Abstract
Disruption of the bone morphogenic protein (BMP)-linked signaling pathway has been suggested as an important factor in the development of hereditary spastic paraplegia (HSP). HSP-causing proteins spastin, spartin and NIPA1 were reported to inhibit the BMP pathway. We have previously shown a strong interaction of NIPA1 and atlastin-1 proteins. Hence, we investigated the role of another HSP-associated protein atlastin-1 in this signaling cascade. Endogenous and expressed atlastin-1 showed a strong interaction with BMP receptors II (BMPRII) and analyzed missense, HSP-causing mutations R239C and R495W disrupted BMPRII trafficking to the cell surface. BMPRII does not require the presence of atlastin-1 because knockdown expression of atlastin-1 did not alter endogenous BMPRII cellular distribution. Expression of mutant forms of atlastin-1 also interfered with the signaling response to BMP4 stimulation and reduced phosphorylation of Smad 1/5 proteins. Our results suggest that HSP-causing atlastin-1 mutations exhibit a dominant-negative effect on trafficking of BMPRII, which disrupts the BMP pathway in neurons. This, together with previously demonstrated inhibition of atlastin-1 of BMP pathway, further supports the role of this signaling cascade in axonal maintenance and axonal degeneration, which is seen in various types of HSP.
Keywords: BMP pathway, Atlastin-1, Hereditary spastic paraplegia, BMPRII trafficking
Introduction
Hereditary spastic paraplegia (HSP) is a group of neurodegenerative disorders that leads to severe disability due to spastic weakness of the lower extremities (Salinas et al., 2008; Züchner, 2007). In spite of recent proposals for the function of several proteins associated with HSP, the molecular mechanisms of selective and stereotypical pathological changes caused by mutant forms of these proteins remain mostly unknown. HSP is characterized by relatively isolated axonal degeneration (Deluca et al., 2004; Wharton et al., 2003). Interestingly, while the pathological features of HSP are relatively uniform, mutations in more than 40 genes have been identified thus far, making it one of the most genetically heterogeneous neurodegenerative disorders (Blackstone et al., 2011; Salinas et al., 2008; Züchner, 2007). This implies the existence of common biochemical pathways central to the pathogenesis of HSP. However, it remains unclear which HSP-associated gene products interact directly and how mutations alter these interactions to produce the clinical phenotype (Blackstone et al., 2011).
Mutations in atlastin-1 are the most common cause of autosomal dominant (AD) HSP with an early age of onset (Abel et al., 2004; Durr et al., 2004; Hedera et al., 2004). Atlastin-1 is an integral membrane protein, which belongs to the dynamin superfamily of large GTP-ases (Zhao et al., 2001). It is important for the formation of the tubular endoplasmic reticulum(ER) network, and HSP mutations inhibit tubule interconnections (Hu et al., 2009; Orso et al., 2009). However, whether this is the main pathogenic mechanism remains unknown (Namekawa et al., 2007; Rismanchi et al., 2008; Zhu et al., 2003). We have previously shown that atlastin-1 also strongly interacts with NIPA1 protein, which is mutated in another form of AD HSP (Botzolakis et al., 2011; Rainier et al., 2003; Zhao et al., 2008). One possible mechanism of axonal degeneration caused by NIPA1 mutations is inhibition of bone morphogenic protein (BMP)-linked signaling pathway (Blackstone et al., 2011; Tsang et al., 2009; Wang et al., 2007). We hypothesized that atlastin-1 also interacts with the BMP pathway, which was also recently reported in a zebrafish model of HSP (Fassier et al., 2010). Here, using a combination of confocal microscopy, immunoprecipitation, flow cytometry and knock-down expression using RNA interference, we also show that endogenous and expressed atlastin-1 interacts with the BMP signaling cascade and demonstrate that HSP-associated mutant forms of atlastin-1 interfere with BMP receptors II (BMPRII) trafficking and this disruption is consistent with a dominant-negative effect.
Results
Atlastin-1 and BMPRII are binding partners, and HSP mutations do not alter this interaction
Our previous report of interactions between atlastin-1 and NIPA1, which is a known inhibitor of the BMP pathway, prompted us to examine whether atlastin-1 also plays a role in this biochemical pathway (Botzolakis et al., 2011). We used previously developed BMPRII antibodies to co-immunoprecipitate atlastin-1 and BMPRII from the rat brain tissue to determine whether or not these proteins were in vivo binding partners. We first confirmed the specificity of BMPRII antibodies, which yielded a single band using in an agreement with previous reports (Hamid et al., 2009; Ramos et al., 2006; Yu et al., 2008) and this was absent from pulmonary endothelial tissue obtained from BMPRII conditional knock-out mouse (Hong et al., 2008) (Supplementary Figure).
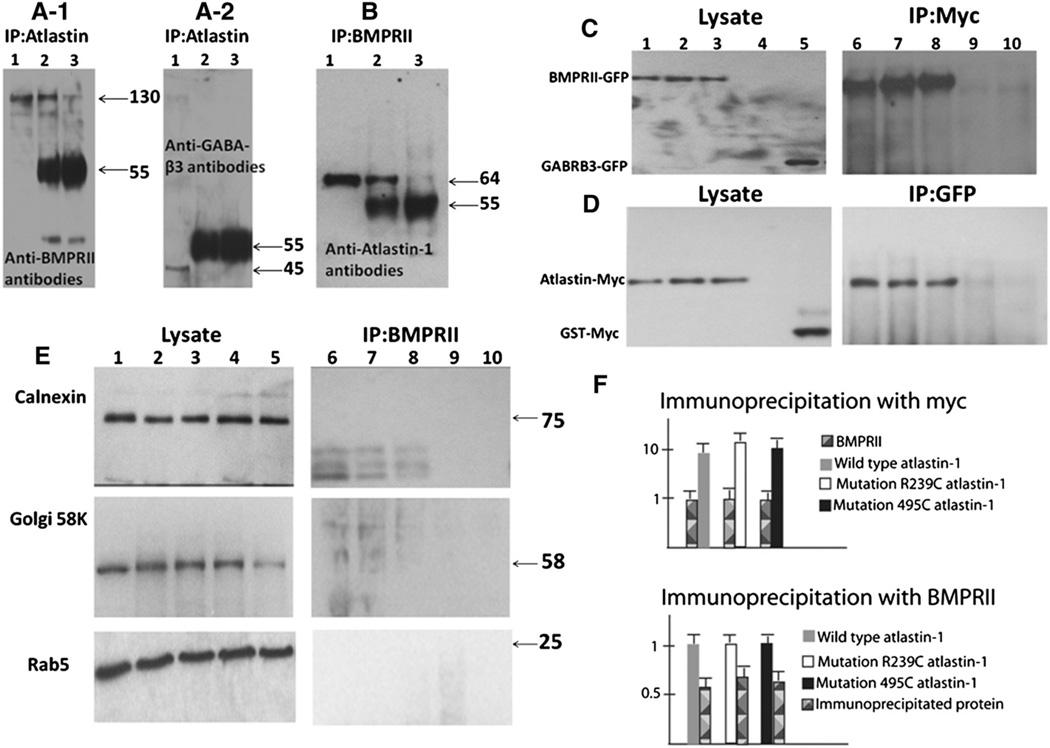
Immunoprecipitation experiments from the whole rat brain demonstrated that these proteins formed hetero-complexes, as both proteins could be pulled down when the other putative binding partner was used as bait (Fig. 1, panels A1, A2 and B). As previously reported, antibodies against BMPRII and immunoprecipitation with atlastin-1 was observed mostly for 130 kDa form of protein (Fig. 1, panel A1). Similarly, we detected atlastin-1 using BMPRII for coimmunoprecipitation (panel B). Neither protein could be pulled down using antibodies to an unrelated membrane protein, the GABAA receptor β3 subunit (GABRB3) (Panel A2), supporting the conclusion that the interaction between BMPRII and atlastin-1 proteins was specific in spite of faint background present in control experiments (panels A2 and B, 3rd lanes).
Fig. 1.
Atlastin-1 and BMPRII are binding partners. The whole rat brain extracts proteins were immunoprecipitated with atlastin-1 antibodies and analyzed by immunoblotting with BMPRII antibodies (panels A1), control GABAA-β3 antibodies (GABRB3) (panel A2), or immunoprecipitated with BMPRII antibodies and immunoblotted with atlastin-1 antibodies (panel B). Lane 1 shows studied proteins from cell lysates used for immunoprecipitation experiments, lane 2 antibodies used as the “bait” (atlastin-1 panels A-1 and A-2, BMPRII panel B) incubated with protein A/G agarose beads and lane 3 control experiments with protein A/G agarose beads alone. We observed 130 kDa band, corresponding to BMPRII (panel A-1, lane 2) and 64 kDa band, corresponding to atlastin-1 (panel B, lane 2), which were immunoprecipitated in the presence of bait antibodies. Non-specific background was observed mostly at the 55 kDa size. Very faint bands were also present in lane 3 in panels A-1 and B, again representing a non-specific background; however, lane 2 with co-immunoprecipitation had more than 100-fold higher activity in both instances. GABRB3 antibodies served as a control protein for the specificity of immunoprecipitation and we did not detect this protein after immunoprecipitation in lanes 2 and 3 (panel A-2). Panels C and D show analysis of HEK239-T cells cotransfected with BMPRII-GFP and WT atlastin-1-myc (lane 1), R239R atlastin-1:myc (lane 2), R459W atlastin-1:myc (lane 3). Control experiments included untransfected control cell lines (lane 4) and cotransfection of WT atlastin-myc with GABRB3-GFP (panel C, lane 5) or BMPRII-GFP and GST-myc (panel D, lane 5). Immunoprecipitation was done using anti-myc antibodies (panel C) or anti-GFP antibodies (panel D). Co-immunoprecipitation was only observed for coexpressed BMPRII-GFP and WT and mutant forms of atlastin-1:myc (panels C and D, lanes 6–8), while control proteins GABRB3 or GST did not interact with atlastin-1 or BMPRII (lanes 9 and 10). Blots from panels C and D were also analyzed with the markers of subcellular organelles where atlastin-1 can colocalize in order to detect membrane segments containing both BMPRII and atlastin-1 (panel E). We used markers for endoplasmatic reticulum (calnexin, upper row), Golgi complex (Golgi 58K, middle row) and early endosomes (Rab5, lower row). No signal was observed for BMPRII protein (right column) while cellular lysates showed a robust signal of used subcellular markers in all five lanes (left column). Introduced atlastin-1 mutations did not modify the interaction with BMPRII. Semiquantitative analysis after normalization of BMPRII signal from cell lysates (upper part of panel E) and after signal normalization from all three studied forms of atlastin-1 (lower part of panel E) did not reveal any statistically significant differences of signal intensity from immunoprecipitated partner proteins.
We also investigated whether studied HSP-causing mutations in the atlastin-1 (SPG3A) gene alter these protein interactions. We selected two missense mutations, R239C and R495W, which were extensively studied previously (Botzolakis et al., 2011; Namekawa et al., 2007). We could only study expressed proteins and we used myc tagging for WT and mutant forms of atlastin-1, and GFP tagged WT BMPRII expressed in HEK293-T cells. Similar levels of co-immunoprecipitation with BMPRII were observed for WT, R239C and R495W mutant forms of atlastin-1 (Fig. 1, panels C and D). The specificity of this interaction was further supported by the absence of immunoprecipitation if control, putatively unrelated proteins were used (GABRB3-GFP instead of BMPRII-GFP, panel C, lanes 5, 10, and glutathione synthetase [GTS-myc] instead of atlastin-1-myc, panel D, lanes 5 and 10). Additionally, we performed a semiquantitative analysis of precipitated three forms of atlastin-1 (WT and both mutations) normalized to BMPRII expression or precipitated amount of BMPRII normalized to expression of studied forms of atlastin-1 (Fig. 1, panel F); no significant differences have been detected, suggesting very similar affinity of BMPRII and WT or mutant forms of atlastin-1.
Atlastin-1 and BMPRII are membrane proteins and we also considered a possibility that both proteins are contained in membrane segments from various subcellular organelles rather than binding directly to each other. However, proteins associated with endoplasmatic reticulum (ER), Golgi complex (GC) and early endosomes, where atlastin-1 was reported to localize (Botzolakis et al., 2011; Hu et al., 2009; Namekawa et al., 2007; Orso et al., 2009; Rismanchi et al., 2008; Zhu et al., 2003), did not identify microsomes after co-immunoprecipitation experiments, supporting a direct interaction between atlastin-1 and BMPRII (Fig. 1, panel E).
Expression of atlastin-1 mutations alters trafficking of the BMPRII to the cell surface
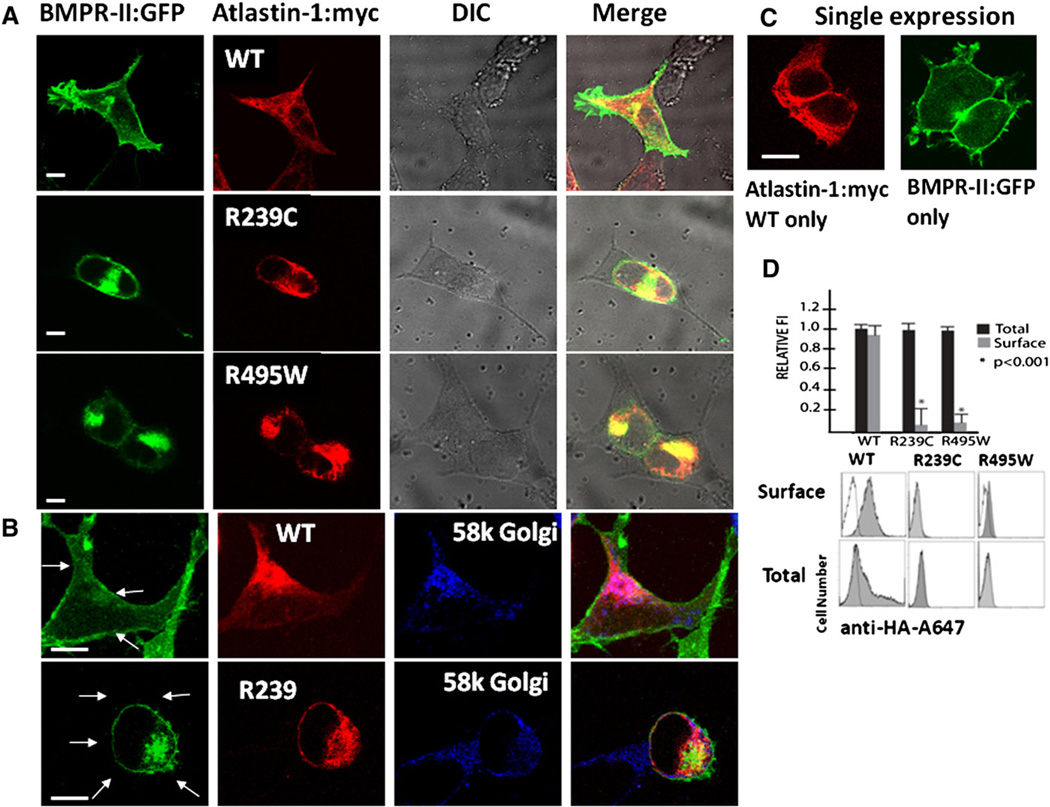
Coexpression of WT atlastin1- and BMPRII in heterologous HEK239-T cells showed a partial overlap of these proteins, including in GC and a robust presence of BMPRII on the cell surface (Fig. 2, panels A and B). The intracellular distribution of the BMPRII was dramatically altered in the presence of studied HSP-causing atlastin-1 mutations. Similarly to our previous report, both mutant forms of atlastin-1 formed thick intracellular bundles and aggregates with retention in GC rather than the reticular and punctuate expression seen with WT atlastin-1 (Botzolakis et al., 2011). Mutant forms of atlastin-1 colocalized more robustly with expressed BMPRII protein, resulting in its retention in GC and essentially a complete absence from the cell surface (Fig. 2, panel A, 2nd and 3rd rows, panel B, lower row). Flow cytometry studies were used to quantify the difference of BMPRII-HA on the cell surface and the expression of studies atlastin-1 mutations almost completely abolished the presence of BMPRII-HA on the cell surface. This was not secondary to changed intracellular expression of BMPRII-HA, as permeabilizing cells prior to staining revealed significant amounts of protein in all three sets of experiments. However, the presence of atlastin-1 was not required for the trafficking of BMPRII to the cell surface (Fig. 2, panel D). Transfected cos-7 cells, which do not express any atlastin-1 or other atlastin isoforms (Zhu et al., 2003) showed the same pattern of BMPRII cell surface expression (Fig. 2, panel C).
Fig. 2.
Mutations in Atlastin-1 disrupt cell surface trafficking of the BMPRII. Coexpression of BMPRII and atlastin-1 in HEK239-T cells (panel A) showed a robust localization of BMPRII on the cell surface only when coexpressed with WT atlastin-1. Expressed BMPRII in the presence of mutant forms of atlastin-1 colocalized strongly with atlastin-1 with its retention in the Golgi complex (panel B) and no BMPRII-GFP was present on the cell surface membrane, which is indicated by arrows. HSP-causing atlastin-1 mutations completely abolished the trafficking of the BMPRII to the cell surface. BMPRII did not require the presence of atlastin-1 for its transport to the cell surface as shown on Cos-7 cells, which do not express any endogenous atlastin-1, after a single expression of BMPRII (panel C). Panel D shows plotting of the surface and total expression of HA tagged BMPRII in the presence of WT atlastin-1, R239C and R495W atlastin-1 mutations. Surface staining (without membrane permeabilization) is depicted by gray columns and total cellular staining (after membrane permeabilization) is depicted as black columns. Co-transfection of WT atlastin-1myc and BMRII-HA resulted in a robust presence of BMPRII on the cell surface and the expression of both HSP-causing mutations significantly reduced its trafficking to the cell surface (p<0.001). Representative flow cytometry frequency histograms of Alexa-647 (A647) fluorescence intensity for cells expressing BMPRII-HA, mock transfected cells (white) are also shown. Histogram was overlaid on each of the histograms from positively transfected cells (grey). The x-axis indicates arbitrary fluorescence units (log scale) and the y-axis indicates the number of cells. Scale bar = 5 µm.
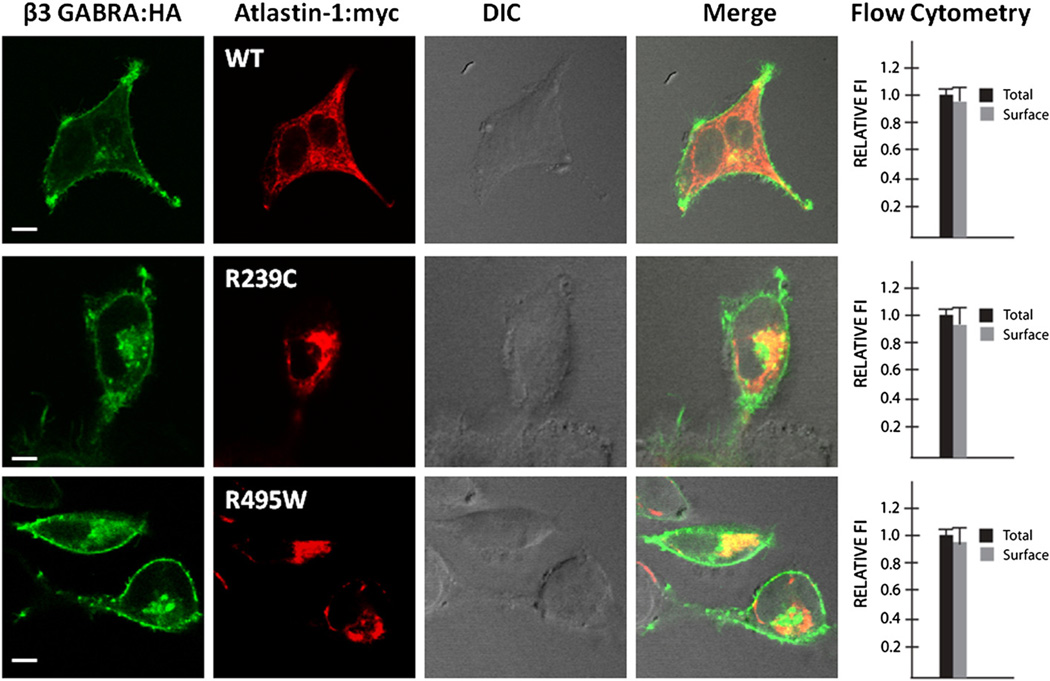
The disruption of BMPRII trafficking to the cytoplasmatic membrane could also result from a non-specific disruption of the secretory pathway given the putative role of atlastin-1 in the GC organellogenesis. That is why we used flow cytometry to compare the cell surface trafficking of BMPRII with β3 subunit of the GABAA receptor, which is also transported through the secretory pathway. The presence of both mutant forms of atlastin-1 did not change the distribution of the β3 subunit of the GABAA receptor on the cell surface, indicating specific interactions of atlastin-1 and BMPRII rather than a non-specific interference with an intracellular transport due to the disruption of secretory pathway (Fig. 3).
Fig. 3.
Intracellular trafficking of unrelated proteins is not affected by the presence of mutant forms of atlastin-1. We also evaluated the trafficking of an unrelated protein β3 subunit of the GABAA receptor (GABRB3), which is transported through the secretory pathway to exclude a possibility of a non-specific disruption of intracellular trafficking in the presence of mutant forms of atlastin-1. Flow cytometry was used to assay the proportion of surface signal (non-permeabilized cells) and total signal (permeabilized cells) from GABRB3. Plotting of the surface (gray bar) and total expression (black bar) of HA tagged β3 GABAA receptor showed the same proportion of GABRB3 on the cell surface in the presence of WT or studied atlastin-1 HSP mutations (91% WT, 90.5% R239C and 92% for R459W, differences not significant). Scale bar=5 µm.
Mutant forms of atlastin-1 also alter the distribution of endogenous BMPRII in cultured rat cortical neurons
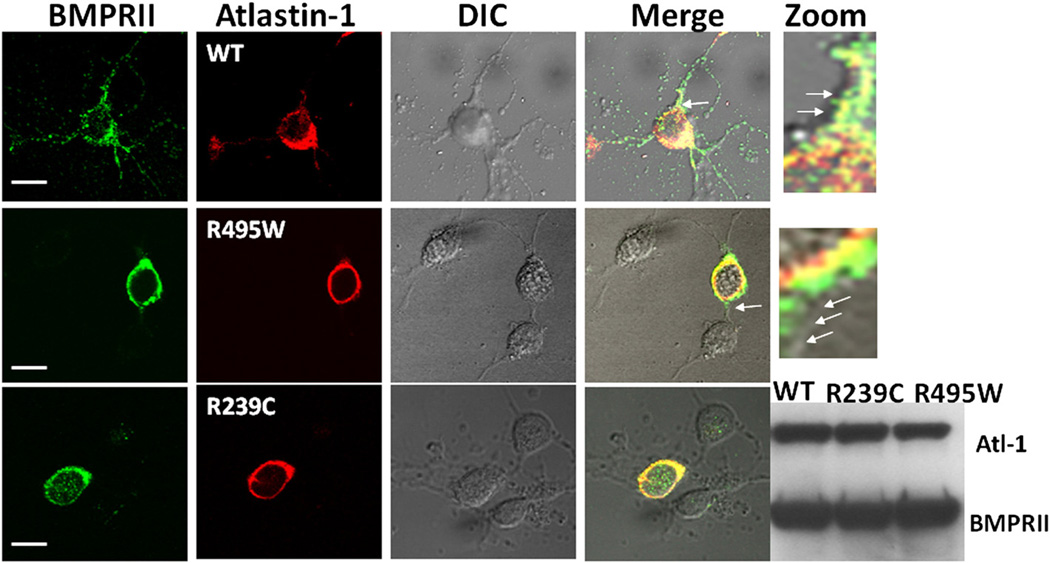
We also investigated the effect of HSP-causing atlastin-1 mutations on the intracellular distribution of the endogenous BMPRII protein in cultured cortical rat neurons. Endogenous BMPRII and both expressed and endogenous WT atlastin-1 showed a similar pattern of colocalization and distribution in cultured rat cortical neurons with a robust presence of the BMPRII on the cell surface (Fig. 4, first row), which was comparable with the pattern seen in heterologous cells (Fig. 2). Furthermore, BMPRII was present in neuronal processes in neurons expressing WT atlastin-1, and overexpression of WT atlastin-1 did not alter the distribution of BMPRII in axons and dendrites. Introduction of two studied atlastin-1 mutations resulted in disruption of trafficking of endogenous BMPRII. This was most obvious in the absence of BMPRRII in axons and dendrites, even though the primary defect was consistent with its retention in GC, similar to our findings in heterologous cells (Fig. 4, second and third row, triple staining with Golgi markers not shown). The presence of mutant forms of atlastin-1 also reduced the degree of neuritic branching as was reported previously (Botzolakis et al., 2011).
Fig. 4.
The intracellular distribution of endogenous BMPRII is affected by the presence of HSP-causing atlastin-1 mutations. Endogenous BMPRII in cultured cortical rat neurons was present on the cellular surface, similar to the findings in heterologous cells, and in the neuronal processes. Expression of WT atlastin-1 did not alter the pattern of distribution seen with the endogenous atlastin-1 (upper row). HSP-causing mutations of atlastin-1 interfered with the cell surface trafficking of BMPRII with its retention in the GC (staining for GC not shown). Zoom images from the areas indicated by single arrows in the merge column showed BMPRII on the cell surface membrane when a wild-type atlastin-1 was expressed in neurons (first row), while the mutant forms of atlastin-1 resulted in cell surface membrane, designated by arrows, being devoid of BMPRII (second row). Both studied mutations had very similar effect on the distribution of BMPRII. Scale bar=10 µm. The changes of distribution of endogenous BMPRII were not caused by the inhibition of its synthesis in the presence of mutant forms of atlastin-1, because WT, R329C and R495W mutations did not change the amount of detected BMPRII (2 × 106 GFP-positive cells were analyzed by Western blotting).
We also determined whether changes in the BMPRII intracellular distribution induced by the R239C and R495W atlastin-1 mutations were a result of inhibited synthesis of BMPRII or disruption of its intracellular transport. Assay of the total amount of endogenous BMPRII present in transfected cultured cortical rat neurons, which were selected by flow cytometry, did not show any differences in the presence of WT or mutant forms of atlastin-1, further supporting the disruption of the intracellular transport without any additional inhibition of its transcription or translation (Fig. 4, fifth column from left).
The presence of WT atlastin-1 is not necessary for intracellular trafficking of BMPRII
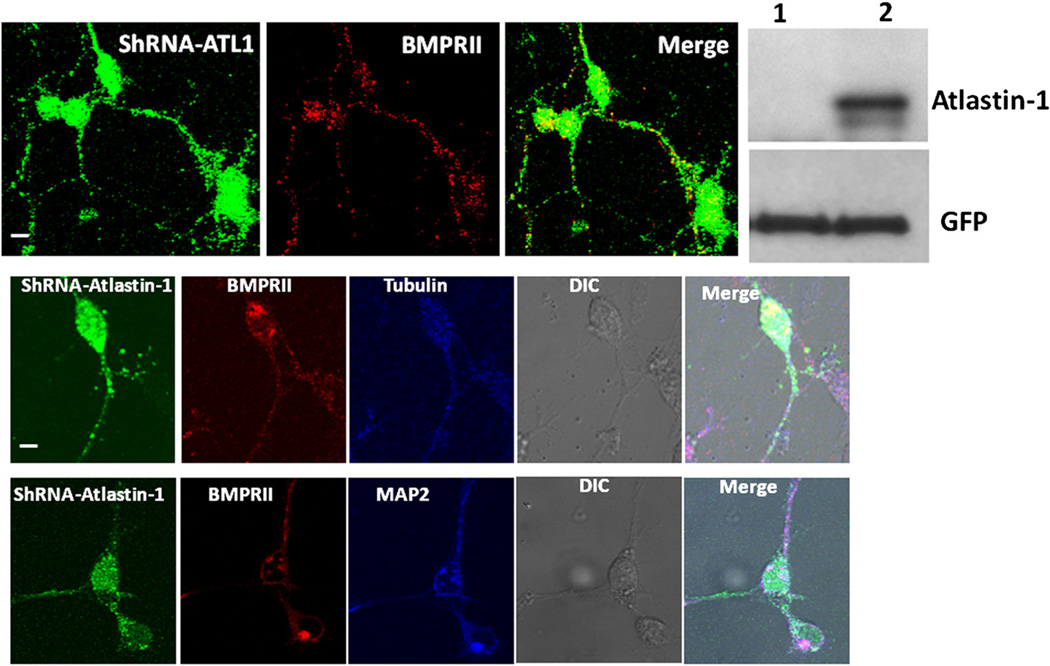
Disruption of surface cell trafficking of BMPRII in the presence of mutant forms of atlastin-1 may be due to a loss or gain of function. We hypothesized that the loss of function mechanism would create similar changes in the absence of endogenous atlastin-1. Knock down expression of atlastin-1, however, did not alter the intracellular distribution of endogenous BMPRII, which was abundantly present on the cell surface and was trafficked into the neuronal processes (Fig. 5, upper row). We also explored a possibility that more selective disruption of BMPRII trafficking to axons or dendrites is the consequence of expression of mutant forms of atlastin-1. The comparison of the presence of endogenous BMPRII in dendrites and axons did not reveal any disparities between these two types of neuronal processes, further arguing against a selective disruption of BMPRII trafficking due to the absence of endogenous atlastin-1 (Fig. 5, middle and lower rows). Thus, this supports the hypothesis that expressed HSP-causing mutations in atlastin-1 exhibit a dominant-negative effect on its binding partner, BMPRII.
Fig. 5.
Trafficking of BMPRII is unaltered after knock down expression of atlastin-1. Cultured cortical rat neurons expressing siRNA oligonucleotide targeting rat atlastin-1, indicated by expression of GFP showed unchanged intracellular distribution of endogenous BMPRII (red) with the strong presence on the cell surface and in neuronal processes. Western blot show the absence of detectable atlastin-1 expression in GFP positive cells with 2 × 106 GFP-positive cells selected by flow cytometry in lane 1, lane 2 control transfected with an empty vector, lower row shows protein loading detected with anti-GFP antibodies. We also analyzed the presence of endogenous BMPRII in axons visualized by tubulin staining (middle row, tubulin detected by Cy5-conjugated antibodies [blue]) and in dendrites visualized by MAP2 staining (lower row, MAP2 detected by Cy5-conjugated antibodies [blue]). We did not detect any observable difference in the presence of BMPRII in these two types of neuronal processes (BMPRII visualized by Cy-3 conjugated antibodies [red]).
Mutations of atlastin-1 reverse the BMP-mediated phosphorylation of Smads1/5
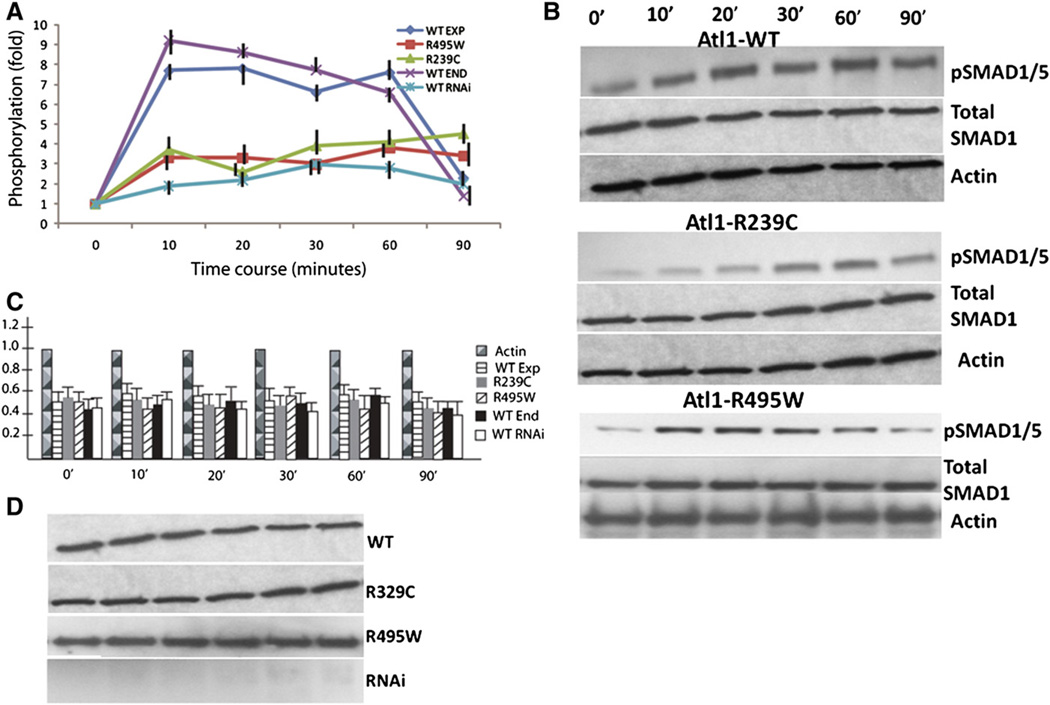
HEK239-T cells transfected with an empty control vector showed up to ten-fold increase of phosphorylated fraction of pSmads1/5 after BMP4 stimulation; this response returned to the baseline in 90 minutes (Fig. 6, panel A). Overexpression of a WT atlastin-1 did not change this induction of pSmads1/5 phosphorylation and the dynamics of the response to BMP4 stimulation was similar to the presence of endogenous atlastin-1 expressed by HEK239-T cells. However, introduction of studied HSP-causing atlastin-1 mutations dramatically reduced the phosphorylated fraction of pSmads1/5 with the maximal 4-fold increase for the R495W mutation. Knockdown expression of endogenous atlastin-1 had very similar dynamics as the presence of the R495W and R239C atlastin-1 mutations (Fig. 6, panel A).
Fig. 6.
Phosphorylation of Smads1/5 after BMP4 stimulation is reduced in the presence of atlastin-1 mutations. BMP4 stimulation of HEK239-T cells expressing endogenous (sham transfection) and overexpressing WT atlastin-1 resulted in increased phosphorylation of Smads1/5 and the difference between these two conditions was not statistically significant (panel A, endogenous atlastin-1 purple, WT dark blue). This was markedly reduced in cell lines expressing studied atlastin-1 mutations (panel A, R495W red, R239C green) and after knock-down of endogenous atlastin-1 (panel A, light blue); there was not statistically significant difference between mutations or RNAi experiments . BMP4 stimulation after transfection with a non-sensical RNA was the same as the sham transfection (data not shown). Each assay was repeated 10-times and phosphorylated Smads1/5 was compared to total Smads1 protein. Data were normalized to an arbitrary zero point and changes were assayed for 90 minutes after the BMP4 stimulation. Induction of phosphorylation by WT or endogenous atlastin-1 and mutations or RNAi was statistically significant (p<0.001). Panel B shows representative western blots for pSmads 1/5 activity, total Smad1 and actin load control for WT overexpression, and R239C, R495C mutations (control [sham] transfection and knock down of atlastin-1 not shown). Total amounts of Smad1 protein were analyzed by comparisons to actin levels, which were arbitrarily normalized to the value of 1; we used the same intervals as for the phosphorylation assays. There were no statistically significant differences in the total amount of Smad1 protein among all five studied conditions (panel C). We also assayed expression levels of atlastin-1 and results of RNAi knock down (panel D). There were no differences between the levels of expression of WT and mutant forms of atlastin-1 and we did not see any changes following the BMP4 stimulation. RNAi experiments showed essentially a complete absence of atlastin-1 protein.
Changing of phosphorylated fraction of pSmad1/5 could be affected by significant changes of the total amount of Smad1/5 proteins. That is why we also analyzed the levels of expression of total Smad1 compared to actin expression and all five analyzed conditions (empty vector transfection, overexpression of WT, R495W and R239C mutations, and knock-down expression of atlastin-1) did not show any statistically significant differences when compared to actin levels during the analyzed 90 minutes intervals (Fig. 6, panel C).
Discussion
Our understanding of the pathogenesis of axonal degeneration, which is a hallmark feature of HSP is only emerging; however, recent progress suggested abnormalities in cytoskeletal assembly, abnormal ER morphogenesis, defects in oxidative phosphorylation, and disruption of axonal transport as the most common mechanisms leading to HSP (Blackstone et al., 2011; Salinas et al., 2008; Züchner, 2007).Microtubular dynamics and axonal transport are regulated by multiple signaling pathways, including the BMP cascade, which is also important for axonal development and regulates several axonal inhibitory factors, such as the Nogo pathway (Hartung et al., 2006; Matsuura et al., 2008; Shi et al., 2007).
Several studies have demonstrated that NIPA1 plays an important inhibitory role in the BMP signaling. NIPA1 directly interacts with BMP receptors II (BMPRII), which then activate BMP receptors I (BMPRI), resulting in phosphorylation of intracellular signaling molecules Smad 1, 5 and 8 (Tsang et al., 2009; Wang et al., 2007). Binding of NIPA1 to BMP receptors also promotes endocytosis and lysosomal degradation, an effect impaired by HSP-causing mutations in NIPA1. HSP-associated mutations of NIPA1 altered the trafficking of BMPRII and reduced their degradation and recycling (Tsang et al., 2009). Furthermore, mutations in the Drosophila NIPA1 ortholog spichthyin resulted in a distal axonal abnormalities with synaptic overgrowth at the neuromuscular junction (Wang et al., 2007). Thus, BMP signaling may represent a possible common final pathway in HSP because spastin and spartin proteins, which are also mutated in different forms of HSP, interact with NIPA1 and have a similar effect on the downstream BMP cascade (Blackstone et al., 2011; Tsang et al., 2009).
We have recently demonstrated that NIPA1 and atlastin-1 bind together (Botzolakis et al., 2011). This would further suggest that atlastin-1 may also play an important role in the BMP signaling pathway. In addition to previously demonstrated direct interaction of atlastin-1 and BMPRI, we now also show that atlastin-1 interacts with BMPRII receptors using immunoprecipitation and cytohistochemistry methods. There was a striking contrast in the intracellular trafficking of BMPRII in the presence of WT and mutant forms of atlastin-1. WT atlastin-1 is not absolutely necessary for trafficking of these receptors because we have observed a strong presence of BMPRII on the cell surface even after knockdown expression of atlastin-1. Similarly, expression of BMPRII in Cos-7 cells, which do not express any endogenous atlastin-1, atlastin-2 or atlastin-3 (Zhu et al., 2003), showed the same robust presence of BMPRII on the cell surface. However, the presence of two studied mutant forms of atlastin-1 resulted in a complete absence of BMPRII receptors from the cytoplasmic membrane and its clustering in the cytoplasm. Diminished response to BMP signaling in cells expressing these mutant forms of atlastin-1 may also be related to this trafficking defect, resulting in the absence of BMPRII receptors on the cell surface. This suggests a dominant-negative effect of HSP causing atlastin-1 mutations. Alternatively, this may be accentuated by overexpression of mutant forms of atlastin-1, even though the overexpression alone cannot account for all of these findings because overexpressed WT atlastin-1 did not induce any observable consequences on BMPRII trafficking. The role of HSP-causing proteins in receptor trafficking is only emerging and their contribution to the HSP pathogenesis remains unknown. It is intriguing that mutations in spartin, causing SPG20, selectively disrupt the degradation and internal trafficking of the epidermal growth factor receptor (EGFR) (Bakowska et al., 2007). Spartin is a multifunctional protein, which also inhibits the BMP pathway and the impact of disrupted trafficking of EGFR and other types of receptors on the pathogenesis of axonal degeneration will require further studies (Tsang et al., 2009).
There is also additional support for the interaction of atlastin-1 and the BMP pathway. Atlastin-1 also appears to be enriched in neuronal growth cones, and knockdown of atlastin-1 expression impaired axonal elongation (Zhu et al., 2006). Previous work using Drosophila animal model suggested that atlastin-1 may be important for synapse formation (Bayat et al., 2010; Lee et al., 2009). Interestingly, this role was dependent upon the ability of atlastin-1 to interact directly with the microtubule-severing protein spastin, which also inhibits BMP signaling. Null allele atlastin-1 Drosophila homolog larvae have clustered synaptic boutons with satellite boutons and denser microtubular network (Lee et al., 2009). Overexpression of various BMP pathways members resulted in similar abnormalities of Drosophila neuromuscular junction, suggesting that atlastin-1 also interacts with this signaling pathway. However, the first direct evidence of the role of atlastin-1 in the BMP signaling pathway was recently presented using knock down and overexpression of atlastin-1 homolog in zebrafish animal model. Reduced expression of this protein in Danio resulted in severe reduction of larval mobility due to abnormal branching of spinal motor neurons, which also expressed higher levels of pSmads1/5 proteins; however, inhibition of BMP signaling rescued this zebrafish motor phenotype (Fassier et al., 2010).
Atlastin-1 plays an important role in the development of both the ER and GC, where it interacts with REEP1, another HSP-causing protein, and spastin and coordinates ER morphogenesis and microtubule dynamics (Hu et al., 2009; Namekawa et al., 2007; Orso et al., 2009; Park et al., 2010; Rismanchi et al., 2008; Zhu et al., 2003).Moreover, recently identified mutations in the reticulon-2 gene, coding another ER shaping protein, as a cause of SGP12 further emphasize the role of abnormal ER morphogenesis as a cause of HSP (Montenegro et al., 2012). Indeed, abnormalities in tubular ER shaping and network interactions have been suggested to be the main mechanism of axonal degeneration in HSP caused by atlastin-1 mutations (Park and Blackstone, 2010). Contrary to this, our results, together with the previously reported analysis of manipulation of atlastin-1 in zebrafish model also suggest that atlastin may regulate BMP receptor trafficking. However, this data are not necessarily mutually exclusive and there are two possible explanations for these observations. Several HSP-causing proteins have multiple putative functions and thus, may contribute to axonal degeneration by several mechanisms (Salinas et al., 2008; Züchner, 2007). Indeed, atlastin-1 may have additional putative roles, as outlined above. Alternatively, the interaction of atlastin-1 and BMPR receptors may mechanistically be a part of the ER morphogenesis. Atlastin-1was previously shown to interact with spastin, which has several functions, including microtubule severing protein, inhibition of the BMP pathway, endosomal sorting and most recently, ER morphogen (Blackstone et al., 2011; Evans et al., 2006; Salinas et al., 2008; Sanderson et al., 2006; Züchner, 2007). Regulation of microtubules by the largerM1 isoform of spastin is a necessary step for the ER morphogenesis (Park et al., 2010; Renvoise and Blackstone, 2010). Thus, the additional regulation of the microtubular system by the BMP pathway may be also important for this process and further studies will be necessary to clarify this question.
In summary, we have shown that mutant forms of atlastin-1 impair trafficking of BMPRII to the cell surface, suggesting a dominant-negative effect. This data, together with previously demonstrated inhibition of atlastin-1 if BMP pathway, further support the role of this signaling cascade in axonal maintenance and axonal degeneration, which is seen in various types of HSP.
Experimental methods
Eukaryotic DNA expression constructs
The full length of atlastin-1 construct was a gift from Dr. Blackstone (Zhu et al., 2003). The R239C and R495W missense mutations in the atlastin-1 construct were introduced using a site-directed mutagenesis kit (QuickChange, Stratagene, La Jolla, CA) and verified by sequencing (Botzolakis et al., 2011). BMPRII-GFP constructs were obtained as a gift from Dr. N. Morrell (Rudarakanchana et al., 2002).We also modified this construct and replaced GFP tag with HA epitope for flow cytometry studies. All sequences were verified.
Cell lines and transfection
HEK-293 T and COS7 cell lines were purchased from American Type Culture Collection (ATCC, Manassas VA), cultured in Dulbecco's modified Eagle's medium (Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, 100 IU/ml penicillin, and 100 µg/ml streptomycin (Invitrogen, Carlsbad, CA), and maintained at 37 °C in humidified 5% C02/95% air. Cells were seeded at moderate density (approximately 50–60% confluent) in 12-well plates, and transfected the next day with the respective combination of constructs using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's protocol. The primary cortical neurons, prepared from embryonic day 18 (E18) rat embryos, were electrically transfected with the respective constructs using the nucleofactor 1 device and the manufacturer's optimized protocol for the rat neuron nucleofactor kit (Amaxa Inc, Gaithersburg, MD).
Immunocytochemistry and confocal microscopy
Atlastin-1 antibodies (H-37, sc-67232) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), BMPRII antibodies (catalog #612292) from BD Transduction Laboratories (San Jose, CA). Specificity of BMPRII antibodies were verified using two different approaches. We immunoblotted pulmonary endothelial tissue from conditional knockout mouse R26CreER/+;BMPRII2f/2P and wild type control R26CreER/+;BMPRII1f/1f (both provided by Dr. Oh from University of Florida, Gainesville) (Hong et al., 2008). Confirmation of specificity of this antibody for immunohistochemistry was performed by immunogen preabsorption experiments and this was done by performing Western blot analysis and immunohistochemistry of cultured rat cortical neurons. For these experiments we synthesized BMPRII peptide corresponding to aa residues 891–920, which are absolutely conserved, because the originally used mouse peptide aa 803–996was not available for purchase. For peptide immunogen preabsorption experiments we incubated 20 µg of peptide and 1 µg of BMPRII antibodies. Other used antibodies were purchased from Sigma-Aldrich, St. Louis, MO, Abcam, Cambridge, MA and Rockland, Gilbertsville, PA as reported before (Botzolakis et al., 2011; Zhao et al., 2008). Assessment of axonal and dendritic distribution of BMPRII was done using tubulin and MAP2 antibodies (Montani et al., 2009; Paspalas et al., 2009). Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences, Fort Washington, PA) and blocked with 2% bovine serum albumin in 1 × PBS containing 0.5% Triton X-100. Cells were incubated with the specific antibody at a dilution of 1:800. After 2 h of incubation, cells were washed twice with 1 × PBS and incubated with the secondary antibody conjugated to Cy3 or Cy5 for 1 h. After three washes, coverslips were mounted onto microscope slides and fluorescent images acquired with a Zeiss LSM510 META laser-scanning confocal microscope and processed with LSM Image software. Transfected cells were evaluated in a blind fashion in regards of the presence of WT, mutant forms or sham transfections; we evaluated between 50 and 100 cells for each construct and specific numbers can be found in the Results section. Images were visualized using a confocal microscope and the LSM images were photographed at 20×, 40×, 60×, and 100× magnification.
Immunoprecipitation and Western blot analysis
For immunoprecipitation of endogenous proteins, rat brain tissue lysates were prepared from adult Wistar male rat brain. The brain tissue was homogenized in M-PER protein extraction reagent (Pierce) with the protease inhibitor cocktail (Roche). The lysate was then centrifuged at 15000 rpm/4c/10 min. The supernatant was either incubated with 6 µg of anti-atlastin-1 antibodies or the same concentration of control IgG derived from the same species, and 30 µl protein A/G agarose beads (Thermo Scientific), or with 6 µg of anti-BMPRII, or the same concentration of control IgG, and 30 µl protein A/G agarose beads at 4 °C over-night. The beads were washed by wash buffer and the proteins were resolved by SDSPAGE and immunoblotted with specific antibodies.
For immunoprecipitation of expressed tagged proteins, HEK-293 cells were transfected with a mixture of atlastin-1(WT)-myc and BMPRII-GFP, atlastin-1(R239C)-myc or atlastin-1(R495W)-myc mutations with BMPRII-GFP, an empty vector and GTS-myc and GABRB3-GFP as a control for myc and GFP immunoprecipitation. The success rate of transfection of heterologous cells was >85%. After 36 h of transfection, cells were lysated and immunoprecipitation was done using anti-myc antibodies and antibody-coupled agarose eliminating any antibody contamination (Thermo Scientific). Immunoprecipitation with GFP antibodies (Ab290, Abcam) was done as described above using protein A/G agarose beads. Cell lysates were used as protein input control of IP, together with actin immunoblotting as a loading control.
Western blot analysis of transfected neurons, including RNAi experiments was done only after selection of transfected neurons by flow cytometry. Samples were run on a LSR II flow cytometer (BD Biosciences) and 48 h after transfection we performed cell sorting where 2 × 106 GFP-positive cells were selected for each tested condition and control experiments.
Atlastin-1 protein depletion by siRNA knock-down expression
siRNA oligonucleotides targeting rat atlastin-1 nucleotides 1138–1156 (directed against sequence GenBank accession no. AY581896) were created as previously described using the primers: 5′-gatccccCCGAGAGCCTAGATATTAAttcaagagaTTAATATCTAGGCTCTCGGttttt-3′ and 5′-agctaaaaaCCGAGAGCCTAGATATTAAtctcttgaaTTAATATCTAGGCTCTCGGggg-3′ and cloned into pGFP-V-RS plasmid with eGFP reporter [30]. As a negative control we used plasmid-A (sc-108060), encoding a scrambled shRNA sequence that will not lead to the specific degradation of any cellular message, also obtained from Santa Cruz Biotechnology. The efficacy of atlastin-1 protein depletion was tested 48 h after transfection. We performed cell sorting by flow cytometry and 2 × 106 GFP-positive cells were selected for each tested siRNA and control experiments. The cells were lysed and Western blot with atlastin-1 antibodies was performed as described above to confirm atlastin-1 protein depletion.
Flow cytometry
Cells were harvested 24 h after transfection using 37 °C trypsin/EDTA (Invitrogen) and placed immediately in 4 °C FACS buffer composed of PBS (Mediatech), 2% fetal bovine serum (FBS) (Invitrogen), and 0.05% sodium azide (VWR). Cells were then transferred to 96-well plates, where they were washed twice in FACS buffer (i.e., pelleted by centrifugation at 450 ×g, vortexed, and resuspended). For surface protein staining, cells were incubated in antibody-containing FACS buffer for 1 h at 4 °C, washed in FACS buffer three times, and resuspended in 2% w/v paraformaldehyde (PFA) (Electron Microscopy Sciences). For total protein staining, samples were first fixed and permeabilized using Cytofix/Cytoperm (BD Biosciences) for 15 min. After washing twice with Permwash (BD Biosciences) to remove residual fixative, cells were resuspended in antibody-containing Permwash for 1 h at 4 °C. Following incubation with antibody, samples were washed four times with Permwash and twice with FACS buffer before resuspension in 2% PFA. The HA antibody (clone 16B12) was obtained from Covance as an Alexa-647 conjugate and used at a 1:250 dilution for surface staining and a 1:500 dilution for total protein staining.
Samples were run on a LSR II flow cytometer (BD Biosciences). For each staining condition, 50,000 cells were analyzed. Nonviable cells were excluded from analysis based on forward- and side-scatter profiles (data not shown), as determined from staining with 7-aminoactinomycin D (7-AAD) (Invitrogen). The Alexa-647 fluorophore was excited using a 635 nm laser and detected with a 675/20 band-pass filter. Data were acquired using FACSDiva (BD Biosciences) and analyzed off-line using FlowJo 7.1 (Treestar). To compare surface and total expression levels of BMPRII-HA in the presence of wild-type (WT) and mutant proteins, the mean fluorescence intensity of mock transfected cells was subtracted from the mean fluorescence intensity of each positively transfected condition. The remaining fluorescence was then normalized to that of the WT condition, yielding a relative fluorescence intensity (“Relative FI”). Control experiments with GABR3 were performed using the same methods. Statistical significance was determined using a one-sample t-test using a hypothetical mean of 1 (since data in each condition were normalized to WT expression). Data were expressed as mean ± SEM.
pSMAD1/5 assays
BMP4 stimulation of HEK239-T cells transfected with shRNA targeting atlastin-1, WT, mutant forms of atlastin-1, and transfection with an empty vector were performed as previously described (Shi et al., 2007). Cells were washed with PBS and then placed in serum-free medium overnight. BMP4 stimulation was performed with 4 ng/ml or 20 ng/ml of BMP4 (R&D systems). Cells were washed in ice-cold PBS, and harvested on ice in lysis buffer containing 1% Triton X-100, in the presence of a complete protease inhibitor cocktail (Roche). Lysates were then centrifuged at 10,000 ×g for 5 min at 4 °C and the supernatant removed. Samples containing equivalent protein masses were heated to 98 °C for 5 min before running on SDS–PAGE and subsequent immunoblotting. For pSmad1/5 immunoblotting, membranes were blocked in PBS with 5% powdered milk and 5% bovine serum albumin for 3 h at room temperature, followed by incubation overnight at 4 °C with the primary antibody in 5% BSA in Tris-buffered saline with 0.1% Tween (TBS-T), followed by secondary antibodies. Each experiment was repeated 6-times and data was normalized to an arbitrary zero point and changes were assayed for 90 minutes after the BMP4 stimulation. Each membrane was assayed for pSMAD1/5, total SMAD1, actin and atlastin-1-myc expression and this was done sequentially with stripping of membranes for previously used antibodies; this was necessary because of frequent interference of secondary antibodies. Exposure times were normalized and always the same for different conditions. We also assayed the levels of overexpressed WT and mutant forms of atlastin-1, and levels of endogenous atlastin-1 present in HEK239-T cells. Quantification of Western blots for the determination of expressed levels of endogenous NIPA1 protein was performed with ChemiImager 5500 (Alpha Innotech) using Alpha Ease Fc software (Alpha Innotech).
Statistical analysis
Levels of protein expressions were compared using Student t-test.
Supplementary Material
Acknowledgements
This work was supported by K02NS057666 (NIH/NINDS) to PH. We gratefully acknowledge the use of the Vanderbilt University Medical Center Cell Imaging Core Resource. The Vanderbilt University Medical Center Cell Imaging Core Resource is supported by National Institutes of Health grants CA68485 and DK20593.We appreciate Dr. Oh's generous gift of BMPRII knock-out mouse pulmonary endothelial tissue.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mcn.2012.10.005.
References
- Abel A, Fonknechten N, Hofer A, Dürr A, Cruaud C, Voit T, Weissenbach J, Brice A, Klimpe S, Auburger G, et al. Early onset autosomal dominant spastic paraplegia caused by novel mutations in SPG3A. Neurogenetics. 2004;5:239–423. doi: 10.1007/s10048-004-0191-2. [DOI] [PubMed] [Google Scholar]
- Bakowska JC, Jupille H, Fatheddin P, Puertollano R, Blackstone C. Troyer syndrome protein spartin is mono-ubiquitinated and functions in EGF receptor trafficking. Mol. Biol. Cell. 2007;18:1683–1692. doi: 10.1091/mbc.E06-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat V, Jaiswal M, Bellen HJ. The BMP signaling pathway at the Drosophila neuromuscular junction and its links to neurodegenerative diseases. Curr. Opin. Neurobiol. 2010;21:1–7. doi: 10.1016/j.conb.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C, O'Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat. Rev. Neurosci. 2011;12:31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botzolakis EJ, Zhao J, Gurba KN, Macdonald RL, Hedera P. The effect of HSP-causing mutations in SPG3A and NIPA1 on the assembly, trafficking, and interaction between atlastin-1 and NIPA1. Mol. Cell. Neurosci. 2011;46:122–135. doi: 10.1016/j.mcn.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca GC, Ebers GC, Esiri MM. The extent of axonal loss in the long tracts in hereditary spastic paraplegia. Neuropathol. Appl. Neurobiol. 2004;30:576–584. doi: 10.1111/j.1365-2990.2004.00587.x. [DOI] [PubMed] [Google Scholar]
- Durr A, Camuzat A, Colin E, Tallaksen C, Hannequin D, Coutinho P, Fontaine B, Rossi A, Gil R, Rousselle C, et al. Atlastin1 mutations are frequent in young-onset autosomal dominant spastic paraplegia. Arch. Neurol. 2004;61:1867–1872. doi: 10.1001/archneur.61.12.1867. [DOI] [PubMed] [Google Scholar]
- Evans K, Keller C, Pavur K, Glasgow K, Conn B, Lauring B. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. PNAS. 2006;103:10666–10671. doi: 10.1073/pnas.0510863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassier C, Hutt JA, Scholpp S, Lumsden A, Giros B, Nothias F, Schneider-Maunoury S, Houart C, Hazan J. Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nat. Neurosci. 2010;13:1380–1387. doi: 10.1038/nn.2662. [DOI] [PubMed] [Google Scholar]
- Hamid R, Cogan JD, Hedges LK, Austin E, Phillips JA, III, Newman JH, Loyd JE. Penetrance of pulmonary arterial hypertension is modulated by the expression of normal BMPR2 allele. Hum. Mutat. 2009;30:649–654. doi: 10.1002/humu.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung A, Bitton-Worms K, Rechtman MM, Wenzel V, Boergermann JH, Hassel S, Henis YI, Knaus P. Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol. Cell. Biol. 2006;26:7791–7805. doi: 10.1128/MCB.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedera P, Fenichel GM, Blair M, Haines JL. Novel mutation in the SPG3A gene in an African American family with an early onset of hereditary spastic paraplegia. Arch. Neurol. 2004;61:1600–1603. doi: 10.1001/archneur.61.10.1600. [DOI] [PubMed] [Google Scholar]
- Hong K-H, Lee YJ, Lee E, Park SO, Han C, Beppu H, Li E, Raizada MK, Bloch KD, Oh SP. Genetic ablation of the BMPR2 gene in pulmonary endothelium is sufficient to predispose to pulmonary arterial hypertension. Circulation. 2008;118:711–730. doi: 10.1161/CIRCULATIONAHA.107.736801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, Rapoport TA, Blackstone C. A class of dynamin-like GTAases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Paik SK, Lee MJ, Kim YJ, Kim S, Nahm M, Oh SJ, Kim HM, Yim J, Lee CJ, et al. Drosophila Atlastin regulates the stability of muscle microtubules and is required for synapse development. Dev. Biol. 2009;330:250–262. doi: 10.1016/j.ydbio.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Matsuura I, Taniguchi J, Hata K, Saeki N, Yamashita Y. BMP inhibition enhances axonal growth and functional recovery after spinal cord injury. J Neurochem. 2008;150:1471–1479. doi: 10.1111/j.1471-4159.2008.05251.x. [DOI] [PubMed] [Google Scholar]
- Montani L, Gerrits B, Gehring P, Kempf A, Dimou L, Wollscheid B, Schwab ME. Neuronal Nogo-A modulates growth cone motility via Rho-GTP/LIMK1/cofilin in the unlesioned adult nervous system. J Biol. Chem. 2009;284:10793–10807. doi: 10.1074/jbc.M808297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro G, Rebelo AP, Connel J, Allison R, Babalini C, D'Aloia M, Montieri P, Schulle R, Ishiura H, Price J, et al. Mutations in the ER-shaping protein reticulon-2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J. Clin. Invest. 2012;122:538–544. doi: 10.1172/JCI60560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa M, Muriel MP, Janer A, Latouche M, Dauphin A, Debeir T, Martin E, Duyckaerts C, Prigent A, Depienne C, et al. Mutations in the SPG3A gene encoding the GTPase atlastin interfere with vesicle trafficking in the ER/Golgi interface and Golgi morphogenesis. Mol. Cell. Neurosci. 2007;35:1–13. doi: 10.1016/j.mcn.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, McNew JA, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- Park S, Blackstone C. Further assembly required: construction and dynamics of the endoplasmatic reticulum network. EMBO Rep. 2010;11:515–521. doi: 10.1038/embor.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin. Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paspalas CD, Perley CC, Venkitaramani DV, Goebel-Goody SM, Zhang Y, Kurup P, Mattis YH, Lombroso PJ. Major vault protein is expressed along the nucleus-neurite axis and associates with mRNA in cortical neurons. Cereb. Cortex. 2009;19:1666–1677. doi: 10.1093/cercor/bhn203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainier S, Chai JH, Tokarz D, Nicholls RD, Fink JK. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6) Am. J. Hum. Genet. 2003;73:967–971. doi: 10.1086/378817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos M, Lamé MW, Segall HJ, Wilson DW. The BMP type II receptor is located in lipid rafts, including caveolae, of pulmonary endothelium in vivo and in vitro. Vascul. Pharmacol. 2006;44:50–59. doi: 10.1016/j.vph.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Renvoise B, Blackstone C. Emerging themes of ER organization in the development and maintenance of axons. Curr. Opin. Neurobiol. 2010;20:531–537. doi: 10.1016/j.conb.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C. Atlastin GTPase are required for Golgi apparatus and ER morphogenesis. Hum. Mol. Genet. 2008;17:1591–1604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudarakanchana N, Flanagan JA, Chen H, Upton PD, Machado R, Patel D, Trembath RC, Morrell NW. Functional analysis of bone morphogenetic protein type II receptor mutations underlying primary pulmonary hypertension. Hum. Mol. Genet. 2002;15:1517–1525. doi: 10.1093/hmg/11.13.1517. [DOI] [PubMed] [Google Scholar]
- Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- Sanderson CM, Connell JW, Edwards TL, Bright NA, Duley S, Thompson A, Luzio JP, Reid E. Spastin and atlastin, two proteins mutated in autosomaldominant hereditary spastic paraplegia, are binding partners. Hum. Mol. Genet. 2006;5:307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Chang C, Nie S, Xie S, Wan M, Cao X. Endofin acts as a Smad anchor for receptor activation in MBP signaling. J. Cell Sci. 2007;120:1216–1224. doi: 10.1242/jcs.03400. [DOI] [PubMed] [Google Scholar]
- Tsang HT, Edwards TL, Wang X, Connell JW, Davies RJ, Durrington HJ, O'Kane CJ, Luzio JP, Reid E. The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian BMP signaling. Hum. Mol. Genet. 2009;18:3805–3821. doi: 10.1093/hmg/ddp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shaw WR, Tsang HT, Reid E, O'Kane CJ. Drosophila spichthyin inhibits BMP signaling and regulates synaptic growth and axonal microtubules. Nat. Neurosci. 2007;10:177–185. doi: 10.1038/nn1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton SB, McDermott CJ, Grierson AJ, Wood JD, Gelsthorpe C, Ince PG, Shaw PJ. The cellular and molecular pathology of the motor system in hereditary spastic paraparesis due to mutation of the spastin gene. J. Neuropathol. Exp. Neurol. 2003;62:1166–1177. doi: 10.1093/jnen/62.11.1166. [DOI] [PubMed] [Google Scholar]
- Yu PB, Deng DY, Beppu H, Hong CC, Lai C, Hoyng SA, Kawai N, Bloch KD. Bone morphogenetic protein (BMP) type II receptor is required for BMP-mediated growth arrest and differentiation in pulmonary artery smooth muscle cells. J. Biol. Chem. 2008;283:3877–3888. doi: 10.1074/jbc.M706797200. [DOI] [PubMed] [Google Scholar]
- Zhao XP, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, Tukel T, Apak M, Heiman-Patterson T, Ming L, et al. Mutation in a novel GTPase cause autosomal dominant hereditary spastic paraplegia. Nat. Genet. 2001;29:326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- Zhao J, Matthies DS, Botzolakis EJ, Macdonald RL, Blakely RD, Hedera P. Hereditary spastic paraplegia-associated mutations in the NIPA1 gene and its C. elegans homolog trigger neural degeneration in vitro and in vivo through a gain of function mechanism. J. Neurosci. 2008;28:13938–13951. doi: 10.1523/JNEUROSCI.4668-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu PP, Patterson A, Lavoie B, Stadler J, Shoeb M, Patel R, Blackstone C. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J. Biol. Chem. 2003;278:49063–49071. doi: 10.1074/jbc.M306702200. [DOI] [PubMed] [Google Scholar]
- Zhu PP, Soderblom C, Tao-Cheng JH, Stadler J, Blackstone C. SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum. Mol. Genet. 2006;15:1343–1353. doi: 10.1093/hmg/ddl054. [DOI] [PubMed] [Google Scholar]
- Züchner S. The genetics of hereditary spastic paraplegia and implications for drug therapy. Expert Opin. Pharmacother. 2007;8:1433–1439. doi: 10.1517/14656566.8.10.1433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.