Abstract
Time-lapse or longitudinal fluorescence microscopy is broadly used in cell biology. However, current available time-lapse fluorescence microscopy systems are bulky and costly. The limited field-of-view (FOV) associated with microscope objective necessitates mechanical scanning if a larger FOV is required. Here we demonstrate a wide FOV time-lapse fluorescence self-imaging Petri dish system, termed the Talbot Fluorescence ePetri, which addresses these issues. This system’s imaging is accomplished through the use of the Fluorescence Talbot Microscopy (FTM). By incorporating a microfluidic perfusion subsystem onto the platform, we can image cell cultures directly from within an incubator. Our prototype has a resolution limit of 1.2 µm and an FOV of 13 mm2. As demonstration, we obtained time-lapse images of HeLa cells expressing H2B-eGFP. We also employed the system to analyze the cells’ dynamic response to an anticancer drug, camptothecin (CPT). This method can provide a compact and simple solution for automated fluorescence imaging of cell cultures in incubators.
Keywords: Time-lapse, longitudinal, wide field-of-view, fluorescence microscopy, microfluidics
Introduction
Time-lapse or longitudinal fluorescence microscopy can reveal dynamics of biological processes, and is of crucial importance to many studies in cell biology, such as clarifying gene function by RNA interference,1 visualizing cell apoptosis,2 and aiding drug discovery.3 Time-lapse imaging generally requires a wide field-of-view (FOV) in order to view and track a large number of migrating and dividing cells over extended periods of time. However, traditional fluorescence microscopes use an objective lens and have a limited FOV. Microscopes equipped with motorized stages are available in the market, which are capable of imaging multiple areas and stitching them together to create a larger FOV.2,4 Unfortunately, these systems are bulky and costly.
Over the past few years, significant efforts have been made to build a wide FOV on-chip fluorescence imaging system with reduced size and cost. Most of these efforts 5,6,7 concentrated on using an image sensor chip (either CCD or CMOS-based) to directly collect fluorescence signals from the samples that are placed in proximity to the chip (the descriptor of ‘on-chip’ for characterizing this line of work refers to the proximity of sample to the sensor chip). This design choice eliminates the microscopy objective and achieves an effective FOV that is approximately equal to the sensor chip’s active sensor area. These methods span a range of resolution. The simplest method 6 inserts an absorption fluorescence filter between the sample and a CCD sensor chip. In the demonstration experiment, the distance between sample and sensor was ~100 µm. The sample was excited by uniform illumination and the emitted fluorescence would spread on the CCD. Based on point-spread function (PSF), such a system has an image resolution of ~100 µm. (The authors claimed a compressive sensing resolution of ~10 µm. However, this parameter is only applicable to sparsely distributed sample and the same method can be applied to conventional microscopy to claim improved resolution for sparse samples.) In another approach, Arpali et al. used a spatial light modulator (SLM) to generate an array of Gaussian-shaped spots on the sample surface.7 The sample was loaded onto the CCD surface through a channel made of adhesive tapes, and the spot array laterally scanned the sample to get a sequence of images. These images were then merged to generate the final image. Based on PSF, this demonstrated resolution should be ~200 µm – the width of the Gaussian spots. Neither of these approaches can provide enough resolution for subcellular features (typically on a scale of several microns), and thus are not adequate for longitudinal cell fluorescence imaging.
Recently, we developed a new on-chip fluorescence imaging technique, the Fluorescence Talbot Microscope (FTM).8 The method uses a uniformly laser-illuminated lenslet grid to generate a grid of tightly focused laser spots (pitch = d). By the Talbot self-imaging effect,9 this light grid propagates forward and regenerates itself into focused light spots at distances of z=2nd2/λ, where n is an integer, λ is the wavelength, and ZT =2d2/λ is defined as the Talbot length. This focused grid can then be used to perform high-resolution scanning (~1 µm) of the sample (see system setup and also our previous work 8 for detailed description).
This approach has several advantages for wide FOV time-lapse fluorescence imaging. First, its ~1 µm resolution is sufficient for imaging sub-cellular features such as cell membranes and nuclei. Second, the use of Talbot effect affords a large working distance (~3.7 mm in our case) between the optical illumination elements and the sample – this spacing is vital for allowing the incorporation of microfluidic components for perfusion culture purposes. This advantage also distinguishes the FTM method from a related non-Talbot approach which has a much shorter demonstrated working distance (~40 µm).10 Third, the FOV is directly scalable by using a larger lenslet grid to create more focal spots and by employing a larger sensor chip for signal collection. This advantage stands in contrast with conventional microscopy where scaling up the FOV requires broader mechanical scanning range and therefore longer image acquisition time. Fourth, our key components are a MEMS mirror, a lenslet grid, and a webcam CMOS sensor for detection. All of them are low cost, mass-producible and compact. In this study, we further demonstrate that an FTM system in combination with microfluidic cell culture and temperature control components can be applied to perform wide FOV time-lapse fluorescence imaging of cell cultures from within an incubator. To our knowledge, this is the first demonstration of automated wide FOV and high-resolution fluorescence imaging for longitudinal cell culture studies that does not rely on conventional microscopy methods.
The new imaging modalities that can simply and effectively monitor cell cultures from within an incubator is a recent scientific development. To date, we have developed a brightfield wide FOV on-chip imaging method that provides a compact wide FOV imaging capability for cell culture applications (termed ePetri).11 This current work extends this line of development by introducing fluorescence imaging capability. We believe that this approach can significantly streamline the work flow of Petri dish based assays by reducing labor. With its wide FOV and ~1 µm resolution, our demonstrated method can potentially offer a labor-saving, low-cost and mass-producible solution for drug screening, dynamic cell imaging, and medical diagnostics.
In the next section, we will describe the system setup (hereby termed Talbot Fluorescence ePetri) in detail and explain the imaging approach. Next we will demonstrate our longitudinal fluorescence imaging of HeLa cells expressing histone 2B-tagged eGFP (H2B-eGFP), where cells in the sequence of images are automatically counted and tracked. Finally, we will apply our system to investigate the response of cells to an anti-cancer drug, camptothecin.
Results
System setup
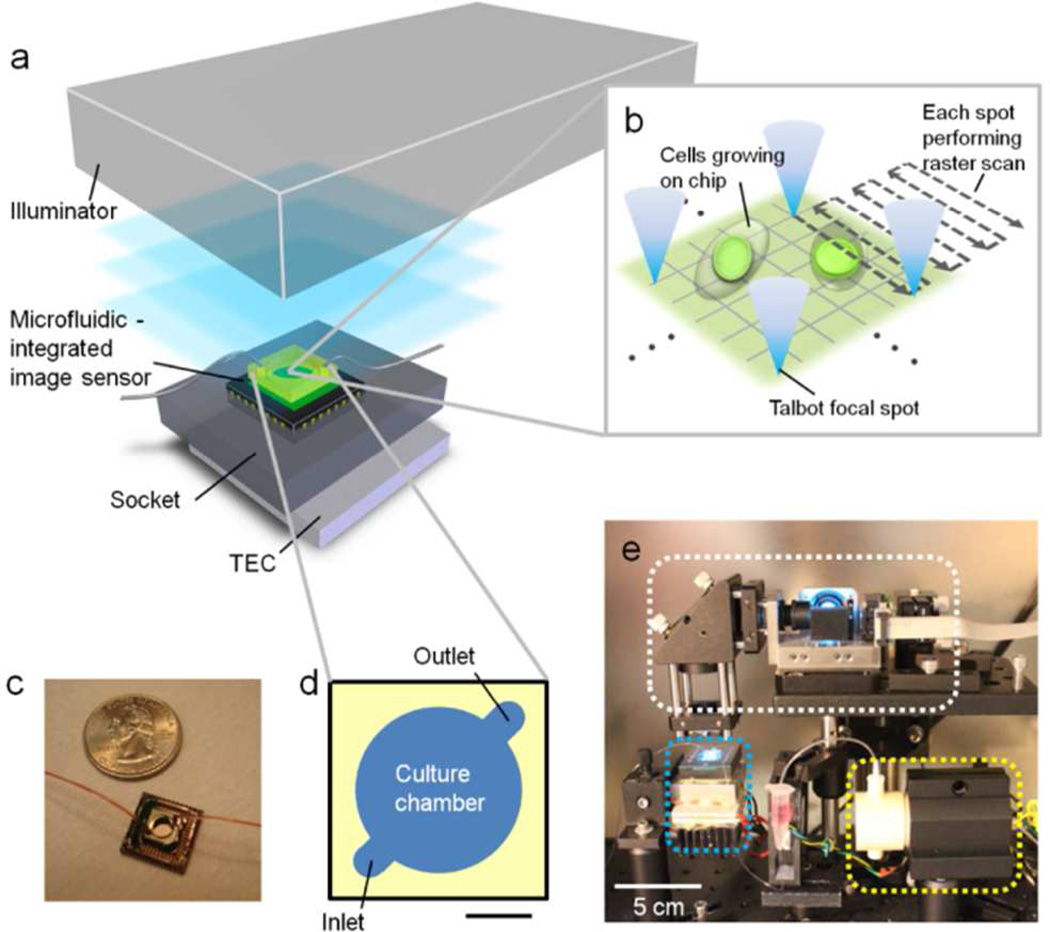
Our Talbot Fluorescence ePetri system is depicted in Figure 1a. The illuminator is described in detail in our previous work.8 Briefly, the illuminator used a 488 nm 30 mW fiber coupled laser (FiberTec II, Blue Sky Research) as its light input. The input light was collimated and was then reflected off a MEMS mirror (Mirrorcle). The reflected light transmitted through a lenslet grid (18-00407, SUSS) with pitch d = 30 µm, area = 225 mm2 and focal distance Zf = 90 µm, to create a grid of focused laser spots at a distance Zf after the lenslet grid. This grid of focused spots has the same pitch d as the lenslet grid. Due to the Talbot effect,9 this grid repeated itself at distance Zf+nZT, where n is an integer and ZT = 2d2/λ (λ is the wavelength). For our experiments, we chose to use the n = 1 refocused Talbot spot grid to serve as our scanning grid (samples will be placed in that plane for imaging). This afforded us with a large working distance of ~3.7 mm between the illuminator and the sample. By angularly tilting the input light field using the MEMS mirror, we can laterally shift the scanning grid. By this means, we can raster scan the scanning grid across the sample. An angular tilt of the input light field by 8 mrad would laterally shift the scanning grid by 30 µm. Therefore, by this approach, we could raster scan each Talbot focal spot over an area of 30 µm × 30 µm (Figure 1b). Since the laser spot pitch was equal to the lenslet grid pitch (30 µm), this scanning approach allowed us to fully cover the whole area of interest.
Figure 1.
The Talbot Fluorescence ePetri system. (a) System setup. The system consists of an illuminator, a microfluidic-integrated image sensor, a socket and a thermoelectric cooler (TEC). (b) The raster scan of the focal spots on the cells cultured on the sensor surface. (c) The picture of an assembled microfluidic-integrated image sensor. (d) The design of the microfluidic cell culture chamber. The scale bar is 2 mm. (e) The system (white rectangle: the illuminator; blue rectangle: the microfluidic-integrated image sensor, socket and TEC) and the micro-pump (yellow rectangle) placed inside an incubator.
The heart of the sensing platform consisted of a CMOS sensor chip (MT9M001C12STM, Aptina Imaging) (top glass window removed) with pre-coated absorption fluorescence filter (Aptina green1, Aptina, thickness = ~10 µm, optical density = 6) 5 and a microfluidic component (Figure 1c). The cell culture chamber in the microfluidic component had a diameter of 5 mm and a height of 400 µm, with two channels as the inlet and outlet (Figure 1d). The ceiling of the chamber was a glass coverslip. The microfluidic component was fabricated by following a series of standard micro-molding processes with modifications 12 (Figure S-1). This microfluidic-integrated sensor chip was mounted onto a camera board (SV9M001, EPIX) via a customized sensor socket for signal readout. A thermoelectric cooler (TEC, 89413, American Science & Surplus) was attached to the socket to protect cells from the heat generated by the sensor circuit. The other side of the TEC was cooled by a CPU fan.
To provide an adequate cell culture environment, the imaging platform was placed inside a humidified 5% CO2 incubator at 37 °C (Figure 1e). We turned on the TEC (power input = 13W) during imaging to prevent the sensor surface temperature from exceeding 37 °C (Figure S-2). The microfluidic component was connected with a self-priming micro-pump (EW-73120-64, Cole-Parmer), and fresh medium was loaded into the microfluidic culture chamber every 10 min. The maximum flow speed was set below 50 µm/s, which is within the suggested range for perfusion culture.13 The illumination, image data acquisition, and micro-pump were all automatically controlled by a customized LabVIEW program.
We have established that the filter-coated sensor chip’s sensitivity is ~105 fluorophores/µm2 for Alexa Fluor 488 under 488 nm laser illumination with 10 mW/cm2 intensity and 15 ms integration time (It can be derived that the sensitivity for eGFP is also on the order of 105 fluorophores/µm2). Each Talbot focal spot has a full width at half maximum (FWHM) of 1.28 µm.8 The resolution limit of the imaging system has been established to be 1.2 µm in our previous work by imaging a resolution mask patterned on a sensor’s surface.8 During imaging, the power delivered by each Talbot focal spot was ~10−4 mW, therefore the power density on each fluorescence detection area (3 pixels × 3 pixels, which is 15.6 µm × 15.6 µm) is ~40 mW/cm2.
In summary, this prototype was able to acquire a full image at a maximum rate of 1 frame per 115 second, an FOV of 13 mm2 and a resolution of 1.2 µm. The full dimensions of the complete system were ~20 × 20 × 20 cm3 – we note that the size can be significantly reduced for future versions.
Wide FOV time-lapse imaging from within the incubator
All the following experiments were conducted within a humidified 5% CO2 incubator at 37 °C. The data was streamed from the incubator via a CAT-6 Ethernet cable.
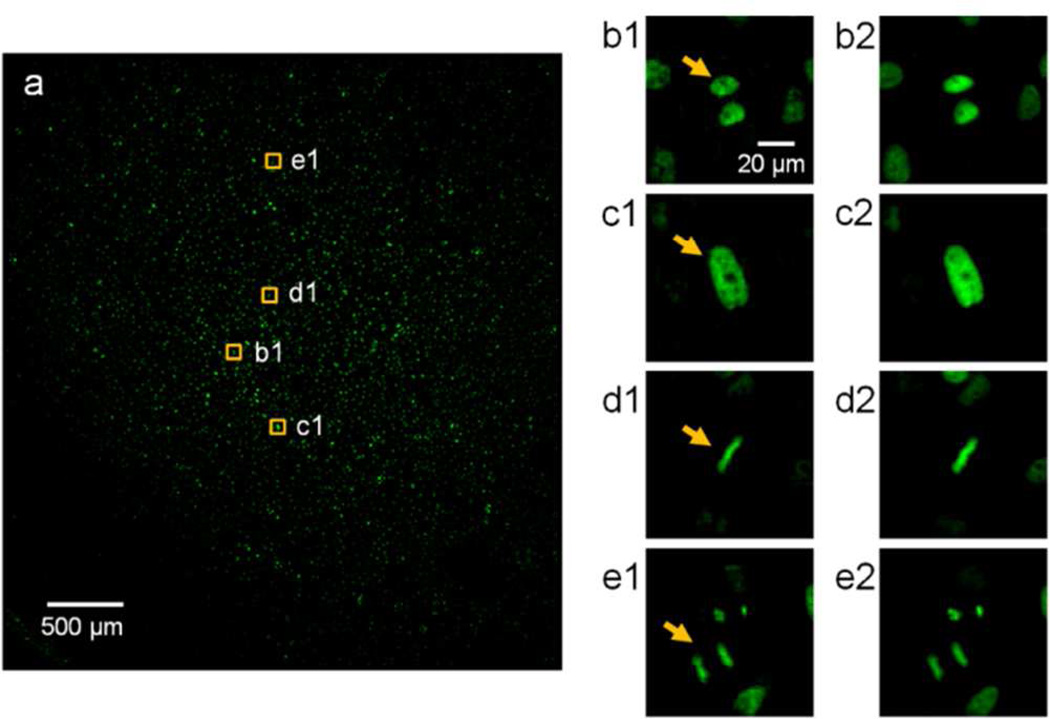
As an initial demonstration, we chose HeLa cells expressing H2B-eGFP in their nuclei as our imaging targets. The cells were cultured on the image sensor surface, and a 3.7 mm × 3.5 mm FOV was imaged (Figure 2a). The scan step size was set at 0.6 µm. It took 85 s to scan the entire FOV, and 30 s to perform data readout and storage. Typical cells in G1, G2, metaphase, and anaphase stages could be recognized in this image (Figure 2b1,c1,d1,e1). The nucleoli were also recognizable as the dark regions inside the nuclei, where the H2B proteins were absent (Figure 2b1,c1). For comparison, the same cells were imaged under a standard microscope with a 20x/0.4 NA objective (Figure 2b2,c2,d2,e2). The resolution of the FTM system was adequate for us to distinguish the four different stages (G1, G2, metaphase, and anaphase) in the cell cycle.
Figure 2.
Wide FOV fluorescence imaging of the GFP cells. (a) The 3.7 mm × 3.5 mm image. (b1, c1, d1, e1) Cropped images of typical cells in (a), including G1 (b1), G2 (c1), metaphase (d1), and anaphase (e1) (arrows). (b2, c2, d2, e2) The same cells as imaged by a conventional microscope with a 20x/0.4 NA objective.
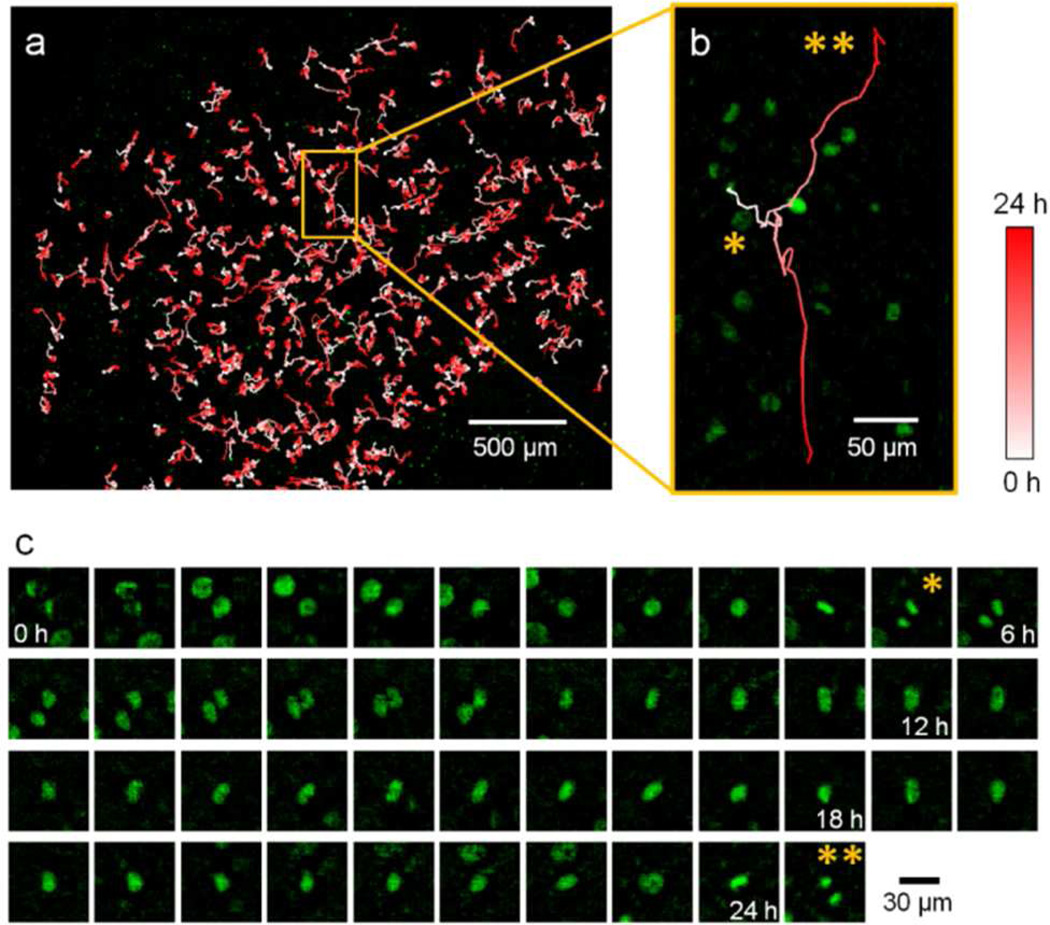
To demonstrate the system’s longitudinal study capability, we employed it to acquire a sequence of images at 33 min intervals over a total duration of 24.8 hours (Movie S-1). After reconstructing the image sequence, we extracted the outline of each nucleus from the background by thresholding and the application of watershed segmentation algorithm 14 (Figure S-3). We first evaluated the segmentation accuracy by processing six 1 mm × 1 mm reconstructed sample images, and compared the results with manual segmentation. Among the segmented objects, the algorithm had an accuracy of 96.1±2.2% (1304 objects; 3.3±1.7% oversegmented (falsely cut) and 0.7±0.5% undersegmented (falsely merged)). There were 5.8±2.3% cells that were not bright enough to be recognized by the algorithm. We then used the nearest-neighbor tracking method on the segmented images,15 which finds the next position of each cell by searching its neighborhood in the next frame. A threshold for the neighbor range was defined according to the image sequence to optimize the tracking results. Newly divided cells were given a new tag.
The trajectories for each tracked cell over 24.8 h were then plotted (Figure 3a). Figure 3b shows a zoomed-in vignette illustrating our capacity to perform single cell tracking. This cell migrated in the bottom-right direction, divided at 5.5 h (marked by *), and the two daughter cells started to migrate in opposite directions. At 24.8 h, the daughter cell which was moving upwards divided again (marked by **). By using the trajectory information, we can also automatically crop each cell out of the original wide FOV image sequence (Figure 3c).
Figure 3.
Longitudinal imaging of the GFP cells. (a) Trajectories of cells over 24.8 hours. The position of a cell at each time point is illustrated by its color. (b) The trajectory showing a single cell which divided at 5.5 h (*), with one of the daughter cells that moved upwards dividing again at 24.8 h (**). (c) Longitudinal images of the cell in (b) as it migrated and divided. The asterisks indicate the cells in anaphase at 5.5 h (*) and 24.8 h (**) during the division events.
Quantitative study of camptothecin treatment
Drug study is an excellent longitudinal study format for which an inexpensive, high-resolution and wide FOV fluorescence imaging system can significantly improve the efficiency. As a demonstration of such an application, we performed the following experiment with camptothecin (CPT) - a well-characterized anti-cancer drug. CPT inhibits DNA and RNA replication and synthesis by targeting the nuclear enzyme topoisomerase I.16 Analogues of CPT have been successfully used in cancer chemotherapy.17
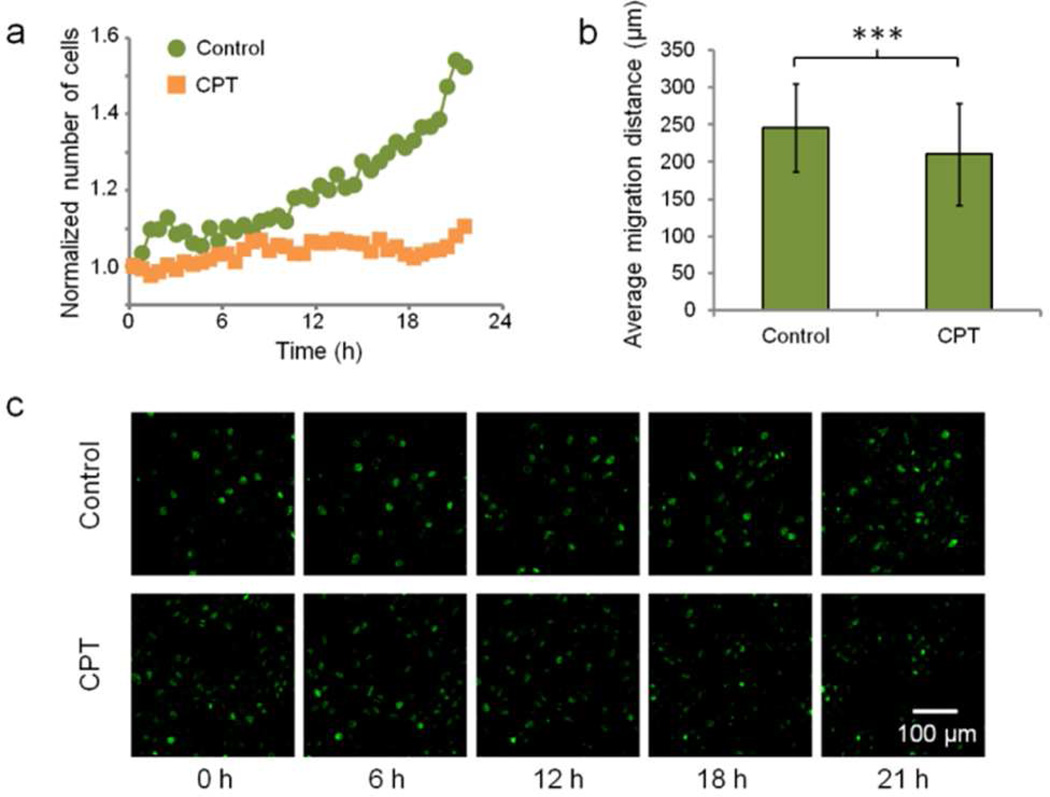
Here we used our system to investigate HeLa cell division and migration behaviors in response to CPT. In this longitudinal study, we prepared two cell culture groups: the CPT treatment group and the control group. For the CPT group, cell culture medium with 1 µM CPT was loaded into the microfluidic culture chamber at 0 h. The control group was loaded with the medium without CPT. Wide FOV images were taken at 33 min intervals over a total duration of 21.5 hours. We performed watershed segmentation to count the number of cells in each image. At 0 h, the control and CPT group were counted to have 1158 and 1297 cells respectively. At 21.5 h, the cell count for control group increased 52.4% while the cell count for CPT group increased 10.5% (Figure 4a). We then tracked 373 and 422 cells in the control and CPT groups, respectively, and calculated the migration distance of each cell over the 21.5 hours. The cells in the control and CPT groups had average migration distances of 246.0±59.2 µm and 210.3±68.5 µm, respectively, showing a significant difference by the Student’s t-test (P<0.001) (Figure 4b). Representative time-lapse images are shown in Figure 4c. From these data, we can conclude that CPT inhibits cell proliferation and migration, and the Talbot Fluorescence ePetri system can be used for automate drug testing and cell behavior analysis.
Figure 4.
Effect of camptothecin (CPT) treatment. (a) Cell count over time for the control group and the CPT group. (b) Average cell migration distances for the control group and the CPT group over 21.5 hours (***P<0.001). (c) Representative images for the two groups at 0, 6, 12, 18, and 21 h.
Discussion
The Talbot Fluorescence ePetri approach requires an undistorted projection of the Talbot generated light spots on the sample. As PDMS is porous and vulnerable to deformation, a microfluidic reservoir comprised entirely of PDMS could significantly degrade the achieved resolution. To mitigate this, we developed a fabrication method for implementing a glass/PDMS hybrid microfluidic component, as described in the Experimental section. Here we incorporated a glass cover as the top of the reservoir to preserve the quality of the Talbot light spots. For our demonstration experiment, the current reservoir design consisted only of a center culture chamber and two ports. It is possible to integrate more sophisticated microfluidic functions into the system. For example, it would be possible to implement multiple branch channels to generate a stable chemical gradient 18 to study the response of cells to different concentrations of chemical signals. We can also include microvalves 19 to modulate the amount of drug delivery over time and investigate the resulting cell dynamics.
Our current scanning time for a 13 mm2 image is 85 s and the data storage upload time is 30 s, both of which are limited by the number of scan steps and the amount of data collected. We refer to the total time as frame acquisition time. We set the imaging interval to be 33 min. This interval can be shortened as desired (to the limit of 2 min) without significant changes to our current prototype. The frame acquisition time can be significantly shortened by upgrading the camera board and the data acquisition pipeline. However, we note that, while there is no significant technical barrier against acquiring more fluorescence images per time period, users need to keep in mind that fluorophore bleaching will set a hard limit on the total number of fluorescence images they can take for a given experiment.
Next, we would like to further emphasize the scalability of the FOV for this imaging design. This imaging approach can directly tackle a larger area by simply employing a larger lenslet grid and a larger sensor chip. Furthermore, unlike with conventional scanning microscopes, the total scanning time would remain the same rather than scaling linearly with the area covered.
We observed that the fluorescence intensity of the H2B-eGFP varied as a cell went through different stages in the cell cycle: it was very bright during mitosis when the chromosomes were highly condensed, but became very dim in early interphase (such as G1 phase) when H2B was diffuse in the nuclei. This may explain the fact that 5.8±2.3% of the cells cannot be recognized by our segmentation algorithm. We tracked ~30% of the brightest cells of the whole population throughout the time-lapse imaging process in our CPT treatment experiment to avoid errors in our statistics for the migration speed. In this study, the main focus was on the demonstration of the Talbot Fluorescence ePetri platform, and we expect that more advanced algorithms can be used to track a larger percentage of cells. For example, we can implement trajectory repair processes in our tracking algorithm, which connects the broken trajectories if the positions where a cell disappears and reappears are within a certain neighborhood. Furthermore, the typical cell cycle stages (such as metaphase, anaphase, G1, and G2) can be automatically recognized by introducing machine vision techniques such as the support vector machine (SVM).14 The cell annotation over a wide FOV is likely useful for a host of bioscience studies.
Conclusions
In this work, we demonstrated the utility of the Talbot Fluorescence ePetri for live cell imaging and drug study applications. To our knowledge, this is the first implementation of automated on-chip, wide FOV and high-resolution fluorescence imaging for longitudinal cell cultures, in which the conventional microscope objective and its associated shortcomings are eliminated. A glass/PDMS hybrid microfluidic component was developed, and HeLa cells expressing eGFP in nuclei were cultured and continuously imaged with an FOV of 13 mm2 and a resolution limit of 1.2 µm. We further explored the application of our system by observing cellular response to CPT, demonstrating the system’s potential for the study of drug dynamics and anticancer drug discovery. Our on-chip system is simple in implementation, low cost, integratable with microfluidic devices, and compact enough to be placed inside a standard incubator. We believe our Talbot Fluorescence ePetri system can also find significant applications in many emerging fields such as tissue engineering and stem cell differentiation where the longitudinal study of a large number of living samples is needed.
Experimental section
Fabrication of the microfluidic-integrated image sensor
The microfluidic-integrated image sensor consists of a CMOS sensor and a microfluidic component, and was fabricated separately and assembled together. The CMOS image sensors (MT9M001C12STM, Aptina Imaging) have a pixel size of 5.2 µm. The glass cover was removed from each sensor to provide access to the sensor surface. Filter materials were coated onto the sensors by following the same methods in our previous work.5 An additional PDMS layer was spin-coated on top of the filter layer for protection. The microfluidic component was fabricated in seven steps (Figure S-1a-g). Finally, the completed microfluidic component was attached to a coated CMOS sensor with the edges sealed by PDMS (Figure S-1h), and Tygon tubing connected to the inlet and outlet for cell and medium loading (Figure 1c).
Temperature measurement
For measuring the local temperature inside the microfluidic device during the imaging process, the microfluidic device was placed inside the humidified 5% CO2 incubator at 37 °C. A 76 µm-diameter T-type thermocouple (Omega Engineering) was inserted into the cell culture chamber from the inlet, and the temperature was read out by a digital thermometer. The TEC was switched on 1 min before imaging for pre-cooling.
Microfluidic cell culture and time-lapse imaging
HeLa cells (ATCC) were infected at approximately 35% confluence with VSVG-pseudotyped lentivirus to obtain cells ubiquitously expressing eGFP fused to histone-2B (H2B), driven by the chicken beta-actin- CMV (CAG) promoter. Stably infected cells were propagated in DMEM (Cellgro) supplemented with 10% FBS (Cellgro).
The microfluidic-integrated image sensor was treated with Poly-L-lysine solution (Sigma-Aldrich) overnight for better cell attachment. Upon experiment, the cell solution was adjusted to a concentration of 1×106 cells/ml, and loaded into the microfluidic device. After 4 h of incubation to permit the cells to attach to the image sensor surface, the image sensor was mounted onto the socket for imaging.
Supplementary Material
Acknowledgement
We thank Dr. Scott Fraser (Biological Imaging Center, Caltech) for use of his lab facilities. This project is funded by National Institute of Health under the grant 1R01AI096226-01. D.V.B. is supported by NHLBI NRSA fellowship F30 HL110723.
Footnotes
Supporting Information
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Zhong Q, Busetto AG, Fededa JP, Buhmann JM, Gerlich DW. Nat. Methods. 2012;9:711–713. doi: 10.1038/nmeth.2046. [DOI] [PubMed] [Google Scholar]
- 2.Kim YE, Chen J, Chan JR, Langen R. Nat. Methods. 2010;7:67–73. doi: 10.1038/nmeth.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, Cohen L, Danon T, Perzov N, Alon U. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 4.Schroeder T. Nat. Methods. 2011;8:S30–S35. doi: 10.1038/nmeth.1577. [DOI] [PubMed] [Google Scholar]
- 5.Pang S, Han C, Lee LM, Yang CH. Lab Chip. 2011;11:3698–3702. doi: 10.1039/c1lc20654k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coskun AF, Sencan I, Su TW, Ozcan A. PLoS One. 2011;6:e15955. doi: 10.1371/journal.pone.0015955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arpali SA, Arpali C, Coskun AF, Chiang HH, Ozcan A. Lab Chip. 2012;12:4968–4971. doi: 10.1039/c2lc40894e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang S, Han C, Kato M, Sternberg PW, Yang CH. Opt. Lett. 2012;37:5018–5020. doi: 10.1364/OL.37.005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patorski K. Prog. Opt. 1989;27:3–108. [Google Scholar]
- 10.Orth A, Crozier K. Opt. Express. 2012;20:13522–13531. doi: 10.1364/OE.20.013522. [DOI] [PubMed] [Google Scholar]
- 11.Zheng GA, Lee SA, Antebi Y, Elowitz MB, Yang CH. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16889–16894. doi: 10.1073/pnas.1110681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffy DC, McDonald JC, Schueller OJ, Whitesides GM. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 13.Kim L, Toh YC, Voldman J, Yu H. Lab Chip. 2007;7:681–694. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 14.Held M, Schmitz MH, Fischer B, Walter T, Neumann B, Olma MH, Peter M, Ellenberg J, Gerlich DW. Nat. Methods. 2010;7:747–754. doi: 10.1038/nmeth.1486. [DOI] [PubMed] [Google Scholar]
- 15.Hand AJ, Sun T, Barber DC, Hose DR, MacNeil S. J. Microsc. 2009;234:62–79. doi: 10.1111/j.1365-2818.2009.03144.x. [DOI] [PubMed] [Google Scholar]
- 16.Hsiang YH, Hertzberg R, Hecht S, Liu LF. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 17.Garcia-Carbonero R, Supko JG. Clin. Cancer Res. 2002;8:641–661. [PubMed] [Google Scholar]
- 18.Figueroa XA, Cooksey GA, Votaw SV, Horowitz LF, Folch A. Lab Chip. 2010;10:1120–1127. doi: 10.1039/b920585c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung BG, Lin F, Jeon NL. Lab Chip. 2006;6:764–768. doi: 10.1039/b512667c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.