophilic (3-fold) ion channels, the hollow nanocavities of different sizes, the sequence variations among the ferritin protein cages from different organisms, and the different thermal and chemical stabilites of ferritin protein cages are all features of ferritin nanocages to be used in nanochemistry and technology. During the last several decades ferritin nanocages have been used in number applications, briefly summarized here; detailed descriptions are in [41–44]. The first use of ferritin cages was as templates for nanoparticles/materials synthesis [45–47]. Other uses include the synthesis and delivery of magnetic resonance imaging (MRI) contrast agents [48–52], drug delivery and catalysis [44, 53–56].
Ferritin cages from both mesophilic and thermophilic bacteria are stable to relatively harsh conditions[57]. In aqueous solution, ferritin cages remain intact at least up to 80°C, 6 M urea or guanidine (at pH 7.0, room temperature), or 1% sodium dodecyl sulfate (SDS)[38], as neutral values of pH. The ferritin cage can be stabilized in organic solvents, such as dichloromethane, ethyl acetate and toluene, with long chain hydrocarbons (C9, C12, C14) by derivatizing carboxylates on the external surface of the cage [58–63]. The disassembly/assembly of ferritin in aqueous solution at low pH/ high pH has produced different sized nanoparticles (<8nm) entrapped inside ferritin cages [64].
The natural metal binding sites present throughout ferritin protein cages can be used for nanomaterial synthesis and metallocatalysts. In the past many studies used commercially- available ferritin from horse spleen is used [55]. Such ferritins have many fewer functional iron binding sites because they lack oxidoreductase centers and mineral nucleation channels that guide the growth of the natural iron biominerals [7]. Using H ferritins with such channels could be exploited in extruding materials for growth in the cavity under controlled conditions. New functional iron-binding sites are being discovered in ferritin [7, 8]; they remain to be explored as the potential sites of metal binding for catalysis and protein-controlled nanoparticle synthesis.
4.1 Nanoparticle and Nanomaterial Synthesis
Inside the cell, ferritin nanocages selectively uptake the Fe2+ and forms ferric oxide nano mineral cores inside the nanocavity[38]. However, in vitro, purified ferritin cages can incorporate a variety of metal ions (Fe, Au, Pd, Rh, Pt, Ni, Cr, Cd, Ti, Tb, Co, Cu and Zn), most likely through hydrophilic 3-fold ion channel [28, 43, 47] discussed in section 3. When ferritin protein cages are used as a template, the uniform nanoparticles are limited by the interior cavity of the protein cage. Uniform nanoparticle sizes, with diameters < 8 nm, are produced with maxi-ferritin cages and < 5 nm in diameter with mini-ferritin cages as templates. Therefore, ferritin provides a platform for constrained size and shape nanomaterial synthesis. Thus in addition to the narrow size distribution conferred, the protein cage stabilize the nanoparticles for some applications [65].
The caged hydrated ferric oxide minerals synthesized by adding solutions of Fe2+ to solutions of ferritin protein are similar to the iron oxide core formed naturally in ferritin[66]. Use of natural ferritin nanocages as templates has produced CdS (semiconductors) and uniform FeO (MRI) nanoparticles [67, 68]. A series of metallic nanoparticles such as gold, silver, lead, copper and nickel also have been prepared by introducing these metal ions inside the cavity followed by reducing them with external reductants [43]. The creation of additional metal binding sites has depended on protein engineering of ferritin protein cages(Figure 3B), where the introduction of more negatively charged amino acid residues on internal surfaces enhanced silver and gold nanoparticle templating [69]. To achieve a smaller size nanoparticle, the 12 subunit mini-ferritins (Dps proteins) can be used, exemplified by Listeria innocua Dps protein [43, 70]. The ferritin from thermophilic organisms has been used for high temperature syntheses exemplified by the production of magnetite in ferritin from the archaeon, Pyrococus furious [43]. The variety of different amino acid sequences among ferritin nanocages, which can differ by 80%, different ferritin cage sizes (12 and 24 subunits) and continually emerging natural iron-ferritin interactions all show that the application of ferritin cages to nanopartcles syntheses has barely scratched the surface of possibilities.
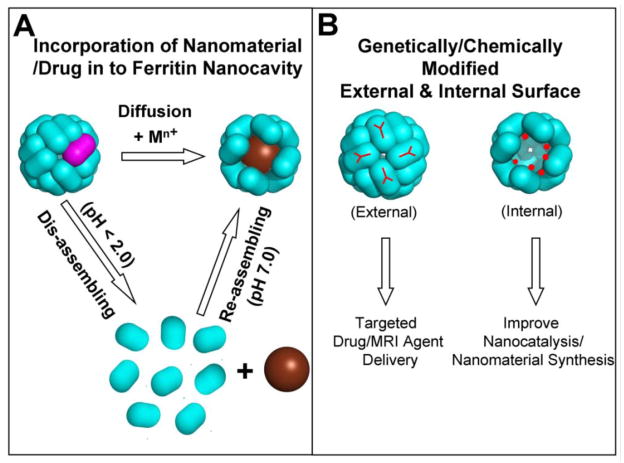
Figure 3. Synthesis and incorporation of nanomaterials/nanoparticles inside the multi-subunit ferritin nano cavity for catalysis and targeted drug delivery.
A. The smaller chemicals, preferably positive charged metal complexes easily diffuse in to the ferritin nanocavity through the 3-fold ion channels by incubation with the intact ferritin cage and nanoparticle synthesis takes place inside the cavity. Bigger nanoparticles (< 8 nm) encapsulated by disassembling/reassembling of multi-subunit ferritin nanocage. B. Both external and internal ferritin cage surfaces modified either by genetically/or chemically to use ferritin nanocage containing nanomaterial for the targeted delivery and to improve catalysis/synthesis. Purple-ferritin polypeptide subunits.
4.2 Targeted Drug Delivery
The large cavity (30% of the cage volume) interface provides the potential to differentially access and exploit the inner and outer surfaces. The use of the internal cavity to trap various chemicals, such as hydrophilic drugs, imaging agents (MRI and fluorophores) [51, 71, 72] (Figure 3) radionuclides or nuclear medicines[44] such as 177Lu3+, 90Y3+, and anti-tumor drugs [54, 73] like deoxyrubicin, cis-platin, carboplatin and ozaloplatin are just early examples. Highly sensitive DNA probes or probes for bioassay and immunoassay have also been entrapped inside ferritin protein cages [44]. The attachment of cell specific ligands/peptides the ferritin external surface, genetically and/or chemically, has great potential for the targeted delivery of these drugs and imaging agents. The design of cell -specific ligands/peptides on the ferritin outer surface (Figure 3B) rests on the many high resolution of ferritin cage structures. The introduction of imaging agents and drugs can use normal diffusion with disassembly of ferritin nanocage at low pH and reassembly at neutral pH (Figure 3A). Recently a magnetite nanocomposite was prepared inside a ferritin nanocage engineered with a peptide RGD-4C (CDCRGDCFC) that is known to bind cell integrins αvβ3 and αvβ5 [51, 71]. The RGD-4C modified ferritins, have been used for imaging vascular inflammation and angiogenesis and influencing metabolism in C32 human melanoma cells providing a dramatic illustration of the potential of ferritin protein nanocages in drug delivery.
The natural ability of living cells to selectively recognize and incorporate exogenous ferritin by ferritin receptors reflect the intrinsic and complex surfaces of ferritin protein cages, understanding is in its infancy [74–77]. The use of selective modifications of ferritin protein cages to attack specific cells in disease cells is a largely untapped source of innovation for nanomedicine.
4.3 Magnetic Resonance Imaging
Magnetic resonance imaging (MRI) has been used to quantify iron levels in different human tissues[50]. It is an indirect measure of tissue iron concentrations that monitors the effect of paramagnetic iron on the proton resonance behavior of the water in tissues. The longitudinal (R1) and transverse (R2) nuclear magnetic relaxation rates are increased when tissue iron interacts with water protons. Increases in the proton relaxation rate can be related to the iron concentration and magnetic properties. The superparamagnetism of ferritin biominerals can be used as an endogenous R2 enhanced MRI contrast agent. The endogenous ferritin is a reporter, in the iron overload associated with hypertranfusion treatments of sickle cell anemia, thalassemia, and in hemochromatosis and Alzheimer disease, to estimate the amount of iron in human tissues such as liver, spleen and brain by MRI [50]. Similar MRI approaches have monitored transgene expression [78]. Ferritin use is limited by the low relaxivity compared to commercially available iron oxide MRI contrasting agents, such as small super paramagnetic iron oxide (SPIO) and ultra small super paramagnetic iron oxide (USPIO)[51, 65]. However, by increasing the ferritin mineral size with iron added in solution (~4000/cage) or by entrapment of superparamagnetic, synthetic magnetite, nanoparticles with higher relaxivity, improved MRI contrasting abilities with ferritin are being achieved [49, 51, 65]. Other contrast agents, trapped inside the ferritin protein cage, by cage disassembly/reassembly, include the fluorescen label Cy5.5 or magnetic nanoparticles. The modified ferritins, used to image vascular macrophages, by MRI or fluorescence and to improve understanding of atherosclerotic plaques and heart disease, are a “proof of principle” for the use of ferritin protein cages in many other imaging applications.
4.4 Nanocatalysis and Nanoelectronics
The inner surfaces of the ferritin nanocages have been used as sites for organometallic catalysts to produce polymers of defined molecular weight, with a narrow size distribution [45, 46, 56]. When palladium and rhuthenium metalloorganic complexes are incubated with ferritin protein, the metal complexes enter the ferritin cavity, possibly during pore unfolding, and bind to specific amino acid ligands such as histidine. The ferritin proteins cages constrain the polymer size and dispersity. In contrast, T synthesis of polymers without protein nanocages, by contrast, leads to polymers with high polydispersity that limits uses in number of applications. Recently, ferritin bound catalysts has also been shown to hydrogenate smaller olefins size selectively. The catalytic activity per cage can be modulated by introducing additional metal binding amino acid residues at the inner surface of the ferritin nanocage and manipulating the ion channels[43]. Ferritin cages as novel templates for iron oxide nanoparticles used in the sustainable productions of olefins using processes such as the Fiescher-Tropsch synthesis [79], remain for future experiments.
Ferritin cages, both 12 and 24 subunit sizes, have been used as templates for metal oxide nanoparticles and metal oxide semiconductors, and also to produce building blocks for the fabrication of electronic devices such as floating nanodot memory device or thin film transistor flash memory[42, 43].
5. Conclusions and Perspective
Ferritins, the complex, highly symmetric, cage structures that synthesize ferric oxo biominerals as metabolic iron concentrates and antioxidants, are found in all kingdoms of life. They share self-assemblying protein cages composed of multiple polypeptides (4-α helix bundles), with multiple embedded oxidoreductase sites, iron entry/exit channels, and a very large (60% by volume) central mineral growth cavity. Ferritins vary in the number of subunits (24 or 12) and size, number of channels, location of the active sites, oxidant (O2 or H2O2), mineral phosphate content and order, and presence or absence of protein-based nucleation channels that guide mineral order.
The enormous progress made in understanding ferritin protein cages has, nevertheless, revealed a number of complex problems that remain to be solved such as: 1. Cytoplamsic delivery and removal of ferritin iron, i.e. identifying ferritin binding partners. 2. Understanding ferritin pore folding and unfolding rates. 3. Mechanisms of diferric oxo release from oxidoreductase sites: the role of protein protonation, O-O bond splitting, etc. 4. Electron transfer and hydration paths during ferritin mineral dissolution. 5. Contributions of ferritin mineral surface area and order to mineral reduction, dissolution. 6. The transit paths between Fe2+ reduced and dissolved from ferritin minerals and the ion channels. 7. Protein based control of mineral crystallinity. 8. Mechanism(s) of movement of Fe3+ O multimers through α-helical bundles of ferritin protein cages
What purposes will be served by solving the many remaining problems in ferritin structure /function? First, fundamental understanding of highly evolved protein cage function will be obtained that is also important in virus protein cage formation and supramolecular protein assembly in general. Second iron chelation therapies will be improved since the iron is ferritin iron biominerals; disease targets include iron overload in transfusional treatments for Sickle Cell disease and Thalassemia and hereditary hemochromatosis and nervous system diseases such as Friedreich’s ataxia. Finally, the ability to manipulate ferritin protein cages more completely in nanotechnology such as the use of both inner and outer surfaces of the ferritin nanocages as templates in homogeneous nanomaterial synthesis, nanocatalysts for sustainable of hydrocarbons, carrier in targeted drug delivery, synthesis and encapsulation of imaging agents, nanocatalysis and nanoelectronics, and possibly will be enhanced by the ability to admit lager catalytic centers, control nanomaterial order for new applications.
HIGHLIGHTS.
Ferritins synthesize of caged hydrated ferric oxide nanominerals as metabolic iron concentrates and antioxidants, trapping Fe from damaged proteins O.
Multiple (24 or 12 polypeptides), folded in 4-α-helix bundles, self -assemble to form ferritin cages.
Fe2+ enters and exit water soluble ferritin through ion channels, similar to membrane ion channels,
Fe2+ and O react at diiron oxidoreductase sites to initiate mineral synthesis with diferric oxo complexes and in some ferritins larger, protein-based, ferric multimers.
Ferrtin proteins cages care are being developed for nanomatierals, nanocatalysts, nanodevices and tissue imaging
Acknowledgments
The work of the authors cited here was supported in part by the NIH-NIDDK (ECT, RKB, and TT), JSPS (TT) and CHORI Partners (ECT, RKB).
ABBREVIATIONS
- DFP
diferrc peroxo
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Theil EC. Current opinion in chemical biology. 2011;15:304–311. doi: 10.1016/j.cbpa.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bou-Abdallah F. Biochim Biophys Acta. 2010;1800:719–731. doi: 10.1016/j.bbagen.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 3.St Pierre T, Tran KC, Webb J, Macey DJ, Heywood BR, Sparks NH, Wade VJ, Mann S, Pootrakul P. Biol Met. 1991;4:162–165. doi: 10.1007/BF01141308. [DOI] [PubMed] [Google Scholar]
- 4.Theil EC, Le Brun NE. In: Comprehensive Inorgannic Chemistry II. Reedijk J, Poeppelmeier K, editors. Elsevier; 2012. [Google Scholar]
- 5.Zhang Y, Orner BP. Int J Mol Sci. 2011;12:5406–5421. doi: 10.3390/ijms12085406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz JK, Liu XS, Tosha T, Theil EC, Solomon EI. Journal of the American Chemical Society. 2008;130:9441–9450. doi: 10.1021/ja801251q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turano P, Lalli D, Felli IC, Theil EC, Bertini I. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:545–550. doi: 10.1073/pnas.0908082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haldar S, Bevers LE, Tosha T, Theil EC. The Journal of biological chemistry. 2011;286:25620–25627. doi: 10.1074/jbc.M110.205278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stillman TJ, Hempstead PD, Artymiuk PJ, Andrews SC, Hudson AJ, Treffry A, Guest JR, Harrison PM. Journal of molecular biology. 2001;307:587–603. doi: 10.1006/jmbi.2001.4475. [DOI] [PubMed] [Google Scholar]
- 10.Johnson E, Cascio D, Sawaya MR, Gingery M, Schroder I. Structure. 2005;13:637–648. doi: 10.1016/j.str.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Tatur J, Hagen WR, Matias PM. J Biol Inorg Chem. 2007;12:615–630. doi: 10.1007/s00775-007-0212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Brun NE, Crow A, Murphy ME, Mauk AG, Moore GR. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbagen.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Macedo S, Romao CV, Mitchell E, Matias PM, Liu MY, Xavier AV, LeGall J, Teixeira M, Lindley P, Carrondo MA. Nature structural biology. 2003;10:285–290. doi: 10.1038/nsb909. [DOI] [PubMed] [Google Scholar]
- 14.Crow A, Lawson TL, Lewin A, Moore GR, Le Brun NE. Journal of the American Chemical Society. 2009;131:6808–6813. doi: 10.1021/ja8093444. [DOI] [PubMed] [Google Scholar]
- 15.Chiancone E, Ceci P. Biochim Biophys Acta. 2010;1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Rosenzweig AC, Nordlund P, Takahara PM, Frederick CA, Lippard SJ. Chemistry & biology. 1995;2:409–418. [PubMed] [Google Scholar]
- 17.Logan DT, Su XD, Aberg A, Regnstrom K, Hajdu J, Eklund H, Nordlund P. Structure. 1996;4:1053–1064. doi: 10.1016/s0969-2126(96)00112-8. [DOI] [PubMed] [Google Scholar]
- 18.Lindqvist Y, Huang W, Schneider G, Shanklin J. The EMBO journal. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 19.Langlois d’Estaintot B, Santambrogio P, Granier T, Gallois B, Chevalier JM, Precigoux G, Levi S, Arosio P. Journal of molecular biology. 2004;340:277–293. doi: 10.1016/j.jmb.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 20.Toussaint L, Bertrand L, Hue L, Crichton RR, Declercq JP. Journal of molecular biology. 2007;365:440–452. doi: 10.1016/j.jmb.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Ha Y, Shi D, Small GW, Theil EC, Allewell NM. J Biol Inorg Chem. 1999;4:243–256. doi: 10.1007/s007750050310. [DOI] [PubMed] [Google Scholar]
- 22.Pereira AS, Small W, Krebs C, Tavares P, Edmondson DE, Theil EC, Huynh BH. Biochemistry. 1998;37:9871–9876. doi: 10.1021/bi980847w. [DOI] [PubMed] [Google Scholar]
- 23.Moenne-Loccoz P, Krebs C, Herlihy K, Edmondson DE, Theil EC, Huynh BH, Loehr TM. Biochemistry. 1999;38:5290–5295. doi: 10.1021/bi990095l. [DOI] [PubMed] [Google Scholar]
- 24.Hwang J, Krebs C, Huynh BH, Edmondson DE, Theil EC, Penner-Hahn JE. Science (New York, NY. 2000;287:122–125. doi: 10.1126/science.287.5450.122. [DOI] [PubMed] [Google Scholar]
- 25.Jameson GN, Jin W, Krebs C, Perreira AS, Tavares P, Liu X, Theil EC, Huynh BH. Biochemistry. 2002;41:13435–13443. doi: 10.1021/bi026478s. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Jin W, Theil EC. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3653–3658. doi: 10.1073/pnas.0636928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arosio P, Ingrassia R, Cavadini P. Biochim Biophys Acta. 2009;1790:589–599. doi: 10.1016/j.bbagen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Tosha T, Ng HL, Bhattasali O, Alber T, Theil EC. Journal of the American Chemical Society. 2010;132:14562–14569. doi: 10.1021/ja105583d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tosha T, Hasan MR, Theil EC. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18182–18187. doi: 10.1073/pnas.0805083105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ilari A, Latella MC, Ceci P, Ribacchi F, Su M, Giangiacomo L, Stefanini S, Chasteen ND, Chiancone E. Biochemistry. 2005;44:5579–5587. doi: 10.1021/bi050005e. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz JK, Liu XS, Tosha T, Diebold A, Theil EC, Solomon EI. Biochemistry. 2010;49:10516–10525. doi: 10.1021/bi101346c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tosha T, Behera RK, Ng H-L, Bhattasali O, Alber T, Theil EC. J Biol Chem. 2012 doi: 10.1074/jbc.M111.332734. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellapadrona G, Stefanini S, Zamparelli C, Theil EC, Chiancone E. The Journal of biological chemistry. 2009;284:19101–19109. doi: 10.1074/jbc.M109.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Honarmand Ebrahimi K, Hagedoorn PL, Hagen WR. J Biol Inorg Chem. 2010;15:1243–1253. doi: 10.1007/s00775-010-0682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takagi H, Shi D, Ha Y, Allewell NM, Theil EC. The Journal of biological chemistry. 1998;273:18685–18688. doi: 10.1074/jbc.273.30.18685. [DOI] [PubMed] [Google Scholar]
- 36.Liu XS, Patterson LD, Miller MJ, Theil EC. The Journal of biological chemistry. 2007;282:31821–31825. doi: 10.1074/jbc.C700153200. [DOI] [PubMed] [Google Scholar]
- 37.Hasan MR, Tosha T, Theil EC. The Journal of biological chemistry. 2008;283:31394–31400. doi: 10.1074/jbc.M806025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Theil EC. Accounts of chemical research. 2005;38:167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 39.Bevers LE, Theil EC. In: Progress in Molecular and Subcellualr Biology. Muller WEG, editor. Springer-Verlag; Berlin-Heidelberg: 2011. p. 18. [Google Scholar]
- 40.Theil EC, Behera RK. The Chemistry of Nature’s Iron BioMinerals In Ferritin Protein Nanocages. In: Watanabe Y, Ueno T, editors. Coordination Chemistry in Protein Cages: Principles, Design, and Applications. Wiley; 2012. [Google Scholar]
- 41.Campan M, Lionetti V, Aquaro GD, Forini F, Matteucci M, Vannucci L, Chiuppesi F, Di Cristofano C, Faggioni M, Maioli M, Barile L, Messina E, Lombardi M, Pucci A, Pistello M, Recchia FA. American journal of physiology. 2011;300:H2238–2250. doi: 10.1152/ajpheart.00935.2010. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita I, Iwahori K, Kumagai S. Biochim Biophys Acta. 2010;1800:846–857. doi: 10.1016/j.bbagen.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Uchida M, Kang S, Reichhardt C, Harlen K, Douglas T. Biochim Biophys Acta. 2010;1800:834–845. doi: 10.1016/j.bbagen.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maham A, Tang Z, Wu H, Wang J, Lin Y. Small. 2009;5:1706–1721. doi: 10.1002/smll.200801602. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z, Takezawa Y, Aoyagi H, Abe S, Hikage T, Watanabe Y, Kitagawa S, Ueno T. Chemical communications (Cambridge, England) 2011;47:170–172. doi: 10.1039/c0cc02221g. [DOI] [PubMed] [Google Scholar]
- 46.Takezawa Y, Bockmann P, Sugi N, Wang Z, Abe S, Murakami T, Hikage T, Erker G, Watanabe Y, Kitagawa S, Ueno T. Dalton Trans. 2011;40:2190–2195. doi: 10.1039/c0dt00955e. [DOI] [PubMed] [Google Scholar]
- 47.Iwahori K, Yamashita I. Nanotechnology. 2008;19:495601. doi: 10.1088/0957-4484/19/49/495601. [DOI] [PubMed] [Google Scholar]
- 48.Sana B, Poh CL, Lim S. Chemical communications (Cambridge, England) 2012;48:862–864. doi: 10.1039/c1cc15189d. [DOI] [PubMed] [Google Scholar]
- 49.Li K, Zhang ZP, Luo M, Yu X, Han Y, Wei HP, Cui ZQ, Zhang XE. Nanoscale. 2012;4:188–193. doi: 10.1039/c1nr11132a. [DOI] [PubMed] [Google Scholar]
- 50.Wood JC. Hematology / the Education Program of the American Society of Hematology American Society of Hematology. 2011;2011:443–450. doi: 10.1182/asheducation-2011.1.443. [DOI] [PubMed] [Google Scholar]
- 51.Terashima M, Uchida M, Kosuge H, Tsao PS, Young MJ, Conolly SM, Douglas T, McConnell MV. Biomaterials. 2011;32:1430–1437. doi: 10.1016/j.biomaterials.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ziv K, Meir G, Harmelin A, Shimoni E, Klein E, Neeman M. NMR in biomedicine. 2010;23:523–531. doi: 10.1002/nbm.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng B, Yamashita I, Uenuma M, Iwahori K, Kobayashi M, Uraoka Y. Nanotechnology. 2010;21:045305. doi: 10.1088/0957-4484/21/4/045305. [DOI] [PubMed] [Google Scholar]
- 54.Xing R, Wang X, Zhang C, Zhang Y, Wang Q, Yang Z, Guo Z. J Inorg Biochem. 2009;103:1039–1044. doi: 10.1016/j.jinorgbio.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 55.Mann S. Nat Mater. 2009;8:781–792. doi: 10.1038/nmat2496. [DOI] [PubMed] [Google Scholar]
- 56.Abe S, Hirata K, Ueno T, Morino K, Shimizu N, Yamamoto M, Takata M, Yashima E, Watanabe Y. Journal of the American Chemical Society. 2009;131:6958–6960. doi: 10.1021/ja901234j. [DOI] [PubMed] [Google Scholar]
- 57.Ramsay B, Wiedenheft B, Allen M, Gauss GH, Lawrence CM, Young M, Douglas T. J Inorg Biochem. 2006;100:1061–1068. doi: 10.1016/j.jinorgbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Liu G, Wang J, Wu H, Lin Y. Anal Chem. 2006;78:7417–7423. doi: 10.1021/ac060653j. [DOI] [PubMed] [Google Scholar]
- 59.Li M, Viravaidya C, Mann S. Small. 2007;3:1477–1481. doi: 10.1002/smll.200700199. [DOI] [PubMed] [Google Scholar]
- 60.Meldrum FC, Heywood BR, Mann S. Science (New York, NY. 1992;257:522–523. doi: 10.1126/science.1636086. [DOI] [PubMed] [Google Scholar]
- 61.Wade VJ, Levi S, Arosio P, Treffry A, Harrison PM, Mann S. Journal of molecular biology. 1991;221:1443–1452. doi: 10.1016/0022-2836(91)90944-2. [DOI] [PubMed] [Google Scholar]
- 62.Wade VJ, Treffry A, Laulhere JP, Bauminger ER, Cleton MI, Mann S, Briat JF, Harrison PM. Biochim Biophys Acta. 1993;1161:91–96. doi: 10.1016/0167-4838(93)90201-2. [DOI] [PubMed] [Google Scholar]
- 63.Wong KK, Colfen H, Whilton NT, Douglas T, Mann S. J Inorg Biochem. 1999;76:187–195. doi: 10.1016/s0162-0134(99)00114-2. [DOI] [PubMed] [Google Scholar]
- 64.Lambert EM, Viravaidya C, Li M, Mann S. Angew Chem Int Ed Engl. 2010;49:4100–4103. doi: 10.1002/anie.201001043. [DOI] [PubMed] [Google Scholar]
- 65.Uchida M, Terashima M, Cunningham CH, Suzuki Y, Willits DA, Willis AF, Yang PC, Tsao PS, McConnell MV, Young MJ, Douglas T. Magn Reson Med. 2008;60:1073–1081. doi: 10.1002/mrm.21761. [DOI] [PubMed] [Google Scholar]
- 66.May CA, Grady JK, Laue TM, Poli M, Arosio P, Chasteen ND. Biochim Biophys Acta. 2010;1800:858–870. doi: 10.1016/j.bbagen.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulte JW, Douglas T, Mann S, Frankel RB, Moskowitz BM, Brooks RA, Baumgarner CD, Vymazal J, Strub MP, Frank JA. J Magn Reson Imaging. 1994;4:497–505. doi: 10.1002/jmri.1880040343. [DOI] [PubMed] [Google Scholar]
- 68.Hosein HA, Strongin DR, Allen M, Douglas T. Langmuir. 2004;20:10283–10287. doi: 10.1021/la0491100. [DOI] [PubMed] [Google Scholar]
- 69.Butts CA, Swift J, Kang SG, Di Costanzo L, Christianson DW, Saven JG, Dmochowski IJ. Biochemistry. 2008;47:12729–12739. doi: 10.1021/bi8016735. [DOI] [PubMed] [Google Scholar]
- 70.Suci PA, Kang S, Young M, Douglas T. Journal of the American Chemical Society. 2009;131:9164–9165. doi: 10.1021/ja9035187. [DOI] [PubMed] [Google Scholar]
- 71.Kitagawa T, Kosuge H, Uchida M, Dua MM, Iida Y, Dalman RL, Douglas T, McConnell MV. Mol Imaging Biol. 2011 doi: 10.1007/s11307-011-0495-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flenniken ML, Uchida M, Liepold LO, Kang S, Young MJ, Douglas T. Curr Top Microbiol Immunol. 2009;327:71–93. doi: 10.1007/978-3-540-69379-6_4. [DOI] [PubMed] [Google Scholar]
- 73.Yang Z, Wang X, Diao H, Zhang J, Li H, Sun H, Guo Z. Chemical communications (Cambridge, England) 2007:3453–3455. doi: 10.1039/b705326f. [DOI] [PubMed] [Google Scholar]
- 74.Zhang L, Fischer W, Pippel E, Hause G, Brandsch M, Knez M. Small. 2011;7:1538–1541. doi: 10.1002/smll.201100238. [DOI] [PubMed] [Google Scholar]
- 75.Li JY, Paragas N, Ned RM, Qiu A, Viltard M, Leete T, Drexler IR, Chen X, Sanna-Cherchi S, Mohammed F, Williams D, Lin CS, Schmidt-Ott KM, Andrews NC, Barasch J. Dev Cell. 2009;16:35–46. doi: 10.1016/j.devcel.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Todorich B, Zhang X, Slagle-Webb B, Seaman WE, Connor JR. Journal of neurochemistry. 2008;107:1495–1505. doi: 10.1111/j.1471-4159.2008.05678.x. [DOI] [PubMed] [Google Scholar]
- 77.San Martin CD, Garri C, Pizarro F, Walter T, Theil EC, Nunez MT. J Nutr. 2008;138:659–666. doi: 10.1093/jn/138.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ono K, Fuma K, Tabata K, Sawada M. Biochemical and biophysical research communications. 2009;388:589–594. doi: 10.1016/j.bbrc.2009.08.055. [DOI] [PubMed] [Google Scholar]
- 79.Galvis HMT, Bitter JH, Khare CB, Ruitenbeek M, Dugulan AI, de Jong KP. Science (New York, NY. 2012;335:834–837. doi: 10.1126/science.1215614. [DOI] [PubMed] [Google Scholar]