Abstract
Objectives
The bulk of spinal imaging is still performed with conventional two-dimensional sequences. This study assesses the suitability of three-dimensional sampling perfection with application-optimised contrasts using a different flip angle evolutions (SPACE) sequence for routine spinal imaging.
Methods
62 MRI examinations of the spine were evaluated by 2 examiners in consensus for the depiction of anatomy and presence of artefact. We noted pathologies that might be missed using the SPACE sequence only or the SPACE and a sagittal T1 weighted sequence. The reference standards were sagittal and axial T1 weighted and T2 weighted sequences. At a later date the evaluation was repeated by one of the original examiners and an additional examiner.
Results
There was good agreement of the single evaluations and consensus evaluation for the conventional sequences: κ>0.8, confidence interval (CI)>0.6–1.0. For the SPACE sequence, depiction of anatomy was very good for 84% of cases, with high interobserver agreement, but there was poor interobserver agreement for other cases. For artefact assessment of SPACE, κ=0.92, CI=0.92–1.0. The SPACE sequence was superior to conventional sequences for depiction of anatomy and artefact resistance. The SPACE sequence occasionally missed bone marrow oedema. In conjunction with sagittal T1 weighted sequences, no abnormality was missed. The isotropic SPACE sequence was superior to conventional sequences in imaging difficult anatomy such as in scoliosis and spondylolysis.
Conclusion
The SPACE sequence allows excellent assessment of anatomy owing to high spatial resolution and resistance to artefact. The sensitivity for bone marrow abnormalities is limited.
Conventional MRI protocols of the spine often consist of T1 weighted and T2 weighted sagittal and axial sequences. The axial images are acquired as blocks or parallel to an intervertebral disc space.
A number of imaging centres use three-dimensional (3D) T2 weighted sequences for imaging of the spine; however, these sequences are used for assessment in the axial plane only [1,2]. With older sequence types there have been compromises in the signal-to-noise ratio, spatial resolution and contrast resolution, although the advantage of the 3D reconstruction ability has been recognised [3,4]. Various sequences have been assessed for suitability, with particular emphasis on the depiction of the intervertebral foramina [1,4,5]. There has also been interest in imaging the whole spine with T2 weighting-based 3D sequences [6].
The 3D sampling perfection with application-optimised contrasts using different flip angle evolutions (SPACE) sequence is a turbo spin-echo T2 weighted 3D sequence using variable flip angles for refocusing instead of the conventional 180° refocusing pulse.
The aim of this study was to assess the suitability of the 3D SPACE sequence for imaging of the spine. The isotropic 3D SPACE sequence was evaluated in its delineation of anatomy and pathology, particularly in anatomically difficult locations. The capacity to replace all other T2 weighted sequences in the routine application was assessed. The aim of the assessment was to prove or disprove equivalency of the SPACE sequence with the conventional sequences for the depiction of anatomy and the susceptibility to artefact in clinical routine.
Methods and materials
Between 29 May 2007 and 25 October 2007, patients attending our institution, The RJAH Orthopaedic Hospital, Oswestry, UK, for MRI of the spine were imaged at random (whenever time allowed it) with our routine protocol and an additional 3D SPACE sequence. This was done as part of our clinical governance protocol when introducing a new imaging protocol/technique into routine practice.
The examinations were performed on a 1.5 T Siemens Avanto (Siemens, Erlangen, Germany). Only integrated spine coils were used. Only patients with a clinical history of back pain, possible nerve root or cord compromise and scoliosis without known cause, and patients being followed up after spinal injury were included. Patients with acute spinal injury or suspected malignancy were not included. These patients are usually imaged with tailored protocols, depending on the exact clinical circumstances.
The routine protocol for the imaged patients comprised T1 weighted and T2 weighted sagittal and block axial T1 weighted and T2 weighted sequences. The 3D SPACE sequence was added. Evaluated were the depiction of anatomy, abnormal anatomy and other pathological features, and in particular which (if any) pathologies are missed using this sequence when compared with conventional sequences. In some cases with unexpected pathology a short tau inversion recovery (STIR) sequence was added by the radiographers; in these cases the STIR sequence was not formally evaluated for depiction of anatomy and image quality. However, note was made of relevant pathology.
The imaging parameters for the cervical spine are presented in Table 1. The block axial images covered routinely the area from the C2/3 to the C7/T1 level. The thoracic spine was imaged with the cervical spine protocol. Block axials were centred on an area of interest as identified on the sagittal sequences. The imaging parameters for the lumbar spine are presented in Table 2. The block axial images covered routinely the area from the L3/4 to the L5/S1 level. If necessary, for example in scoliosis, the slab size of the 3D SPACE sequence was increased. This resulted in a linear increase in imaging time.
Table 1. Sequence parameters for imaging of the cervical and thoracic spine.
| Parameter | Sagittal T1 | Sagittal T2 | Axial T1 | Axial GRE T2 | 3D SPACE |
| TR (ms) | 595 | 4930 | 500 | 1180 | 1500 |
| TE (ms) | 13 | 113 | 12 | 27 | 158 |
| TA (min:s) | 3:39 | 2:57 | 2×1:09 | 2:45 | 5:26 |
| Slice thickness (mm) | 4 | 4 | 5 | 5 | 1.2, isotropic |
| Slice spacing (mm) | 4.8 | 4.8 | 5.5 | 5.5 | – |
| Number of slices | 11 | 11 | 19 | 19 | – |
| Matrix size | 448×224 | 448×224 | 320×210 | 320×180 | 320×317 |
| Field of view (mm) | 380×380 | 380×380 | 230×215 | 200×150 | 380×380×60 |
3D, three-dimensional; GRE, gradient echo; SPACE, sampling perfection with application-optimised contrasts using different flip angle evolutions; TA, acquisition time; TE, echo time; TR, repetition time.
Table 2. Sequence parameters for imaging of the lumbar spine.
| Parameter | Sagittal T1 | Sagittal T2 | Axial T1 | Axial T2 | 3D SPACE |
| TR (ms) | 596 | 3000 | 500 | 5690 | 1500 |
| TE (ms) | 13 | 117 | 12 | 108 | 157 |
| TA (min) | 3:39 | 2:51 | 2×2:05 | 4:37 | 5:26 |
| Slice thickness (mm) | 4 | 4 | 5 | 5 | 1, isotropic |
| Slice spacing (mm) | 4.8 | 4.8 | 5.5 | 5.5 | – |
| Number of slices | 11 | 11 | 19 | 19 | – |
| Matrix size | 512×256 | 512×256 | 320×210 | 384×302 | 320×317 |
| Field of view (mm) | 280×280 | 280×280 | 230×215 | 230×226 | 320×320×60 |
3D, three-dimensional; SPACE, sampling perfection with application-optimised contrasts using different flip angle evolutions; TA, acquisition time; TE, echo time; TR, repetition time.
The images were reviewed by two musculoskeletal radiologists in consensus. A scoring system for the assessment of anatomical detail and artefact was used. The 3D SPACE sequence was reviewed first and all relevant observations noted. Immediately after this the conventional sequences were reviewed. Note was made of any further relevant pathological findings not visualised on the 3D SPACE sequence. The images were reviewed on a Leonardo workstation (Siemens) with 3D functionality.
At a later date the images were reassessed by one of the original reviewers and a further external reviewer. The original consensus report was used as a reference standard. The relevant pathological findings were used as a reference standard. The scoring system used is similar to one described previously by Noebauer-Huhmann et al [7].
The delineation of relevant anatomical structures such as spinal cord, nerve roots and osseous structures of the spine (vertebral bodies, neuroforamina, facet joints) was scored using a five-point grading system for delineation: 1, non-existent; 2, poor; 3, acceptable; 4, good; 5, excellent.
The influence of artefact and image noise on image interpretation was assessed using a five-point grading system: 5, absent; 4, minimal; 3, mild; 2, moderate; 1, severe/limiting diagnostic value. The type of artefact (e.g. motion, susceptibility, pulsation, ringing etc.) was not specifically recorded but only the influence on image quality.
The scores of the different sequences were compared using the paired Wilcoxon signed-rank test to compare qualitative scores [8]. Weighted kappa statistics were performed between the individual assessments vs the consensus reading as standard. Note was made of the main diagnosis based on the SPACE sequence alone vs the reference standard. Bonferroni corrections were made to the level of significance, for the multiplicity of comparisons, in the case of both kappa and Wilcoxon tests. Statistical significance was set at p=0.05.
Results
In the time between 29 May 2007 and 25 October 2007, 62 spinal examinations could be included in the study. A post hoc analysis showed a power of 70% for the Wilcoxon test in distinguishing between the SPACE sequence and the other sequences, for both anatomy and artefact, based on an effect size of 0.44.
The cervical spine was imaged 15 times, the thoracic spine 12 times and the lumbar spine 35 times. The average score for the delineation of anatomy and the average artefact score are shown in Table 3. The distribution for the anatomy and artefact scores is graphically represented in Figure 1.
Table 3. Average score for depiction of anatomya and severity of artefactb (higher scores indicate better image quality).
| Parameter | T1 sagittal | T2 sagittal | T1 axial | T2 axial | SPACE |
| Anatomy | 4.26 | 4.18 | 4.18 | 4.21 | 4.77 |
| Artefact | 4.21 | 3.69 | 3.87 | 3.87 | 4.69 |
SPACE, sampling perfection with application-optimised contrasts using different flip angle evolutions.
aGrading system: 1, non-existent; 2, poor; 3, acceptable; 4, good; 5, excellent.
bGrading system: 1, severe/limiting diagnostic value; 2, moderate; 3, mild; 4, minimal; 5, absent.
Figure 1.
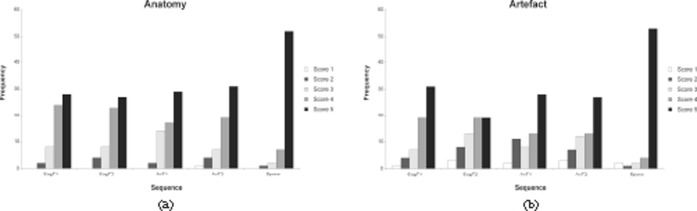
Graphical representation of the depiction of anatomical structures (a) and the influence of artefact and image noise on image interpretation (b). For quality of anatomical representation, as well as for artefact, the 3D sampling perfection with application-optimised contrasts using different flip angle evolutions sequence performs significantly better than any of the other sequences.
The scores for each examination were compared using the paired Wilcoxon signed-rank test. Comparison was made between the sagittal T1 weighted and T2 weighted, and axial T1 weighted and T2 weighted sequences vs the SPACE sequence. The comparison was made for the delineation of anatomy as well as artefact. The values of the z-score and the p-values are shown in Table 4.
Table 4. Comparison of SPACE vs other imaging sequences for anatomy and artefact (z-test statistic and p-value).
| Parameter | Sequence | Sagittal T1 | Sagittal T2 | Axial T1 | Axial T2 |
| Anatomy | Test statistic | 2.68 | 2.75 | 2.87 | 3.13 |
| p-valuea | <0.01b | <0.01b | <0.01b | <0.001b | |
| Artefact | Test statistic | 2.37 | 3.59 | 3.71 | 3.42 |
| p-valuea | <0.01b | <0.001b | <0.001b | <0.001b |
SPACE, sampling perfection with application-optimised contrasts using different flip angle evolutions.
ap<0.01 was set as statistically significant (allowing for multiple comparisons).
bDifference from SPACE sequence.
In cases of significant artefact in a sequence, the depiction of the relevant anatomy was also impaired and the score was lower for both items, as would be expected.
The examination of the anatomical regions separately still resulted in similar outcomes, although the statistical power was less.
The kappa scores for an independent review compared with the consensus reading as standard are shown in Table 5. Both independent reviewers identified all relevant pathology based on the 3D SPACE sequence only compared with the consensus reading as standard. There was good interobserver agreement between the individual assessments and the consensus reading. In the case of the agreement for the SPACE anatomy scores, the results appear paradoxically low. This appears to be the result of not-so-good agreement in a small number of cases (16%) that scored low (<5). In the 84% of cases where the score was maximal, the agreement was very good.
Table 5. Comparison of independent review vs the consensus reading as standard using kappa statistics (with Bonferroni adjustment for multiple tests) and confidence limitsa.
| Table of kappa scores (with confidence limits) |
||||||||||
| Standard vs | Sag T1 anatomy | Sag T1 artefact | Sag T2 anatomy | Sag T2 artefact | Ax T1 anatomy | Ax T1 artefact | Ax T2 anatomy | Ax T2 artefact | SPACE anatomy | SPACE artefact |
| Reviewer 2 | ||||||||||
| Kappa | 0.835 | 0.845 | 0.883 | 0.878 | 0.829 | 0.907 | 0.849 | 0.921 | 0.152 | 0.920 |
| Low CL | 0.620 | 0.671 | 0.719 | 0.748 | 0.671 | 0.783 | 0.687 | 0.835 | −0.324 | 0.782 |
| Upper CL | 1.000 | 1.000 | 1.000 | 1.000 | 0.987 | 1.000 | 1.000 | 1.000 | 0.628 | 1.000 |
| Reviewer 1 | ||||||||||
| Kappa | 0.923 | 0.898 | 0.886 | 0.862 | 0.933 | 0.920 | 0.970 | 0.959 | 0.051 | 0.962 |
| Low CL | 0.780 | 0.752 | 0.709 | 0.718 | 0.837 | 0.831 | 0.899 | 0.896 | −0.424 | 0.889 |
| Upper CL | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 0.526 | 1.000 |
Ax, axial; Sag, sagittal; CL, confidence limit; SPACE, sampling perfection with application-optimised contrasts using different flip angle evolutions.
ap-values for all comparisons except SPACE anatomy are (non-significant) p<0.001.
The following observations were made:
The 3D SPACE sequence is surprisingly resistant to artefact (Figures 2 and 3).
The main indication for the examinations was back pain with or without neural compromise (53/62); in one of these cases, in addition, a suspicion of spondylolysis was indicated. Some patients were sent for the assessment of spinal alignment (9/62), in particular scoliosis (8/9) and (once) kyphosis.
The SPACE sequence was (subjectively) superior in the depiction of spondylolysis, owing to the high spatial resolution and the ability to obtain high-resolution reformats in any imaging plane (Figure 4). There were three cases of spondylolysis; one was clinically suspected; all three cases could be identified on standard sequences, although subjectively the identification was easier on the SPACE sequence. It has since been the authors' experience that with the 3D SPACE sequence spondylolysis can always be demonstrated directly even when it was not confidently identified on standard sequences.
-
The SPACE sequence was superior to the conventional sequences in combination, demonstrating disc herniations that were not visualised with the conventional sequences. These were lesions outside the block axial images and located laterally in the spinal canal. These were seen especially in the thoracic spine; one of these lesions did cause spinal cord deformity, indicating chronic abutment. This lesion was not visible on the conventional sagittal sequences even on secondary review. In cases where the disc herniation was visible on the conventional sagittal sequence the SPACE sequence allowed assessment of these lesions without the need for patient recall or additional axial images; in this the SPACE sequence removes ambiguity from the radiologist's report (Figure 2). The various stages and grades of disc degeneration were equally well seen on the SPACE as well as the conventional T2 weighted sequence.
The signal-to-noise ratio of the 3D SPACE sequence was reduced near the edge of the field of view. This resulted in impaired image quality of the lower thoracic spine when imaging the cervical spine. However, this area was usually not included at all on the conventional sequences.
In the cervical spine in particular the cervical nerve roots were excellently demonstrated with the 3D SPACE sequence, allowing for confident and accurate evaluation (Figure 2).
In three cases, bone marrow oedema was not well demonstrated on 3D SPACE compared with T1 weighted (Figure 5). In two cases of bone marrow lesions due to metastatic disease not all lesions were seen on the 3D SPACE sequence; however, in both cases metastatic disease was seen on the SPACE sequence. In one case a bone marrow lesion was seen only on the 3D SPACE sequence.
In a case of a spinal cord lesion, this was demonstrated on the sagittal T2 weighted and the SPACE sequence, but not on the conventional axial T2 weighted sequence nor on the T1 weighted sequences.
The SPACE sequence was superior for the assessment of patients with scoliosis or complex anatomy for other reasons. The coronal slices enabled easy assessment of the severity of the scoliosis. The ability to angle the axial slices parallel to the endplate was of help in more difficult cases. In most cases this was not necessary because the high spatial resolution allowed sufficiently confident assessment of the anatomical structures in the 3D mode on the workstation (Figure 6). 3D reformatting in the coronal plane allows for easy identification of transitional vertebrae and complex neural anomalies (Figure 7).
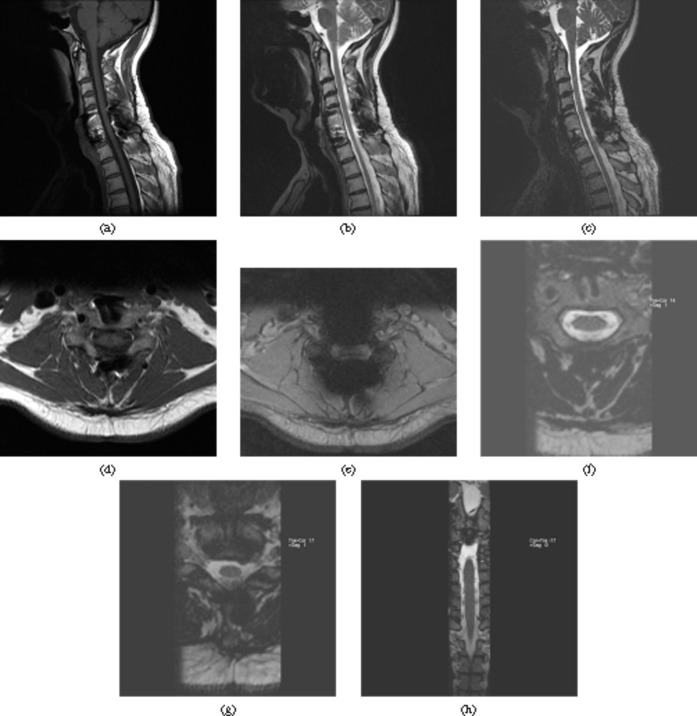
Figure 2.
Previous instrumented fusion of the cervical spine C5–C7. (a) Sagittal T1 weighted, (b) T2 weighted turbo spin-echo and (c) sampling perfection with application-optimised contrasts using different flip angle evolutions (SPACE) images. (d) Axial T1 weighted, (e) Gradient echo T2 weighted and (f, g) SPACE images. (h) Coronal SPACE. The sagittal images demonstrate adequate artefact resistance of the SPACE sequence. On the axial images the SPACE sequence demonstrates the least artefact of all three sequences. The gradient echo T2 sequence is non-diagnostic; a turbo spin-echo sequence would have been more adequate; however, these usually show marked flow artefact in this area. (f) On the axial SPACE reformat the fixation screws of the anterior plate are well depicted. (g) A small disc herniation is well seen on the axial SPACE. (h) Exquisite anatomical detail with depiction of the nerve roots in the spinal canal is seen on the coronal reformat.
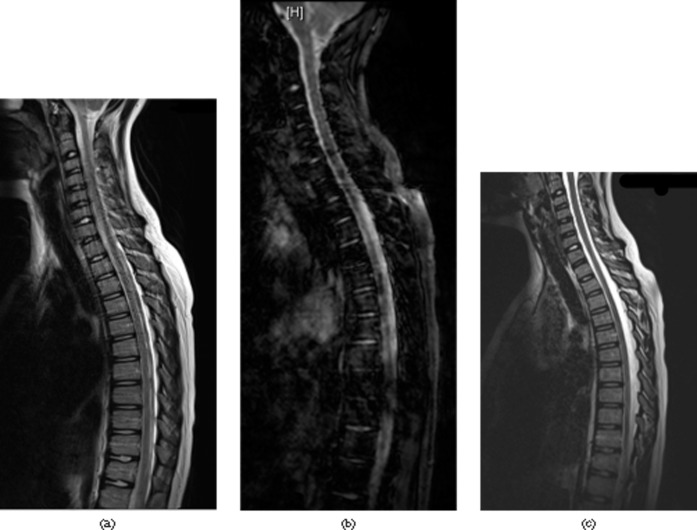
Figure 3.
MR images of cervical and thoracic spine. (a) MRI of cervical and thoracic spine with significant flow and movement artefact on a conventional turbo spin-echo T2 weighted sequence. (b) A gradient echo sequence yielded significantly worse images. (c) The 3D sampling perfection with application-optimised contrasts using different flip angle evolutions sequence results in sharp and artefact free images and is surprisingly resistant to most artefacts.
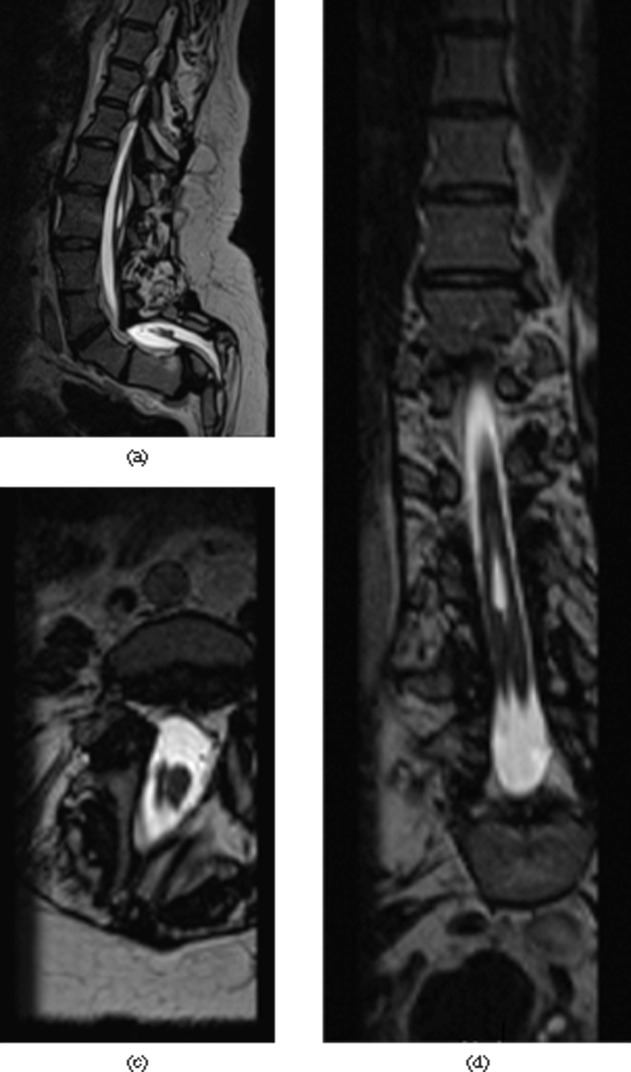
Figure 4.
Pars defect L4. (a) The sagittal T1 weighted and (b) T2 weighted images do not demonstrate the defect well. (c) The defect is best seen on sampling perfection with application-optimised contrasts using different flip angle evolutions (SPACE), sagittal reformat. The SPACE sequence allows for isotropic reconstruction in any plane; the thin slice thickness with still-adequate signal-to-noise ratio allows for better depiction of normal and abnormal anatomy.
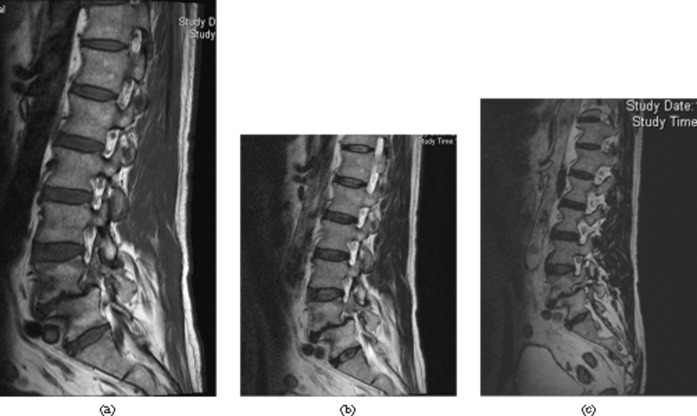
Figure 5.
Collapse of a mid-thoracic vertebral body, T7. Bone marrow oedema is best visualised on the (a) short tau inversion recovery and (b) T1 weighted sequence. (c) The conventional spin echo T2 weighted and (d) the sampling perfection with application-optimised contrasts using different flip angle evolutions demonstrate the fracture well but are not sensitive for bone marrow oedema.
Figure 6.
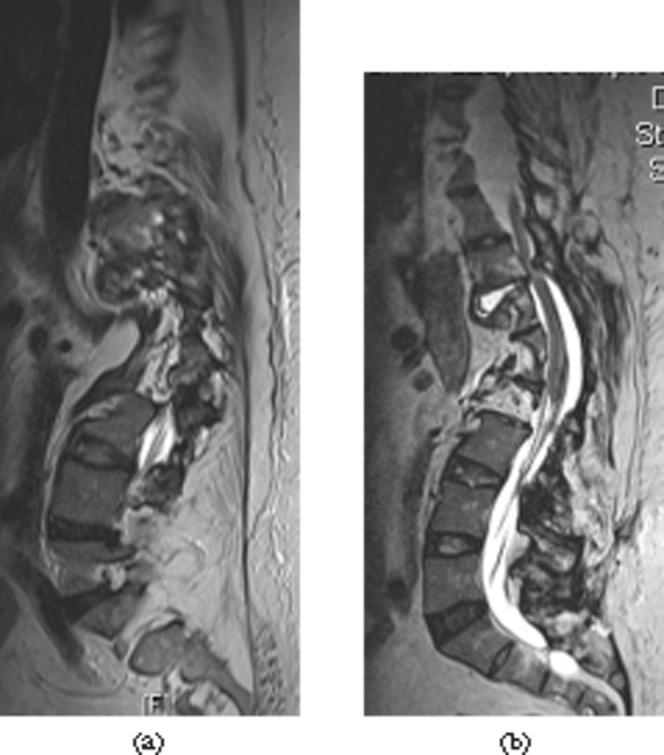
Severe scoliosis of the spine with clinically neurological compromise. (a) Conventional images are difficult to interpret; shown here is the sagittal T2 weighted sequence. (b) The curved sagittal reformat of the sampling perfection with application-optimised contrasts using different flip angle evolutions (SPACE) sequence demonstrates cord compression due to degenerative disease. The ability to reformat in any plane makes the SPACE sequence ideal for any cases with challenging anatomy.
Figure 7.
Complex anatomical developmental anomaly with diastomatomyelia. Reformatting in appropriate planes makes it easy to appreciate this: (a) sagittal, (b) coronal and (c) axial.
Discussion
3D T2 weighted sequences are frequently used for axial imaging of the spine in a number of centres. This is usually done in addition to sagittal conventional T1 weighted and T2 weighted sequences. The display is frequently as conventional reformatted axial images, and not with a 3D viewer or reconstruction in three planes.
The authors wanted to examine the suitability of the 3D SPACE sequence for routine imaging of the spine. This is a sequence that benefits from a large field of view and relatively low acquisition times (332 s in the form used for this study). The underlying theory is quite complex and more detail is described elsewhere [9]. Despite the variable flip angles of the refocusing pulses, the sequence can be seen as virtually identical to conventional turbo spin-echo sequences with 150° flip angles. As with all turbo spin echo sequences there is some element of T1 weighting in the images [7].
The use of a 3D SPACE STIR sequence for imaging of the brachial plexus was also described recently [10]. The excellent resolution and ability to view and reformat the images in any plane was stressed. Similarly, its usefulness for the visualisation of the craniocervical ligaments owing to its 3D multiplanar capabilities was recently stressed [11].
The 3D SPACE sequence offers reasonable soft tissue signal intensities and contrast resolution, and is not limited to myelography such as is representation of the spine seen with some other 3D gradient echo sequences [7,10,11].
The results of the study presented here demonstrate a surprising resilience of the 3D SPACE sequence to artefact, especially movement and cerebrospinal fluid flow/pulsation. There was significantly less artefact in this sequence than in all conventional sequences. It is sensitive to artefact due to metal implants, however, especially implants containing ferromagnetic metals.
The anatomy was also significantly better demonstrated compared with all conventional sequences. The poor kappa coefficient for interobserver comparison for depiction of anatomy was due to a small number of cases where depiction was not optimal and there was disagreement about the grade; in the majority of cases (84%) the anatomical depiction was excellent and there was complete agreement on this. The cervical nerve roots especially are excellently visualised and compromise within the spinal canal by disc disease can be directly observed. Thick-sliced conventional axial images may not directly image the roots. These findings are consistent with the observations of Viallon et al in the brachial plexus [10]. High-resolution axial images obtained with other 3D sequences also allow good visualisation of the cervical nerve root, but the isotropic multiplanar reconstruction ability of the 3D SPACE sequence can be of particular use in severe degeneration and root compromise.
The high and isotropic resolution and ability to view any image plane was particularly useful in the imaging of complex anatomy such as in scoliosis and spondylolysis. In spondylolysis the defect can always be visualised directly; with conventional sequences this is not always the case. However, in this study all cases of spondylolysis were directly identifiable on standard sequences.
The entire imaged area can be reviewed in any plane. This proved useful in the assessment of disc herniations at levels not imaged with the conventional axial blocks. Since introducing this sequence into routine practice, the authors have not had to recall patients for additional axial sequences to assess spinal canal compromise at levels not imaged axially on the original examination. In the more frequent case of small disc herniations seen on the sagittal images, the ability to assess any possible neural compromise confidently on the axial reformats removed ambiguity from the radiologist's report. In addition the SPACE sequence accurately depicts the same MRI features of disc degeneration as conventional MRI sequences.
The main weakness of the 3D SPACE sequence is the poor sensitivity for oedema. Noebauer-Huhmann et al described the oedema sensitivity as similar to conventional two-dimensional (2D) T2 weighted turbo spin-echo sequences and generally good [7]. The study presented here agrees with Noebauer-Huhmann et al that the sensitivity for oedema is similar to the conventional 2D T2 weighted sequences used in this study; however, the sensitivity was not good. In the sequences used for the study presented here the oedema sensitivity is best on conventional T1 weighted sequences, somewhat poorer in the 2D T2 weighted turbo spin-echo sequences used and marginally poorer in the 3D SPACE sequence. However, in conjunction with the sagittal T1 weighted sequence, no bone marrow lesion was missed in the studies examined.
For diagnostic purposes the sagittal T1 weighted sequence plus the 3D SPACE sequence were sufficient to diagnose all relevant pathology. The T2 weighted sequences as well as the axial T1 weighted sequences did not reveal additional relevant information in this study.
There was one spinal cord lesion that was seen only in 3D SPACE and on sagittal T2 weighted images. This was not seen on sagittal T1 weighting. This lesion had occurred in a patient with an old spinal injury.
Careful windowing is very important when using the 3D SPACE, and monitor reporting has to be strongly favoured. In the authors' experience the window setting should be altered to optimally assess different anatomical structures of the spine. This is also true for conventional sequences, however.
The time saved using the sagittal T1 weighted and 3D SPACE sequence compared with the conventional protocol only (545 s vs 752 s) would allow one to add a STIR (or similar) sequence without increasing the total imaging time. It is now the practice of the authors to perform spinal imaging with a SPACE, T1 weighted and STIR sequence for all clinical scenarios. The authors use a conventional, highly water-sensitive STIR sequence (rather than a so-called T1-STIR) to guarantee high sensitivity for oedema, resulting in an imaging time of 757 s. The authors have since seen spinal cord lesions not identified on the T1 weighted or SPACE sequence and for this reason recommend the use of a STIR or similar sequence.
In conclusion, the 3D SPACE sequence in conjunction with a sagittal T1 weighted sequence is sufficient to image the spine in routine applications, combining time saving with increased diagnostic confidence. Adding a STIR sequence enhances confidence in the diagnosis of bone marrow oedema and spinal cord signal changes. The 3D SPACE sequence combines high spatial and reasonable contrast resolution with a high resilience to artefact, and allows high-resolution isotropic reformats with a large field of view. It performs very well in otherwise difficult circumstances such as imaging of complex anatomy (i.e. scoliosis, spondylolysis) and imaging of the thoracic spine (flow artefact resistance, complete axial coverage), and always allows direct visualisation of the nerve roots.
References
- 1.Mahmutyazicioglu K, Ozdemir H, Savranlar A, Ozer T, Erdem O, Erdem Z, et al. Comparison of three-dimensional gradient echo, turbo spin echo and steady-state gradient echo sequences in axial MRI examination of the cervical spine. [In Turkish.] Tani Girisim Radyol 2003;9:432–8 [PubMed] [Google Scholar]
- 2.Baskaran V, Pereles FS, Russell EJ, Georganos SA, Shaibani A, Spero KA, et al. Myelographic MR imaging of the cervical spine with a 3D true fast imaging with steady-state precession technique: initial experience. Radiology 2003;227:585–92 [DOI] [PubMed] [Google Scholar]
- 3.Maldjian C, Adam RJ, Akhtar N, Bonakdarpour A, Boyko OB. Volume fast spin-echo imaging of the cervical spine. Acad Radiol 1999;6:84–8 [DOI] [PubMed] [Google Scholar]
- 4.Maldjian C, Adam RJ, Akhtar N, Maldjian JA, Bonakdarpour A, Boyko O. Volume (three-dimensional) fast spin-echo imaging of the lumbar spine. Acad Radiol 1999;6:339–42 [DOI] [PubMed] [Google Scholar]
- 5.Muhle C, Ahn JM, Biederer J, Schafer FK, Frahm CH, Mohr A, et al. MR imaging of the neural foramina of the cervical spine. Comparison of 3D-DESS and 3D-FISP sequences. Acta Radiol 2002;43:96–100 [DOI] [PubMed] [Google Scholar]
- 6.Rodegerdts EA, Boss A, Riemarzik K, Lichy M, Schick F, Claussen CD, et al. 3D imaging of the whole spine at 3T compared to 1.5T: initial experiences. Acta Radiol 2006;47:488–93 [DOI] [PubMed] [Google Scholar]
- 7.Noebauer-Huhmann IM, Glaser C, Dietrich O, Wallner CP, Klinger W, Imhof H, et al. MR imaging of the cervical spine: assessment of image quality with parallel imaging compared to non-accelerated MR measurements. Eur Radiol 2007;17:1147–55 [DOI] [PubMed] [Google Scholar]
- 8.Tello R, Ptak T. Statistical methods for comparative qualitative analysis. Radiology 1999;211:605–7 [DOI] [PubMed] [Google Scholar]
- 9.Lichy MP, Wietek BM, Mugler JP, 3rd, Horger W, Menzel MI, Anastasiadis A, et al. Magnetic resonance imaging of the body trunk using a single-slab, 3-dimensional, T2-weighted turbo-spin-echo sequence with high sampling efficiency (SPACE) for high spatial resolution imaging: initial clinical experiences. Invest Radiol 2005;40:754–60 [DOI] [PubMed] [Google Scholar]
- 10.Viallon M, Vargas MI, Jlassi H, Lovblad KO, Delavelle J. High-resolution and functional magnetic resonance imaging of the brachial plexus using an isotropic 3D T2 STIR (short term inversion recovery) SPACE sequence and diffusion tensor imaging. Eur Radiol 2008;18:1018–23 [DOI] [PubMed] [Google Scholar]
- 11.Baumert B, Wortler K, Steffinger D, Schmidt GP, Reiser MF, Baur-Melnyk A. Assessment of the internal craniocervical ligaments with a new magnetic resonance imaging sequence: three-dimensional turbo spin echo with variable flip-angle distribution (SPACE). Magn Reson Imaging 2009;27:954–60 [DOI] [PubMed] [Google Scholar]